Abstract
The linear chromosome of the bacterium Streptomyces exhibits a remarkable genetic organization with grossly a central conserved region flanked by variable chromosomal arms. The terminal diversity co-locates with an intense DNA plasticity including the occurrence of large deletions associated to circularization and chromosomal arm exchange. These observations prompted us to assess the role of double strand break (DSB) repair in chromosome plasticity following. For that purpose, DSBs were induced along the chromosome using the meganuclease I-SceI. DSB repair in the central region of the chromosome was mutagenic at the healing site but kept intact the whole genome structure. In contrast, DSB repair in the chromosomal arms was mostly associated to the loss of the targeted chromosomal arm and extensive deletions beyond the cleavage sites. While homologous recombination occurring between copies of DNA sequences accounted for the most part of the chromosome rescue events, Non Homologous End Joining was involved in mutagenic repair as well as in huge genome rearrangements (i.e. circularization). Further, NHEJ repair was concomitant with the integration of genetic material at the healing site. We postulate that DSB repair drives genome plasticity and evolution in Streptomyces and that NHEJ may foster horizontal transfer in the environment.
Introduction
The soil bacterium Streptomyces is renowned for its biotechnological capabilities, most notably its ability to synthesize a wide variety of bioactive secondary metabolites used in medicine as antiproliferative agents (antibiotics, anticancer and antifungal drugs)1. Furthermore, this group of bacteria possesses unique genome characteristics among which a high GC content (circa 72%) and a large linear chromosome (6–12 Mb) ended by inverted repeats. Besides, the chromosome is highly compartmentalized, with a conserved central region flanked by variable arms2,3. The areas located between the conserved and the specific regions show a progressive loss of synteny ensuing from an accumulation of insertion/deletion events4. One very intriguing phenomenon which has been known for decades is the high genetic instability in this genus. Thus, during culture, Streptomyces species offsprings present between 0.1 and 1% of spontaneous mutants defective in morphological and physiological differentiation including secondary metabolic pathways5. This phenotypic instability correlates with the formation of large deletions affecting the chromosomal arms. Deletions have been found to be associated with chromosomal circularization6,7, chromosomal arm exchange8,9 and fusion of chromosomal ends10. High copy number DNA amplification was also associated with the formation of deletions11–13. Moreover, homologous recombination (HR) and illegitimate recombination (IR) are known to be involved in such chromosome rearrangements8,12. These recombination events could also bring into play plasmid/chromosome exchanges resulting in hybrid chromosome formation14,15.
Most of the key actors of HR in many model eubacteria (e.g. RecA) have orthologues in Streptomyces16,17. Yet, there are some striking discrepancies; for example, the presence of AdnAB as the main helicase nuclease activity instead of RecBCD or the presence of alternative post-synaptic actors to RuvABC and RecG18,19. However, HR is effective in Streptomyces, and as in many other bacteria, is assumed to contribute to genome stability as an error free repair mechanism20. On the other hand, HR can occasionally lead to major chromosome structure alteration when occurring between ectopic homologous copies8,9.
Non Homologous End Joining (NHEJ), as an IR mechanism, does not need any intact template to resolve Double-Strand Break (DSB) repair and can directly ligate the broken ends together after a facultative end processing step. This repair mechanism can lead to major chromosome rearrangements and is potentially mutagenic at the recombination site21.
The NHEJ pathway was long considered as the hallmark of eukaryotes, suggesting that DSB repair in prokaryotes fully relies on HR. However, early bioinformatics studies led to the identification of orthologues of a key NHEJ actor (i.e. Ku proteins) in several phylogenetically distant bacterial genomes22,23. While the NHEJ pathway was deciphered in model bacteria such as B. subtilis or M. tuberculosis24,25, we have recently shown that Streptomyces possess a large set of NHEJ-like genes with some of these being conserved (the ‘core’ NHEJ gene set) between species and strains26. Surprisingly, the involvement of all NHEJ-like genes, including the ‘variable’ set, in DNA damage response in Streptomyces suggests that in these organisms, a more complex pathway than in Mycobacterium or Bacillus is present.
Here we aimed to assess the role of DSB repair mechanisms (HR and NHEJ) in the formation of large DNA rearrangements and in chromosomal structure evolution in Streptomyces. To achieve this, we induced the formation of chromosomal DSBs using an I-SceI mediated system and surveyed the chromosomal structure of the survivals.
Results
DSB repair in the central part of the chromosome is mutagenic
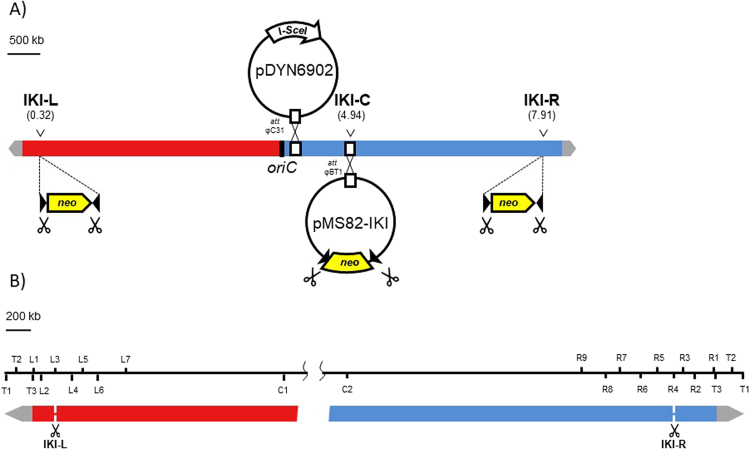
To monitor the consequences of DSB repair along the Streptomyces chromosome, we developed a DSB inducible system based on the meganuclease I-SceI heterologous expression (see Methods). A target cassette named IKI consisting of the neo gene, which confers resistance to kanamycin, flanked by two I-SceI sites, was introduced in the central region of the chromosome (IKI-C locus, position 4.94 Mb) of Streptomyces ambofaciens ATCC 23877 (Fig. 1). I-SceI endonuclease was constitutively expressed from an integrating plasmid in the chromosome. The functional activity of the endonuclease was proved by the loss of the target IKI cassette or the I-SceI gene in at least 79% of the progeny when selective pressure was lifted (Table S1).
Figure 1.
