Abstract
Pigment epithelium-derived factor (PEDF), a noninhibitory member of the serpin superfamily, is the most potent inhibitor of angiogenesis in the mammalian ocular compartment. It also has neurotrophic activity, both in the retina and in the central nervous system, and is highly up-regulated in young versus senescent fibroblasts. To provide a structural basis for understanding its many biological roles, we have solved the crystal structure of glycosylated human PEDF to 2.85 Å. The structure revealed the organization of possible receptor and heparin-binding sites, and showed that, unlike any other previously characterized serpin, PEDF has a striking asymmetric charge distribution that might be of functional importance. These results provide a starting point for future detailed structure/function analyses into possible mechanisms of PEDF action that could lead to development of therapeutics against uncontrolled angiogenesis.
Pigment epithelium-derived factor (PEDF) is found as an extracellular component of the retinal interphotoreceptor matrix (1), as well as in the vitreous and aqueous humors in the adult eye (2, 3). PEDF mRNA is, however, also found in most tissues (4). The ≈50-kDa protein is a member of the serpin superfamily, but does not inhibit proteinases (5). PEDF was initially identified as a product secreted by cultured pigment epithelium cells from fetal human retina that possessed potent neurotrophic activity in retinoblastoma cells (6). The PEDF gene was independently identified as one that is posttranscriptionally and very markedly (>100-fold) up-regulated (7) during G0 phase in young but not senescent cultured fibroblasts (8, 9), giving rise to an alternative name of “early population doubling level cDNA” (EPC-1), suggesting additional roles in both the cell cycle and senescence.
Addition of PEDF at nanomolar concentrations induces a neuronal phenotype in both cultured human retinoblastoma Y79 and Weri cells (10). PEDF promotes neuronal survival of the cerebellar granule (11) and both survival and differentiation of developing spinal motor neurons (12). It prevents death of cerebellar granule neurons (13), spinal motor neurons (14), and developing primary hippocampal neurons (15) caused by glutamate cytotoxicity. It prevents hydrogen peroxide-induced apoptosis of retinal neurons (16), delays death of photoreceptors in the mouse model of retinitis pigmentosa (17), supports both normal development of the photoreceptor neurons and opsin expression after removal of the retinal pigment epithelium (18), and inhibits microglial growth (19).
Notwithstanding the various reports of the ability of PEDF to affect the growth and differentiation of both retinoblastoma cells and normal neurons, perhaps the most exciting findings are the recent reports that PEDF is the most potent inhibitor of angiogenesis in the mammalian ocular compartment as well as a potent proliferation inhibitor in various cell types that acts against a broad range of angiogenic/proliferative inducers (20, 21), probably through an apoptotic mechanism (22). Neovascularization of the retina leads to ischemic retinopathies, such as proliferative diabetic retinopathy and age-related macular degeneration (23), currently the leading causes of blindness in the Western world, and the failure to exclude blood vessels from the cornea leads to loss of visual acuity, opacification, and abnormal healing (24, 25). The neurotrophic and anti-angiogenic properties of PEDF argue in favor of its possible therapeutic value for pathologic ocular neovascularization and retinoblastoma treatment and perhaps more widely for control of neovascularization associated with tumor growth.
Although initial studies on the mechanism(s) of action of PEDF as a neurotrophic agent have been carried out based on a hypothetical model of the molecule (26, 27), a more detailed approach has been greatly hindered by the absence of a three-dimensional structure. We have now determined the crystal structure of glycosylated human PEDF at 2.85 Å resolution. This structure shows the location of a peptide previously identified as having some of the neurotrophic properties of intact PEDF, but suggests that only restricted portions of this peptide are likely to be candidates for receptor interactions. It is unknown whether the various functions of PEDF reside in different regions and involve binding to more than one receptor. Here it may be significant that the structure shows a strikingly asymmetric charge distribution that, in addition to better identifying a proposed heparin-binding site that might play a role in PEDF localization, suggests another possible site of interaction with cofactor proteins or other receptor molecules.
Methods
Protein Expression and Purification.
Glycosylated human PEDF was expressed and secreted by stably transfected BHK cells and initially purified from the cell medium by ammonium sulfate fractionation and SP-Sepharose ion-exchange chromatography as described (3, 28). The protein sample was dialyzed against 50 mM Tris⋅HCl, pH 8.0/20 mM NaCl and loaded onto a pre-equilibrated Q-Sepharose column. PEDF was eluted with a 0.1–0.4 M NaCl gradient at a rate of 2 ml⋅min−1 during 60 min. The purity of the PEDF sample was ascertained by SDS/12% PAGE. Glycosylation of the protein was checked by treatment of the denatured protein with N-glycosidase F followed by SDS/12% PAGE. The sample was filtered through a 0.22-μm membrane, concentrated to 17 mg⋅ml−1 by using a 10-kDa cutoff membrane, and stored at −80°C until needed.
Crystallization.
Crystallization conditions were identified in PEG-ion Hampton crystallization screen (29). Crystals were obtained at 18°C by using the hanging-drop vapor diffusion method, mixing equal volumes of the protein (17 mg⋅ml−1) and the well solution (0.2 M ammonium fluoride/20% PEG 3,350, pH 6.20). Rod-shaped crystals grew within a month to average dimensions of 0.4 × 0.15 × 0.08 mm. The crystals belong to space group P21212 (a = 176.17 Å; b = 62.51 Å; and c = 45.41 Å) with one molecule per asymmetric unit, and ≈58% solvent content.
Data Collection, Structure Determination, and Refinement.
Diffraction data were collected at room temperature from three crystals on a Rigaku RAXIS II detector, by using focused Cu Kα radiation from a Rigaku RU-H2R rotating anode x-ray generator at a power of 50 kV, 100 mA. The data were indexed with DENZO, and scaled and reduced with SCALEPACK (30). The initial phases were determined by molecular replacement using the native structure of α1-proteinase inhibitor (α1-PI; PDB accession code: 1QLP). The search model did not contain the reactive center loop, loop 170, and the loop around residue 320 that connects s6A with s5A (loop 320). Molecular replacement calculations were performed with crystallography & nmr system (31). The rotation search gave a 6.8σ peak, and the translation search gave a solution with a correlation coefficient of 28%. The oriented search model was then divided into several segments and refined as rigid bodies followed by cycles of simulated annealing giving an Rcryst of 41% (20–3.20 Å). Electron-density maps were interpreted by using the program QUANTA (32). Regions of the model with poor density were deleted and regions of insertions were identified. Ten percent of the data were randomly assigned to an Rfree test for cross-validation (31). The model was progressively refined by using simulated annealing protocols, followed by energy minimization and manual inspection and rebuilding. Resolution was increased stepwise. Once Rcryst dropped below 35%, bulk-solvent and anisotropic B-factor corrections were introduced. Electron density for missing loops as well as for the carbohydrate residue appeared strongly once Rcryst dropped below 28%. However, 15 amino acids at the N terminus and 8 amino acids in the reactive center loop were not visible in the final electron density maps. The final stages of refinement, including individual B-factor refinement, were performed against all data. The final model has an R factor of 18.8% and Rfree of 22.7%. The crystallographic data and statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| Crystal | |
| Space group | P21212 |
| Cell dimensions, Å | a = 176.2; b = 62.5; c = 45.4 |
| Number of crystals | 3 |
| Data collection | |
| Resolution limit, Å | 100–2.85 |
| Wavelength, Å | 1.514 |
| Reflections (observed/unique) | 152, 138/11,241 |
| Completeness/last shell, % | 91.2/65.4 |
| Rmerge (overall/last shell), % | 12.7/37 |
| I/σI | 8.5 |
| Refinement | |
| Number of molecules in asymmetric unit | 1 |
| Resolution, Å | 40.00–2.85 |
| Average B factor, Å2 | 31.4 |
| Number of reflections (work/test) | 10,075/1,166 |
| Number of protein atoms (amino acids) | 2,923 (375) |
| Number of solvent molecules | 45 |
| Number of heterogen atoms (carbohydrate) | 14 |
| Rcryst(|F| > 0σ) | 18.8 |
| Rfree(|F| > 0σ) | 22.7 |
| rms deviations from ideality | |
| Bond lengths, Å | 0.007 |
| Bond angles, ° | 1.3 |
Results and Discussion
Overall Structure.