I-SceI DSB induction and chromosomal survey device. (A) I-SceI mediated DSB targeting in S. ambofaciens chromosome. The target sequences for I-SceI meganuclease are carried by the IKI cassette designed with neo gene, conferring resistance to kanamycine, flanked by two convergent I-SceI restriction sites (dark triangle). Red and blue boxes represent left and right replichores of the chromosome. Grey arrows represent the terminal inverted repeats at each chromosome extremity. The IKI cassette was integrated either in the core genome (IKI-C, position 4.94 Mb), in the left chromosomal arm (IKI-L, position 0.32 Mb) or in the right chromosomal arm (IKI-R, position 7.91 Mb). The cassette was inserted at IKI-L or IKI-R loci through homologous recombination. The IKI-C locus corresponds to the attachment site of phage φBT1 (attB φBT1) where pMS82 conjugative plasmid carrying IKI cassette (therefore named pMS82-IKI) was integrated. Meganuclease encoding I-SceI gene was carried by pDYN6902 conjugative plasmid and integrated in the chromosome at the attachment site of phage φC31 (attB φC31). Scissors symbolize the DSB (I-SceI restriction sites) triggered by the meganuclease. (B) Distribution of the loci used for chromosome mapping after DSB induction. T stands for loci included in the TIRs. L, C and R for left, central and right loci, respectively. The data analysis is schematically represented on Fig. 4.
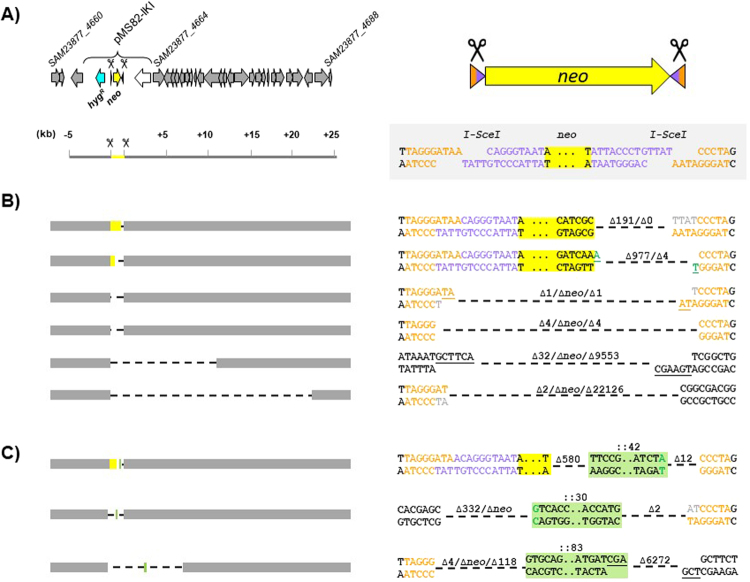
We surveyed the fate of DSBs in the central region by assessing the occurrence of KanS clones which were expected to result from the unfaithful repair of I-SceI cuts at one or both sites of the IKI cassette. To test this hypothesis, fifty independent wild-type lineages harbouring the IKI cassette and expressing I-SceI were selected and their progeny was tested for kanamycin sensitivity (Fig. 2). An overall average of 1.1% of KanS clones was observed (82 KanS clones out 7,420 colonies); each lineage produced between 0 and 10 KanS colonies. The spontaneous KanS frequency in a strain carrying the IKI locus but not expressing I-SceI was significantly lower (<0.02%; 0 out of 5,768 colonies tested, P < 0.01). In order to characterize the repair features, we mapped the IKI locus in each KanS clone by amplifying the whole region and determining the nucleotide sequence of the scar. In all but two cases, the amplification patterns differed from the WT reference and dramatically varied in size revealing deletions estimated to expand between 191 bp and 23,591 bp. In the last two KanS clones, no amplification pattern could be obtained indicating that the deletion was even larger. Given the selective pressure applied to maintain the IKI cassette carrying vector (hygromycin resistance), deletions could not extend beyond the hygR gene located about 500 bp upstream the 5′ I-SceI site (Fig. 3A). Thus, downstream the 3′ I-SceI site, the deletion size reflected the efficiency of the nuclease activity and revealed the dispensability (under laboratory conditions) of the genes included in that chromosomal region (Fig. 3B). Scar sequencing showed that repair events modified one or both I-SceI sites, except for two clones in which both I-SceI sites remained intact despite the deletion of the neo gene. Repair appears to be mediated by micro-homologies of between 1 and 6 nucleotides in 50% of the scars. A nucleotide addition was observed in 40% of the scars and corresponded to a single non-templated nucleotide addition on blunt ends or to a fill-in synthesis of 1 to 4 nucleotides at the 5′ recessed ends. In summary, the scars displayed signatures of a classical NHEJ, as observed in Mycobacterium tuberculosis27. This study provides the first argument in favor of a functional NHEJ repair pathway in Streptomyces.
Figure 2.
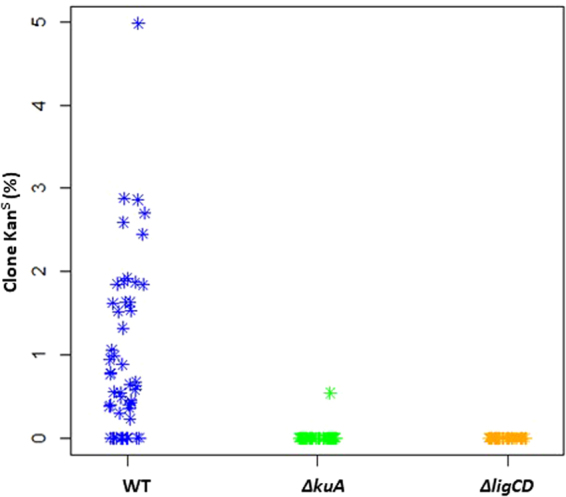
Kanamycin sensitive (KanS) clones appearance after DSB generation at IKI-C. Proportion of KanS clones obtained in each independent lineage (symbolized by a flake) of WT, ∆kuA and ∆ligCD contexts. For each of the 50 lineages, an average of 230 clones was analyzed by replica-plating with and without kanamycin.
Figure 3.
DNA repair features after DSB generation at IKI-C. (A) IKI-C locus: the left panel shows the 30 kb region surrounding the insertion site of pMS82-IKI at IKI-C locus. Scale is in kilobases. Locus_tags of the CDS delimiting the region are indicated according to Thibessard et al.31. The right panel focuses on the IKI cassette structure and sequence. Since the I-SceI recognition site is a non palindromic sequence, the resulting restricted ends are distinguished by two different colors (orange and purple). The neo gene is colored in yellow while hygromycin resistance gene (hygR) is colored in cyan. All DNA repair event examples are represented at these two different scales (see B). (B) Examples of deletion events occurring at IKI-C at the kilobase (left panel) and nucleotide (right panel) scales. Grey boxes represent the region flanking IKI-C cassette. Deleted regions are symbolized by dotted lines. The ∆ symbol indicates the number of deleted nucleotides at each restricted end. When the ∆ symbol is followed by neo, it indicates a complete deletion of the kanamycin resistance gene. (C) DNA integration cases. Nucleotides added by fill-in synthesis are colored in grey. Nucleotides added by non-template addition are colored in green. DNA fragments integrated during repair are boxed in green. Microhomologies present at the repair scar are underlined.