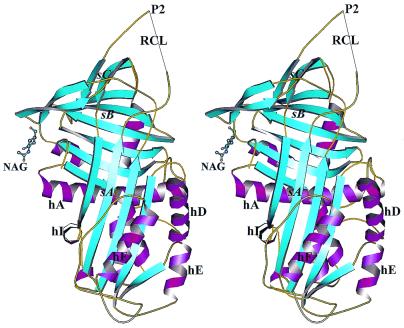
The PEDF crystals contain one molecule per asymmetric unit. The x-ray structure of native glycosylated human PEDF is shown in Fig. 1. With the exception of the extreme 15 residues at the N terminus (residues 1–15) and 8 residues in the reactive center loop (residues 353–360), all of the molecule backbone is well ordered in the final crystal structure (Rcryst 18.8% and Rfree 22.7%; Table 1). Additionally, 11 side chains are disordered (T16, R79, K126, K127, R174, K177, E178, D181, E182, K228, and T352), mainly in the loop 170 that connects helix F (hF) with strand 3A (s3A).
Figure 1.
Stereo view ribbon diagram of glycosylated PEDF in the native form in the typical serpin orientation. β-Strands are cyan, α-helices are magenta, coils and turns are gold, and the carbohydrate residue is represented as ball-and-sticks. Strands and helices are labeled as in the text. The reactive center loop is labeled as RCL, and N-acetylglucosamine is labeled as NAG (produced in MOLSCRIPT; ref. 33).
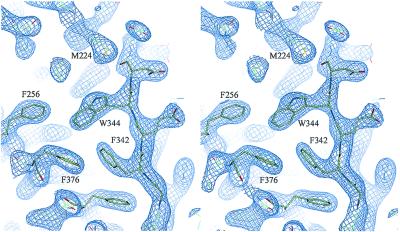
The recombinant human PEDF used in these studies is glycosylated. Glycosylation was confirmed by N-glycosidase-F treatment followed by SDS/PAGE analysis (data not shown). The electron density for the first carbohydrate residue, N-acetylglucosamine (NAG in Fig. 1) appears strongly in the Fourier difference map at position N265. This is in agreement with an earlier proposed glycosylation site (28). Analysis of the crystal packing suggests that the sugar chain is not involved in the crystal contacts. Comparison of the PEDF structure with known structures of other native serpins (α1PI, ovalbumin, etc.), shows a high level of structural conservation despite the relatively low sequence identity among these family members (20–27% for these serpins). The rms deviation between native PEDF and native α1PI was found to be 1.36 Å for 147 α-carbons from β-sheets A, B, and C, but was higher for the helices. Also, 41 of 51 mainly hydrophobic residues, which have been previously shown to be highly conserved throughout the serpin superfamily (34), are present in PEDF and appear to perform the same roles in structural stabilization. Representative electron density of the hydrophobic core is shown in Fig. 2. The N and C termini of the reactive center loop are well defined, although the central 8 residues give no electron density despite being present in the molecule, as judged by absence of cleavage within the loop. These ends extend away from the serpin body and ensure that the missing central portion, whatever its structure, will be very exposed and accessible to interact with potential targets. This exposure is greater than in the reactive center loops of any other serpin of known structure.
Figure 2.
Stereo view of the final |2Fo−Fc|αC electron density map of the PEDF hydrophobic core contoured at a level of 1.7σ (produced in QUANTA; ref. 32).
Asymmetric Charge Distribution.
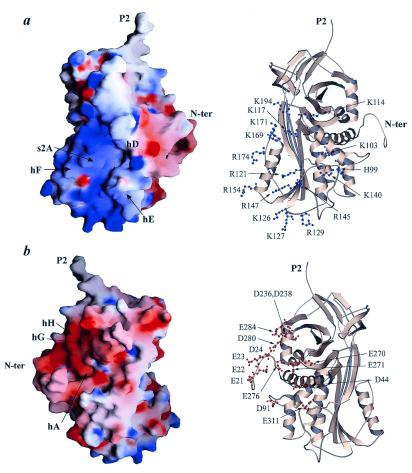
In earlier studies it has been shown that PEDF binds to heparin and other glycosaminoglycans (26, 28), although the interaction is only as strong as that between antithrombin, another serpin, and low-affinity heparin (28). Nevertheless, this interaction may be very important in vivo for surface localization of PEDF. Analysis of our structure reveals a striking asymmetric charge distribution (Fig. 3). The basic residues are concentrated on helices D, E, and F, on strands 1, 2, and 3 of β-sheet A, and the loop 170 (Fig. 3a). We propose that both the heparin- and proteoglycan-binding sites are located at this basic surface of PEDF. This result is only partly in agreement with earlier biochemical studies and homology modeling of PEDF (26). The crystal structure reveals a much larger basic site than was suggested by homology modeling, which demonstrates the clear benefit of having real structural data for detailed analysis of function.