In three KanS clones, the scar analysis revealed the presence of a DNA insertion at the repair point (Fig. 3C). In two cases, the inserted sequence (42 or 83 bp) was originating from a region close to IKI-C. In the third case, the 30 bp sequence perfectly matched to the 5′ end of a transposase gene located in the terminal inverted repeats (TIR).
Mutagenic DSB repair in the central part of the chromosome relies on the NHEJ pathway
In order to confirm the involvement of the NHEJ pathway in DSB repair, we transferred the I-SceI device (enzyme and target cassettes) into strains that were defective for the bacterial NHEJ machinery (deficient for the production of KuA, the main Ku protein, or the ATP-dependant ligases LigC/LigD proteins). Hence, we previously showed that these NHEJ-like genes were involved in the response to genotoxic stress26. As for the WT background, fifty independent lineages per mutant were isolated and screened for KanS clones. In contrast to the WT, only one KanS clone was obtained from ∆kuA lineages and no KanS clone was detected in the ∆ligCD context (Fig. 2) which was significantly different from the WT (t test, α < 0.05). These data confirmed the key role of KuA at the expense of KuB and KuC although both were involved in the cellular response to DNA damaging agents26. It also confirmed that the ligases C and/or D are involved in the illegitimate DSB repair. The unique KanS clone isolated from the ∆kuA background revealed the existence of an alternative illegitimate recombination pathway potentially involving the other two Ku proteins (KuB or KuC). Hence, alternative end joining pathways have been reported in E. coli which is devoid of the canonical NHEJ actors Ku and ATP-dependent DNA ligase LigD28, as well as in eukaryotes (from yeast to human,29,30.
DSB repair in the chromosomal arms generates genome wide rearrangements
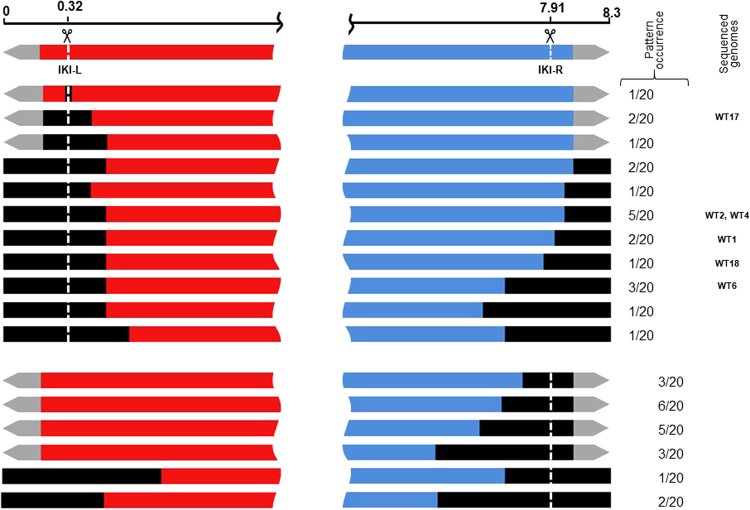
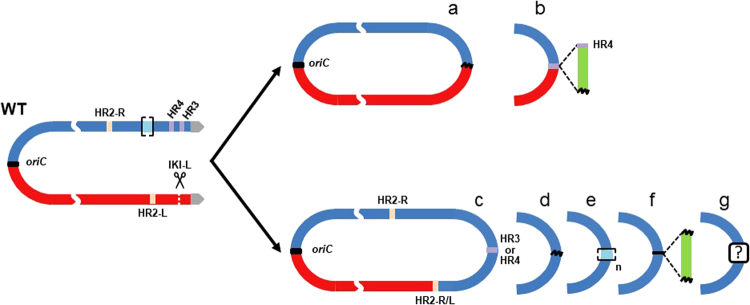
DSBs were generated at two sites in the arms of the WT chromosome: IKI-L (position 0.32 Mb) and IKI-R (position 7.91 Mb; Fig. 1). Twenty lineages were subcloned for each insertion site. Contrasting with DSBs induced in the central part of the chromosome, more than half of the lineages displayed heterogeneous colonial phenotypes at this stage (Fig. S1) and needed further subcloning (up to eight) to reach morphological homogeneity. Once stabilized, the 40 lineages exhibited a wide range of morphologies (Fig. S1C). All but one were KanS. Their chromosomal structures were analyzed through PCR amplification of loci distributed along the chromosome (Fig. 1B). While the loci observed in the centre of the chromosome were systematically present, several loci were simultaneously absent in one or both chromosomal arms suggesting the occurrence of large deletions (Fig. 4). Since chromosome circularization events have frequently been reported in Streptomyces5, deletions affecting both ends were considered to be indicative of such events. Interestingly, circularization appeared more frequent after DSB in the left arm (16 out of 19) than in the right one (3 out of 20, exact Fisher test p < 0.01). As a control, the genome stability of 30 WT lineages carrying the I-SceI gene but not the IKI-cassette was assayed: two out of the 30 strains showed a terminal rearrangement, which is consistent with the previously characterized instability level in S. ambofaciens5. The same procedure (subcloning of 20 lineages and PCR analysis) was applied in the ∆kuA and ∆ligCD contexts after DSB induction at IKI-L. Circularization was indicated in 11 and 15 out of the 20 lineages derived from ∆kuA and ∆ligCD contexts, respectively. At the chromosomal structure level, no significant differences could be observed between WT and NHEJ deficient backgrounds.
Figure 4.
DNA rearrangements occurring in survivors after DSB induction at IKI-L and IKI-R. The chromosome of survivors were mapped using a set of targets for PCR amplication distributed along the chromosome (Fig. 1B). The scale is given in Mb. Grey arrows represent the terminal inverted repeats at each chromosome extremity. The DSB sites (IKI-L or IKI-R) are represented by the vertical white dotted lines. The black boxes represent the deleted areas. The occurrence of each deletion pattern (among 20 strains analyzed per chromosomal arm) and the strains chosen for whole genome sequencing are mentioned on the right.
Six lineages from each of the three genetic backgrounds (WT, ∆kuA, ∆ligCD) cut at IKI-L were chosen according to their diverse PCR patterns for whole genome sequencing (for sequencing data, see Table S2). Sequence reads were aligned on the chromosomal sequence of S. ambofaciens ATCC 23877 as a ref.31. This alignment confirmed that some areas of the chromosome were deleted (i.e. had no read coverage) while other showed an increased coverage level compared to the genome average (by a factor two or more) as illustrated in Fig. 5A. The extent of the deletions can reach several hundreds of kilobases (up to 2.1 Mb) contrasting with that observed in the central part of the chromosome. That could reflect (i) the absence of essential genes in the arms in laboratory growth conditions and (ii) the involvement of a highly efficient exonuclease activity.
Figure 5.
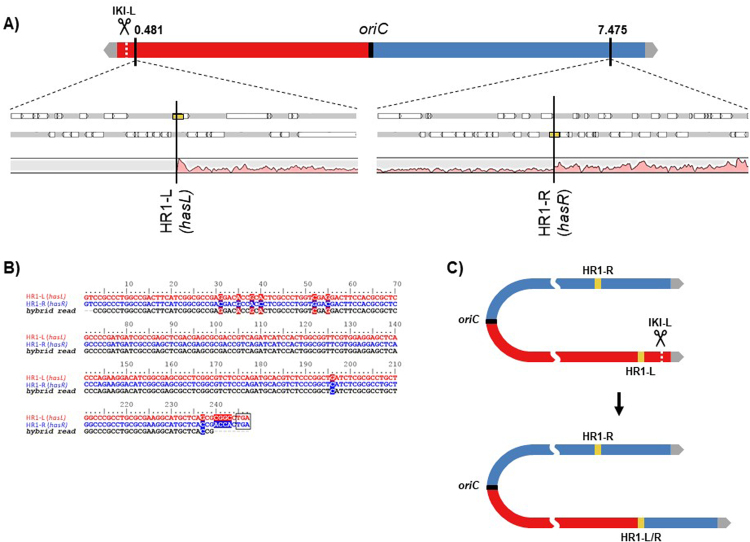
Chromosomal arm replacement by homologous recombination after DSB induction at IKI-L. (A) Analysis of the coverage of sequencing reads along the chromosome of a DSB survivor at IKI-L (WT17). Genome sequencing reads of strain WT17 plotted along the S. ambofaciens ATCC 23877 genome as a reference are shown as a pink area. Yellow boxes symbolize HR1-R and HR1-L (two duplicated copies of a sigma factor encoding gene: hasL, SAM23877_0517, and hasR, SAM23877_6930). (B) Hybrid read resulting from recombination between HR1-L and HR1-R sequences. HR1-L and HR1-R sequences were aligned with one typical hybrid_read found in WT17 sequencing data. The alignment shown covers the 3’ end of the hasL/R sequences (from position 600 to 846). Polymorphism is symbolized with a colored background. The stop codon is boxed. (C) WT17 chromosome structure. The hybrid HR1 sequence is labelled HR1-L/R. The I-SceI cut at IKI-L resulted in the loss of the left chromosomal arm and in a homologous recombination event between HR1-R and HR1-L leading to the duplication of the right chromosomal arm over 825 kb.
In strain WT17 (Fig. 5A) the deleted area extended from the left chromosomal end to the hasL locus (deletion reaching 481 kb) while the area downstream hasR in the right arm had a two-fold coverage reflecting its duplication. Since hasL and hasR share 99% of nucleotide identity over their total length (846 nt,8), we called them HR1-L and HR1-R and suspected a HR event between them, leading to the replacement of the left arm by the right one. The existence of hybrid reads confirmed this hypothesis (Fig. 5B). The genome rearrangement led to the formation of new TIRs of about 829 kb vs. 210 kb in the reference strain (Fig. 5C). This repair event seemed to be the only one that maintained the linear configuration of the chromosome (4 out of 4 linear chromosomes, Table 1). The 14 other sequenced strains harboured a deletion at both chromosomal ends resulting in a circular configuration (Fig. 6). In some cases, circularization occurred by a simple event of fusion between sequences present on the two arms with or without DNA insertion at the fusion point (Fig. 6a,b). Both homologous and illegitimate recombination mechanisms were involved in these fusion events (Table S3). The other cases are typified by a common feature, which consists in the deletion of the left arm until a locus named HR2-L and its recombination with another locus named HR2-R, leading to the formation of a hybrid HR2-R/L sequence. HR2-L and HR2-R correspond to two copies (>99% nucleotide identity) of a 1,672 bp locus carrying two genes (SAM23877_6676 and SAM23877_6677) encoding a sigma factor and a hypothetical protein. The arm replacement resulted in the duplication of the region downstream the recombination point, similarly to the situation described in Fig. 5. However, in these latter cases, chromosomes were invariably circularized. Circularization occurred between sequences belonging to the two copies of the right arm via different molecular events (Fig. 6c,g) involving homologous and/or illegitimate recombination sometimes accompanied by DNA amplification or DNA insertion (Table S3). DNA amplification consisted of several hundred iterations of an amplifiable unit of DNA (AUD). These AUDs ranged from 14.2 to 32.8 kb in size and overlapped the stambomycin biosynthetic gene cluster (from SAM23877_7104 to SAM23877_7128).
Table 1.
Genome features of survival strains after DSB induction at IKI-L in the WT and NHEJ- contexts.