Figure 3.
Surface charge distribution analysis of PEDF reveals striking asymmetric charge distribution that might be of physiological significance. (Left) GRASP (35) representations are displayed. (Right) Ribbon diagrams of PEDF oriented in the same way as the respective surface charge diagrams, with basic (blue) and acidic (red) side chains presented as ball-and-stick (scale, −15 kT/e to +15 kT/e). (a) The basic region that covers parts of hD, hE, s1A, and s2A is displayed. This is the putative heparin-binding site. The view is rotated ≈90°clockwise about the vertical axis relative to Fig. 1. (b) The acidic region consists of side chains from the extreme N terminus, hA, s6B, hG, and hH. The view is rotated ≈90° counterclockwise about the vertical axis relative to Fig. 1.
An interesting and somewhat surprising feature of the PEDF charge distribution is the highly acidic region located around the N-terminal part of the molecule (Fig. 3b), at almost the opposite side from the basic region. Side chains from the loop containing residue 20 (loop 20) and helices A, G, and H contribute to this site. Comparison with other serpin structures emphasizes the unique asymmetric charge distribution of PEDF. This unusual concentration of aspartic and glutamic acid side chains might be of physiological importance, and might play a role in binding with receptors or other cofactor proteins.
Neurotrophic Activity of PEDF.
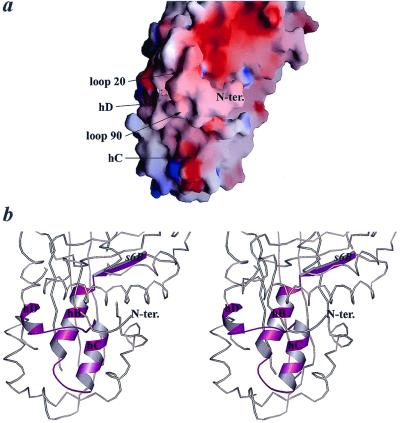
PEDF has neurotrophic activity that has been localized to the N-terminal region from limited peptide studies (5, 27). These studies showed that the region from V58 to T101 is very important for the neurotrophic properties of PEDF. In addition, recent radioligand-binding studies have suggested that the same site of PEDF is involved in binding to a putative PEDF receptor present in the membrane of both the cerebellar granule neurons and the retinoblastoma cells (27). Here we present the structural organization of the earlier-proposed receptor-binding site (Fig. 4). It is clear that the binding site is likely to be far smaller than the one suggested by peptide studies. The peptide V58–T101 used in those studies includes the complete secondary structural elements s6B, hB, and hC as well as one turn of hD. Both s6B and hB are buried in the interior of PEDF, making it highly unlikely that these parts would play any role in receptor binding (Fig. 4b) unless there were a major conformational rearrangements upon binding. On the other hand, a solvent-accessible surface is formed by helices C and D and the loop around residue 90 that connects them (loop 90). The loop 20 also forms part of the same face of the molecule (Fig. 4a). Because it is likely that the identified PEDF–receptor interaction on the membranes of cerebellar neurons leads to the observed neurotrophic changes, we predict from our structure that only the exposed parts of helices C and D and the loop 90 are important for the binding to the putative receptor and for neurotrophic activity. Because this short region is mostly unstructured, it would explain why earlier studies on much larger peptide fragments (27) have fortuitously found them to have biological activity despite not having the correct fold of the intact protein.
Figure 4.
The putative receptor-binding site based on the peptide–receptor binding assays (27). (a) Surface charge distribution representation of the binding site. (b) Stereo view of the binding site. The molecule is in approximately the same orientation as in Fig. 4a. The binding site based on the peptide studies is in magenta with its secondary structure elements displayed as ribbons. The rest of the molecule is represented as a coil (produced in MOLSCRIPT; ref. 33).