| Strain | Conformationa | Deletion (kb) | Duplication (kb) | Insertion (kb) | Amplificationb | Predicted genome size (Mb)c | ||
|---|---|---|---|---|---|---|---|---|
| Size of the AUD (kb) | Estimated copy number | Estimated total size (kb) | ||||||
| WT1 | Circular (6 f) | 876 | 767 | 9.7 | — | 8.20 | ||
| WT2 | Circular (6c) | 785 | 864 | — | — | 8.38 | ||
| WT4 | Circular (6d) | 827 | 761 | — | — | 8.23 | ||
| WT6 | Circular (6e) | 1,140 | 498 | — | 14.7 | 590 | 8,7 | ~16.40 |
| WT17 | Linear | 481 | 829 | — | — | 8.65 | ||
| WT18 | Circular (6d) | 940 | 701 | — | — | 8.06 | ||
| ∆kuA1 | Circular (6 g) | 984 | 665 | — | — | 7.98 | ||
| ∆kuA6 | Circular (6a) | 683 | — | — | — | 7.62 | ||
| ∆kuA12 | Circular (6e) | 1,140 | 498 | — | 14.7 | 840 | 12,3 | ~20.00 |
| ∆kuA13 | Linear | 481 | 829 | — | — | 8.65 | ||
| ∆kuA14 | Circular (6c) | 800 | 783 | — | — | 8.28 | ||
| ∆kuA17 | Linear | 481 | 829 | — | — | 8.65 | ||
| ∆ligCD6 | Circular (6b) | 1,297 | — | 18.8 | — | 7.02 | ||
| ∆ligCD7 | Circular (6e) | 1,097 | 522 | — | 32.8 | 325 | 10,7 | ~18.40 |
| ∆ligCD9 | Circular (6e) | 1,140 | 498 | — | 14.7 | 820 | 12,1 | ~20.00 |
| ∆ligCD11 | Linear | 481 | 829 | — | — | 8.65 | ||
| ∆ligCD16 | Circular (6c) | 785 | 864 | — | — | 8.38 | ||
| ∆ligCD19 | Circular (6a) | 2,116 | — | — | — | 6.18 | ||
aThe chromosomal conformation referring to Fig. 6 is indicated into brackets.
bAUD stands for Amplifiable unit of DNA. The copy number of the amplification was estimated considering the average coverage of sequencing reads of the amplified region relative to that of the whole genome.
cThe genome size of the mutants carrying an amplification of DNA cannot be precisely evaluated.
Figure 6.
Chromosomal circularization after DSB induction at IKI-L. HR2-L and HR2-R are two homologous regions sharing over 99% nucleotide identity along 1,672 bp and are symbolized as salmon pink boxes. The hybrid HR2 sequence is labelled HR2-L/R. HR3 is a region including two identical transposase encoding genes (SAM23877_7417 and SAM23877_7422) in opposite direction. Similarly, HR4 is a region including two identical transposase encoding genes (SAM23877_7326 and SAM23877_7404) in opposite direction. HR3 and HR4 are symbolized as purple boxes. Illegitimate recombination scars are represented by black wave lines. Green boxes represent inserted DNA stretches. Amplified sequences are marked out by light blue boxes between brackets. In some cases, circularization occurred by simple event of fusion between sequences present on both arms with (a) or without (b) DNA insertion. The other cases are typified by a common feature, which consists in the deletion of the left arm until the HR2-L locus and its recombination with its homologous region on the right arm (HR2-R), leading to the formation of a hybrid HR2-R/L sequence. This arm replacement is concomitant with circularization through different molecular event: HR involving HR3 or HR4 (c), IR (d) sometimes accompanied by DNA amplification (e) or DNA insertion (f). For case e, the read coverage level of the amplified region prevents the identification of the fusion sequence and thus the nature of the recombination event. For case (g), the circularization may have occurred through recombination between two short homologous sequences (41 bp) in opposite direction.
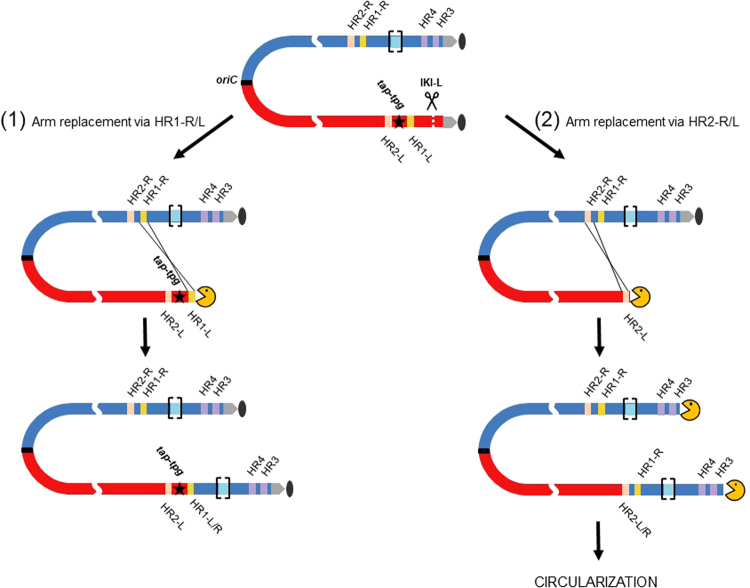
A striking difference distinguished chromosome salvage events involving HR1 loci from those involving HR2 loci. While all the repaired chromosomes recombined through HR1 loci remained linear (4 out of 4), all the events involving HR2 loci became circular (11 out of 11). The location of genes encoding the terminal proteins Tpg and Tap involved in telomere maintenance (SAM23877_0536/0537) between both HR1 and HR2 recombination sites provides the most probable explanation for these contrasting situations (Fig. 7). Hence, arm replacement involving the HR2 loci remove the tap/tpg genes from the chromosome; in consequence, linearity can no longer be maintained and the only way out is circularization. In contrast, when arm replacement involved the HR1 loci, the tap/tpg locus remained present and chromosomal linearity was kept. This is consistent with the report of the circularization of Streptomyces rochei chromosome triggered by the loss of linear plasmids encoding the Tap-Tpg functions32.
Figure 7.
Fates of the chromosome structure following DSB and arm replacement. Red and blue boxes represent left and right replichores of the chromosome respectively. Grey arrows and the grey oval represent the terminal inverted repeats and the terminal proteins attached to them. HR2-L and HR2-R are symbolized as salmon pink boxes. The hybrid HR1 and HR2 sequences are labelled HR1-L/R and HR2-L/R respectively. HR3 and HR4 are symbolized as purple boxes. The AUD90 locus, susceptible to DNA amplification is marked out by light blue boxes between brackets. The tap-tpg locus encoding the terminal associated protein and the terminal protein respectively is represented by a black star. The yellow PacMan symbolizes the accessibility of the chromosome end to nuclease activities. After DSB induction at IKI-L, DNA resection progresses towards the inner parts of the chromosome concomitantly with the loss of the distal part of the arm. In case (1), DNA resection does not reach the tap-tpg locus, and arm replacement occurs by HR between HR1-R and HR1-L. Since tap-tpg locus is preserved, the linear configuration of the rearranged chromosome is stable. In case (2), DNA resection gets past tap-tpg locus and arm replacement is initiated by HR between HR2-R and HR2-L. The tap-tpg being deleted, this chromosome is unstable and irremediably leads to stabilization through circularization by different ways involving either HR3, or HR4 or an amplified sequence as previously shown.
Overall, these rearrangements led to genome size ranging from 6.18 Mb up to c. 20 Mb (reached in case of DNA amplification, Table 1).
Discussion
Deciphering the mechanisms of genome plasticity in Streptomyces is pivotal to understand the biology of this soil bacterium in its competitive ecosystem and to exploit its prolific secondary metabolism. In Streptomyces, the remarkable organization of the chromosome2,3, i.e. central conserved versus terminal variable regions, questions the recombination mechanisms ensuring genome stability. Early genome comparisons achieved in our lab4 showed that variability in the subtelomeres is fueled by the input of exogenous information through horizontal transfer and driven by a very specific recombination pattern along the chromosome, i.e. recombination frequencies growing towards the chromosome ends. Moreover, Streptomyces are well-known as providers of a wide range of bioactive natural products and constitute an untapped source for new treatments against human microbial infections as well as against cancer. Genes responsible for the biosynthesis of metabolites of interest are usually clustered and frequently located in the terminal regions of the chromosome33,34. They are consequently subjected to DNA plasticity. Traces of recombination events are noticeable within the structure and sequence of these gene clusters. Studying genome plasticity can thus give insight into the evolution of the repertoire of secondary metabolism and open the way to its diversification using recombination.
Although HR is known to contribute to genome stability as an error free repair mechanism in all living organisms, our results point out that it is also a main driving force of genome structure plasticity in Streptomyces. DSB repair in the terminal regions was most of the time associated with the loss of the broken arm as well as a deletion towards the inner part of the chromosome. In a vast majority of analyzed genomes (15 of 18), chromosome salvage was achieved by chromosomal arm replacement. Two pairs of DNA repeats (i.e. HR1-L/HR1-R and HR2-L/HR2-R) present on both arms in opposite orientation allows HR to initiate either a crossing over with the opposite arm of another chromosome present in the mycelium compartment or with the opposite arm of the same chromosome. In the former case, recombination leads to the acquisition of an intact arm, the latter could initiate a break-induced replication (BIR) event35. The BIR mechanism is well-known in yeast as a powerful mechanism leading to large segmental duplications of the sub-telomere region. This is also underlying the loss of heterozygosis in mammalian cells36.
The stepwise plasticity associated with the loss of tap/tpg locus also explains the heterogeneity of the progeny observed over several rounds of sporulation. While phenotypic stability was easily reached when the DSB was induced in the right arm, a strong and persisting heterogeneity imposed to subclone several times the lineages derived after a DSB in the left arm. The successive events required for chromosome stabilization could explain the segregation of different colonial phenotypes. This is reminiscent of the hypervariability described in S. ambofaciens and the instability cycle associated with the chromosome fusion10,37.
Out of 18 mutant chromosomes analyzed, four demonstrated extensive DNA amplification (Table 1). All occurred in a locus known as AUD9037 which was later shown to include a large secondary metabolite gene cluster encoding PKSs involved in the biosynthesis of stambomycins38,39. This locus is typified by the presence of numerous repeats corresponding to modular domains of the PKS complex. A close analysis of the boundaries of the amplified unit revealed the presence of short repeated sequences (several hundreds of nucleotides sharing over 90% of identity) that could have been used as substrate for HR. This would support a model proposed by Young and Cullum (1987) in which a replication fork is trapped on the circular amplifiable template formed after recombination between two direct repeats on the chromosome. The implication of HR is also supported by the abolishment of amplification in a recA mutant in S. lividans40. It is interesting to point out that all these amplification cases were associated with an arm replacement event suggesting that the trapped replication fork could have been initiated upstream on the chromosome during a BIR event. Another interesting feature is the circular configuration of the amplified chromosome with large deletion adjacent to the amplification. This implies that the circularization point involves some sequences present in the amplifiable locus. Although no circularization junction could be isolated among the tens of thousands of reads covering this locus in each strain, hindering the identification of the molecular mechanism, the circularization event within this region could be highly frequent because of the high proportion of this amplified DNA in the total genomic DNA. Further, it is not excluded that circularization occurred at several points and by different mechanisms (HR and IR) within the same strain.
Integration of new genetic information can be efficiently achieved through HR between flanking homologous regions. However, HR cannot apply when DNA sequences of donor and receptor are divergent or when incoming DNA does not find any homologous region to recombine with (e.g. specific regions of Streptomyces chromosome). Hence, illegitimate recombination was reported to favor DNA insertion at recombination points in E. coli28 and in eukaryotes41–43. In S. ambofaciens, we identified 5 insertion events at DSB healing sites with size ranging from 30 bp to 18.8 kb. In our experiments, the inserted sequences originated from S. ambofaciens genome and were unambiguously identified thanks to their endogenous origin. We speculate that in presence of exogenous DNA (conducted by conjugational processes for instance) NHEJ could play an active role in the integration of new sequences. Interestingly, a correlation between the capacity of acquisition of foreign DNA and the presence of a NHEJ pathway in prokaryotes was underlined by Popa et al.44. Thus, NHEJ may participate to overcome barriers to horizontal transfer between distantly related organisms.
Methods
Bacterial strains
S. ambofaciens strains were grown at 30 °C on Soya Flour Mannitol (SFM) plates or in Hickey Tresner (HT) medium for liquid cultures. All Streptomyces modified strains derive from our reference strain S. ambofaciens ATCC 2387745. To isolate lineages following DSB induction, a minimum of three subcloning rounds (up to eight rounds until phenotype homogeneity was reached) was realized on SFM plates supplemented with the appropriate selective antibiotic, then spores were harvested as previously described46. The DH5α E. coli strain was used as host for native and recombinant BACs and plasmids. The ET12567 non methylating E. coli strain containing the mobilizing pUZ8002 plasmid was used as donor for all intergeneric conjugation between E. coli and S. ambofaciens. E. coli strains were grown in Luria-Bertani (LB) medium at 37 °C except for the BW25113 pKD20 thermosensitive strain used for PCR targeting, which was grown at 30 °C. When necessary, antibiotics were added to the medium at a concentration of 50 mg/mL for apramycin, kanamycin and hygromycin, and 25 mg/mL for nalidixic acid and chloramphenicol.