Noninhibitory Serpin vs. Inhibitory Serpin.
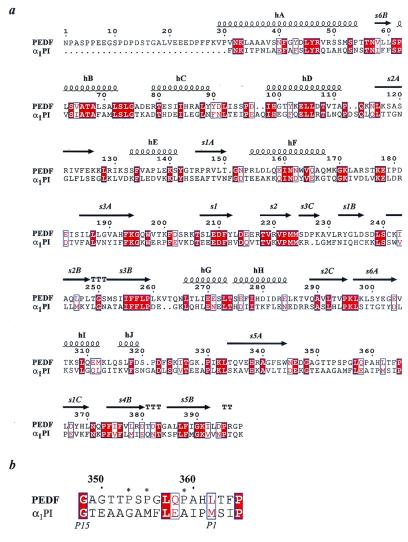
PEDF is a member of the serpin superfamily and has sequence and structural homology with other serpins (Fig. 5a). However, PEDF does not exhibit inhibitory activity against proteinases (5, 28), nor does it undergo the remarkable conformational change upon cleavage of the reactive center loop that occurs in inhibitory serpins as an essential part of the inhibition mechanism (37). These differences from inhibitory serpins might result entirely from the sequence of the reactive center loop, which, unlike inhibitory serpins, has several proline residues preceding what would be the scissile bond (Fig. 5b). These occur at positions P10, P8, and P4, using the nomenclature by which the scissile bond occurs between residues designated P1 and P1′. Because inhibition by serpins requires insertion of the previously exposed reactive center loop into β-sheet A as a new central strand, residues in the loop must be compatible with adopting such β conformation and also not involve burial of charged side chains on such incorporation into β-sheet A. There are examples of pathological single mutations to proline within the reactive center loop of otherwise inhibitory serpins that result in conversion of the serpin from a proteinase inhibitor to a proteinase substrate (38). An alternative explanation for the different properties of PEDF is that the ability of β-sheet A to open up to accommodate the reactive center loop has been lost in PEDF. This is indeed the case with the noninhibitory serpin ovalbumin, where replacement of disadvantageous residues from the reactive center loop still does not allow facile insertion of the cleaved loop into β-sheet A (39). In either case, the result in PEDF is that the reactive center loop is exposed in the native state and does not have a tendency to become buried on proteolytic cleavage, were that to occur. Of the PEDF sequences known (human, mouse, and bovine), all have the reactive center loop sequence SPGL(V)QP, from P9 to P4, almost completely conserved (bovine contains V instead of L at P6). Although the preceding proline at P10 is present only in human and mouse PEDF, a unique possible glycosylation site at P11 in bovine PEDF would also ensure that this portion of the reactive center loop remained exposed.
Figure 5.
Sequence alignment of PEDF and α1PI. (a) Secondary structure elements are based on the PEDF structure; strands are presented as arrows, whereas helices are presented as coils. (b) Excerpt of the alignment of PEDF and α1PI demonstrating the unusually high number of prolines in the reactive center loop of PEDF. Prolines are labeled with *. Identical residues are highlighted in red, whereas neutral changes are labeled with blue boxes. Numbering is based on the PEDF sequence (produced by using MULTALIN; ref. 36).
Although the reactive center loop of PEDF is not completely visible in our crystal structure, both the N-terminal and C-terminal stalks are clearly defined and project outward from the body of the protein, carrying the central portion with them (Fig. 1). Both the limited ability of selected proteinases to cleave within this loop (28) and the integrity of the loop in naturally isolated PEDF suggest that the loop sequence has evolved not to be readily cleaved by most proteinases, but at the same time to give a highly exposed structure. In inhibitory serpins, the result of having exposed reactive center loops is rapid recognition by target proteinases and subsequent inhibition, with initial rates of association that are often diffusion limited (39). In PEDF it is tempting to speculate that the even greater reactive center loop exposure is also designed for rapid binding to a target protein. However, in this case it is unlikely to be a proteinase, for the reasons given above, but may instead be a protein involved in the regulatory functions of PEDF. In this regard, a previous report (5) that reactive center loop-cleaved PEDF still possesses neurotrophic activity does not rule out the involvement of this region in the activity, because the inability of the cleaved loop to insert into β-sheet A would ensure that the cleaved ends would still be highly exposed and available for binding to receptor or cofactor.