I-SceI mediated DSB targeting
The coding sequence of yeast endonuclease I-SceI47 was optimized for expression in Streptomyces and I-SceI gene was cloned under the control of PtipA in a conjugative and integrative vector (pDYN6902,48. The construct, pDYN6902-I-SceI, was transferred from E. coli to S. ambofaciens by conjugation, where it stably integrates in the chromosome by site-specific recombination at bacteriophage φC31 attachment site (at position 4.11 Mb) (Fig. 1). The basic expression of I-SceI gene in S. ambofaciens, i.e. in the absence of the inducer thiostrepton, was checked by semi-quantitative RT-PCR. Hence PtipA is known to enable a basic expression49,50. The I-SceI endonuclease was therefore considered as being expressed in S. ambofaciens as soon as the construct was introduced by conjugation. We constructed a target cassette of the endonuclease consisting of the aminoglycoside 3’-phosphotransferase gene (neo, conferring the resistance to kanamycin), flanked by two I-SceI sites. The two sites were in a convergent orientation in order to avoid the compatibility of the overhanging DNA extremities produced by the endonuclease. This cassette was named IKI for I-SceI/kanR/I-SceI. For the insertion in the central part of the chromosome, the IKI cassette was inserted in the conjugative plasmid pMS8251 that integrates by site-specific recombination at the φBT1 attachment site (position 4.94 Mb). This insertion was called IKI-C, C for central region (Fig. 1). To trigger DSBs in the chromosomal arms, the IKI cassette was inserted by homologous recombination into intergenic spaces (positions 0.32 Mb and 7.91 Mb) to generate the insertion sites IKI-L and IKI-R for left and right arm insertion respectively. Briefly, the IKI cassette was inserted by PCR targeting52 into a recombinant BAC containing the locus of interest. Transfer of the recombinant BAC by intergeneric conjugation E. coli/Streptomyces into S. ambofaciens followed by suitable antibiotic selection (kanamycin resistance for the IKI cassette, sensitivity to apramycin for the removal of the BAC) led to the selection for the insertion of the IKI cassette into the targeted intergenic sequence. For all experiments, the IKI cassette insertion was selected before the introduction of the I-SceI expression construct (pDYN6902- I-SceI).
Molecular analyses
For repair scar or chromosomal structure analyses, Streptomyces genomic DNA was extracted by low binding sulfate salt method as described by Kieser et al.46. Liquid HT medium was used for growth of mycelium before DNA extraction. To determine the nucleotide sequence of the scars after DSB at IKI-C, the locus was amplified by PCR and the amplicon was gel purified then sequenced by the Sanger method (Beckman Coulter Genomics). For chromosomal structure analyses, a set of PCR amplifications was carried out using primer pairs distributed along the WT chromosome (Fig. 1B) on genomic DNA extracted from strains isolated after DSB induction at IKI-L or IKI-R locus (150 isolates).
Whole genome sequencing
Genomic DNA was extracted as described above and paired-ends sequencing of short DNA fragments (average size 760 nt) using an Illumina Genome Analyzer was performed for 18 strains (Table S2). Mapping of sequencing reads on genome reference as well as coverage analyses was carried out using CLC Genome Workbench (version 6.5, Qiagen).
Statistical analyses
Fisher’s exact test was used to test the statistical significance of the different proportions (small samples) throughout the work.
Electronic supplementary material
Acknowledgements
This work was supported by the French National Research Agency program Streptoflux (ANR Blanc 0096_01), by ANR through the Laboratory of Excellence ARBRE (ANR-12- LABXARBRE-01) and by the Région Lorraine.
Author Contributions
Investigation: G.H., C.B., E.P.; Computer analysis: A.T. and P.L.; Methodology: C.B., A.T., P.L.; Conceptualization: C.B., A.T., and P.L.; Writing original draft: G.H., C.B., A.T., and P.L.; Writing-review and editing: C.B., A.T., and P.L.; Resources: P.L.; Funding acquisition: C.B., A.T., and P.L.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23622-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chater KF. Recent advances in understanding. Streptomyces. F1000Research. 2016;5:2795. doi: 10.12688/f1000research.9534.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda H, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 4.Choulet F, et al. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol. Biol. Evol. 2006;23:2361–2369. doi: 10.1093/molbev/msl108. [DOI] [PubMed] [Google Scholar]
- 5.Thibessard, A. & Leblond, P. Subtelomere Plasticity in the Bacterium Streptomyces. in Subtelomeres 243–258 (Springer, Berlin, Heidelberg) 10.1007/978-3-642-41566-1_14 (2014).
- 6.Fischer G, Decaris B, Leblond P. Occurrence of deletions, associated with genetic instability in Streptomyces ambofaciens, is independent of the linearity of the chromosomal DNA. J. Bacteriol. 1997;179:4553–4558. doi: 10.1128/jb.179.14.4553-4558.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue S, Higashiyama K, Uchida T, Hiratsu K, Kinashi H. Chromosomal circularization in Streptomyces griseus by nonhomologous recombination of deletion ends. Biosci. Biotechnol. Biochem. 2003;67:1101–1108. doi: 10.1271/bbb.67.1101. [DOI] [PubMed] [Google Scholar]
- 8.Fischer G, Wenner T, Decaris B, Leblond P. Chromosomal arm replacement generates a high level of intraspecific polymorphism in the terminal inverted repeats of the linear chromosomal DNA of Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA. 1998;95:14296–14301. doi: 10.1073/pnas.95.24.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchida T, Miyawaki M, Kinashi H. Chromosomal arm replacement in Streptomyces griseus. J. Bacteriol. 2003;185:1120–1124. doi: 10.1128/JB.185.3.1120-1124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wenner T, et al. End-to-end fusion of linear deleted chromosomes initiates a cycle of genome instability in Streptomyces ambofaciens. Mol. Microbiol. 2003;50:411–425. doi: 10.1046/j.1365-2958.2003.03698.x. [DOI] [PubMed] [Google Scholar]
- 11.Leblond P, Demuyter P, Simonet JM, Decaris B. Genetic instability and hypervariability in Streptomyces ambofaciens: towards an understanding of a mechanism of genome plasticity. Mol. Microbiol. 1990;4:707–714. doi: 10.1111/j.1365-2958.1990.tb00641.x. [DOI] [PubMed] [Google Scholar]