Conclusions and Implications
PEDF is known to possess a range of fundamentally important biological properties that affect neuronal growth, cellular differentiation and senescence, and angiogenesis. It is also known to have a binding site for heparin and glycosaminoglycans that might be involved in these or other processes. Whether the various biological roles involve the same or different regions of PEDF or involve binding to the same or different receptors is currently unknown. However, the structure of PEDF presented here has provided the necessary structural insight to permit refinement of the likely regions for mediation of neurotrophic activity and for glycosaminoglycan binding and prompted speculation about the possible involvement of the unusual reactive center loop and the highly asymmetric charge distribution in one or other of PEDF's biological activities. The structure will serve as a base for the design of PEDF variants to test the involvement of these different regions in the varied and growing list of biological activities of this fundamentally important regulatory protein.
Acknowledgments
Medium containing human glycosylated PEDF was kindly provided by the laboratory of Dr. Philip Patston (University of Illinois, Chicago). We thank Dr. K. Moffat and coworkers at the BioCars beam line, Advanced Photon Source, Argonne National Laboratory, for access and beam time. Also, we thank Drs. Karen Colley and Steven Olson for suggestions and comments on the manuscript. This work was supported by National Institutes of Health Grants GM47522 (to K.V.), and HL49234 and HL64013 (to P.G.W.G.) and by funds from the Vice Chancellor for Research, University of Illinois, Chicago. M.S. is the recipient of a University fellowship from the University of Illinois, Chicago.
Abbreviations
- PEDF
pigment epithelium-derived factor
- α1PI
α1-proteinase inhibitor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1IMV).
References
- 1.Tombran-Tink J, Chader G G, Johnson L V. Exp Eye Res. 1991;53:411–414. doi: 10.1016/0014-4835(91)90248-d. [DOI] [PubMed] [Google Scholar]
- 2.Ortego J, Escribano J, Becerra S P, Coca-Prados M. Invest Ophthalmol Vis Sci. 1996;37:2759–2767. [PubMed] [Google Scholar]
- 3.Wu Y Q, Becerra S P. Invest Ophthalmol Vis Sci. 1996;37:1984–1993. [PubMed] [Google Scholar]
- 4.Tombran-Tink J, Mazuruk K, Rodriguez I, Chung D, Linker T, Englander E, Chader G. Mol Vis. 1996;2:11–19. [PubMed] [Google Scholar]
- 5.Becerra S P, Sagasti A, Spinella P, Notario V. J Biol Chem. 1995;270:25992–25999. doi: 10.1074/jbc.270.43.25992. [DOI] [PubMed] [Google Scholar]
- 6.Tombran-Tink J, Johnson L. Invest Ophthalmol Visl Sci. 1989;30:1700–1709. [PubMed] [Google Scholar]
- 7.Coljee V W, Rotenberg M O, Tresini M, Francis M K, Cristofalo V J, Sell C. J Cell Biochem. 2000;79:442–452. doi: 10.1002/1097-4644(20001201)79:3<442::aid-jcb90>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 8.Pignolo R J, Cristofalo V J, Rotenberg M O. J Biol Chem. 1993;268:8949–8957. [PubMed] [Google Scholar]
- 9.Tombran-Tink J, Shivaram S M, Chader G J, Johnson L V, Bok D. J Neurosci. 1995;15:4992–5003. doi: 10.1523/JNEUROSCI.15-07-04992.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seigel G M, Tombran-Tink J, Becerra S P, Chader G J, Diloreto D A, del Cerro C, Lazar E S, del Cerro M. Growth Factors. 1994;10:289–297. doi: 10.3109/08977199409010995. [DOI] [PubMed] [Google Scholar]
- 11.Taniwaki T, Becerra S P, Chader G J, Schwartz J P. J Neurochem. 1995;64:2509–2517. doi: 10.1046/j.1471-4159.1995.64062509.x. [DOI] [PubMed] [Google Scholar]
- 12.Houenou L J, D'Costa A P, Li L, Turgeon V L, Enyadike C, Alberdi E, Becerra S P. J Comp Neurol. 1999;412:506–514. [PubMed] [Google Scholar]
- 13.Taniwaki T, Hirashima N, Becerra S P, Chader G J, Etcheberrigaray R, Schwartz J P. J Neurochem. 1997;68:26–32. doi: 10.1046/j.1471-4159.1997.68010026.x. [DOI] [PubMed] [Google Scholar]
- 14.Bilak M M, Corse A M, Bilak S R, Lehar M, Tombran-Tink J, Kuncl R W. J Neuropathol Exp Neurol. 1999;58:719–728. doi: 10.1097/00005072-199907000-00006. [DOI] [PubMed] [Google Scholar]
- 15.DeCoster M A, Schabelman E, Tombran-Tink J, Bazan N G. J Neurosci Res. 1999;56:604–610. doi: 10.1002/(SICI)1097-4547(19990615)56:6<604::AID-JNR6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 16.Cao W, Tombran-Tink J, Chen W, Mrazek D, Elias R, McGinnis J F. J Neurosci Res. 1999;57:789–800. [PubMed] [Google Scholar]
- 17.Cayouette M, Smith S B, Becerra S P, Gravel C. Neurobiol Dis. 1999;6:523–532. doi: 10.1006/nbdi.1999.0263. [DOI] [PubMed] [Google Scholar]
- 18.Jablonski M M, Tombran-Tink J, Mrazek D A, Iannaccone A. J Neurosci. 2000;20:7149–7157. doi: 10.1523/JNEUROSCI.20-19-07149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugita Y, Becerra S P, Chader G J, Schwartz J P. J Neurosci Res. 1997;49:710–718. doi: 10.1002/(SICI)1097-4547(19970915)49:6<710::AID-JNR5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 20.Dawson D W, Volpert O V, Gillis P, Crawford S E, Xu H, Benedict W, Bouck N P. Science. 1999;285:245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 21.Chader G J. Proc Natl Acad Sci USA. 2001;98:2122–2124. doi: 10.1073/pnas.061024098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stellmach V, Crawford S E, Zhou W, Bouck N. Proc Natl Acad Sci USA. 2001;98:2593–2597. doi: 10.1073/pnas.031252398. . (First Published January 23, 2001; 10.1073/pnas.031252398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schachat A, Murphy R. Retina. Baltimore: Mosby; 1994. [Google Scholar]
- 24.Klintworth G. Corneal Angiogenesis: A Comprehensive Critical Review. New York: Springer; 1991. [Google Scholar]
- 25.Kaminska G, Niederkorn J. Invest Ophthalmol Vis Sci. 1993;34:222–230. [PubMed] [Google Scholar]
- 26.Alberdi E, Hyde C C, Becerra S P. Biochemistry. 1998;37:10643–10652. doi: 10.1021/bi9802317. [DOI] [PubMed] [Google Scholar]
- 27.Alberdi E, Aymerich M S, Becerra S P. J Biol Chem. 1999;274:31605–31612. doi: 10.1074/jbc.274.44.31605. [DOI] [PubMed] [Google Scholar]
- 28.Stratikos E, Alberdi E, Gettins P G, Becerra S P. Protein Sci. 1996;5:2575–2582. doi: 10.1002/pro.5560051220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jancarik J, Kim S H. J App Crystallogr. 1991;24:409–411. [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:461–472. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Molecular Simulations, Inc. quanta. San Diego; 1997. [Google Scholar]
- 33.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 34.Huber R, Carrell R. Biochemistry. 1989;28:8951–8966. doi: 10.1021/bi00449a001. [DOI] [PubMed] [Google Scholar]
- 35.Nicholls A, Sharp K, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 36.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stratikos E, Gettins P. Proc Natl Acad Sci USA. 1999;96:4808–4813. doi: 10.1073/pnas.96.9.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gettins P G W, Patston P A, Olson T A. Serpins: Structure, Function and Biology. Austin, TX: Landes; 1996. [Google Scholar]
- 39.Huntington J A, Fan B, Karlsson K E, Deinum J, Lawrence D A, Gettins P G. Biochemistry. 1997;36:5432–5440. doi: 10.1021/bi9702142. [DOI] [PubMed] [Google Scholar]