- 12.Birch A, Häusler A, Rüttener C, Hütter R. Chromosomal deletion and rearrangement in Streptomyces glaucescens. J. Bacteriol. 1991;173:3531–3538. doi: 10.1128/jb.173.11.3531-3538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanai K, Murakami T, Bibb M. Amplification of the entire kanamycin biosynthetic gene cluster during empirical strain improvement of Streptomyces kanamyceticus. Proc. Natl. Acad. Sci. USA. 2006;103:9661–9666. doi: 10.1073/pnas.0603251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamasaki M, Kinashi H. Two chimeric chromosomes of Streptomyces coelicolor A3(2) generated by single crossover of the wild-type chromosome and linear plasmid scp1. J. Bacteriol. 2004;186:6553–6559. doi: 10.1128/JB.186.19.6553-6559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandza S, et al. Recombination between the linear plasmid pPZG101 and the linear chromosome of Streptomyces rimosus can lead to exchange of ends. Mol. Microbiol. 1998;28:1165–1176. doi: 10.1046/j.1365-2958.1998.00877.x. [DOI] [PubMed] [Google Scholar]
- 16.Rocha EPC, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1:e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang T-W, Chen CW. A recA null mutation may be generated in Streptomyces coelicolor. J. Bacteriol. 2006;188:6771–6779. doi: 10.1128/JB.00951-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, et al. The adnAB locus, encoding a putative helicase-nuclease activity, is essential in Streptomyces. J. Bacteriol. 2014;196:2701–2708. doi: 10.1128/JB.01513-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff G, Bertrand C, Piotrowski E, Thibessard A, Leblond P. Implication of RuvABC and RecG in homologous recombination in Streptomyces ambofaciens. Res. Microbiol. 2017;168:26–35. doi: 10.1016/j.resmic.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Michel, B. & Sandler, S. J. Replication Restart in Bacteria. J. Bacteriol. 199 (2017). [DOI] [PMC free article] [PubMed]
- 21.Glickman, M. S. Double-Strand DNA Break Repair in Mycobacteria. Microbiol. Spectr. 2 (2014). [DOI] [PubMed]
- 22.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doherty AJ, Jackson SP, Weller GR. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 2001;500:186–188. doi: 10.1016/S0014-5793(01)02589-3. [DOI] [PubMed] [Google Scholar]
- 24.Weller GR, et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 25.Della M, et al. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 26.Hoff G, et al. Multiple and Variable NHEJ-Like Genes Are Involved in Resistance to DNA Damage in Streptomyces ambofaciens. Front. Microbiol. 2016;7:1901. doi: 10.3389/fmicb.2016.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta R, Barkan D, Redelman-Sidi G, Shuman S, Glickman MS. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 2011;79:316–330. doi: 10.1111/j.1365-2958.2010.07463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chayot R, Montagne B, Mazel D, Ricchetti M. An end-joining repair mechanism in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2010;107:2141–2146. doi: 10.1073/pnas.0906355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 30.Sinha S, et al. Microhomology-mediated end joining induces hypermutagenesis at breakpoint junctions. PLoS Genet. 2017;13:e1006714. doi: 10.1371/journal.pgen.1006714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thibessard A, et al. Complete genome sequence of Streptomyces ambofaciens ATCC 23877, the spiramycin producer. J. Biotechnol. 2015;214:117–118. doi: 10.1016/j.jbiotec.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Nindita Y, et al. The tap-tpg gene pair on the linear plasmid functions to maintain a linear topology of the chromosome in Streptomyces rochei. Mol. Microbiol. 2015;95:846–858. doi: 10.1111/mmi.12904. [DOI] [PubMed] [Google Scholar]
- 33.Aigle B, et al. Genome mining of Streptomyces ambofaciens. J. Ind. Microbiol. Biotechnol. 2014;41:251–263. doi: 10.1007/s10295-013-1379-y. [DOI] [PubMed] [Google Scholar]
- 34.Xu M-J, et al. Deciphering the streamlined genome of Streptomyces xiamenensis 318 as the producer of the anti-fibrotic drug candidate xiamenmycin. Sci. Rep. 2016;6:18977. doi: 10.1038/srep18977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.So A, Le Guen T, Lopez BS, Guirouilh-Barbat J. Genomic rearrangements induced by unscheduled DNA double strand breaks in somatic mammalian cells. FEBS J. 2017;284:2324–2344. doi: 10.1111/febs.14053. [DOI] [PubMed] [Google Scholar]
- 37.Leblond P, et al. Hypervariability, a new phenomenon of genetic instability, related to DNA amplification in Streptomyces ambofaciens. J. Bacteriol. 1989;171:419–423. doi: 10.1128/jb.171.1.419-423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aigle B, et al. An amplifiable and deletable locus of Streptomyces ambofaciens RP181110 contains a very large gene homologous to polyketide synthase genes. Microbiol. Read. Engl. 1996;142(Pt 10):2815–2824. doi: 10.1099/13500872-142-10-2815. [DOI] [PubMed] [Google Scholar]
- 39.Laureti L, et al. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volff JN, Viell P, Altenbuchner J. Artificial circularization of the chromosome with concomitant deletion of its terminal inverted repeats enhances genetic instability and genome rearrangement in Streptomyces lividans. Mol. Gen. Genet. MGG. 1997;253:753–760. doi: 10.1007/s004380050380. [DOI] [PubMed] [Google Scholar]
- 41.Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 43.Ricchetti M, Dujon B, Fairhead C. Distance from the chromosome end determines the efficiency of double strand break repair in subtelomeres of haploid yeast. J. Mol. Biol. 2003;328:847–862. doi: 10.1016/S0022-2836(03)00315-2. [DOI] [PubMed] [Google Scholar]
- 44.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinnert-Sindico S. [A new species of Streptomyces producing antibiotics Streptomyces ambofaciens n. sp., cultural characteristics] Ann. Inst. Pasteur. 1954;87:702–707. [PubMed] [Google Scholar]
- 46.Kieser, T., Bibb, M., Buttner, M., Chater, K. & Hopwood, D. Practical Streptomyces genetics. (John Innes Foundation, 2000).
- 47.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/S0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 48.Thibessard A, Bertrand C, Hiblot J, Piotrowski E, Leblond P. Construction of pDYN6902, a new Streptomyces integrative expression vector designed for cloning sequences interfering with Escherichia coli viability. Plasmid. 2015;82:43–49. doi: 10.1016/j.plasmid.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Chiu ML, et al. Broad spectrum thiopeptide recognition specificity of the Streptomyces lividans TipAL protein and its role in regulating gene expression. J. Biol. Chem. 1999;274:20578–20586. doi: 10.1074/jbc.274.29.20578. [DOI] [PubMed] [Google Scholar]
- 50.Ali N, Herron PR, Evans MC, Dyson PJ. Osmotic regulation of the Streptomyces lividans thiostrepton-inducible promoter, ptipA. Microbiol. Read. Engl. 2002;148:381–390. doi: 10.1099/00221287-148-2-381. [DOI] [PubMed] [Google Scholar]
- 51.Gregory MA, Till R, Smith MCM. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J. Bacteriol. 2003;185:5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.