Abstract
The pathogenic relevance in Alzheimer’s disease (AD) presents a decrease of cerebrospinal fluid (CSF) amyloid-β42 (Aβ42) burden and an increase in CSF total-tau (T-tau) levels. In this work, we performed genome-wide association study (GWAS) and genome-wide interaction study (GWIS) of T-tau/Aβ42 ratio as an AD imaging quantitative trait (QT) on 843 subjects and 563,980 single nucleotide polymorphisms (SNPs) in ADNI cohort. We aim to identify not only SNPs with significant main effects but also SNPs with interaction effects to help explain “missing heritability”. Linear regression method was used to detect SNP-SNP interactions among SNPs with uncorrected p-value≤0.01 from the GWAS. Age, gender and diagnosis were considered as covariates in both studies. The GWAS results replicated the previously reported AD-related genes APOE, APOC1 and TOMM40, as well as identified 14 novel genes, which showed genome-wide statistical significance. GWIS revealed 7 pairs of SNPs meeting the cell-size criteria and with bonferroni-corrected p-value≤0.05. As we expect, these interaction pairs all had marginal main effects but explained a relatively high-level variance of T-tau/Aβ42, demonstrating their potential association with AD pathology.
Keywords: Cerebrospinal fluid (CSF), Amyloid-β42 (Aβ42), Total tau (T-tau), T-tau/Aβ42 ratio, GWIS, ADNI
1. Introduction
Alzheimer’s disease (AD) is the most common form of dementia and characterized by pathological results at autopsy of the accumulation of amyloid-β (Aβ) protein in senile plaques and hyper-phosphorylated tau in neurofibrillary tangles in brain (Mukaetova-Ladinska et al., 2015). The levels of two measures, cerebrospinal fluid (CSF) amyloid-β42 (Aβ42) and total tau (T-tau), have been shown strong promise as predictive biomarkers of the progression from mild cognitive impairment (MCI, a prodromal stage of AD) to AD (Blennow and Hampel, 2003; Pan et al., 2015). Typically, the pathogenic relevance in AD presents a decrease of CSF Aβ42 burden and an increase in CSF T-tau levels simultaneously (Li et al., 2015).
However, the emerging literatures reported that a group of individuals have never shown clinical symptoms of AD in their lifetime but detected out tauopathies and amyloid plaques at autopsy (Hohman et al., 2014). In addition, some normal cognitive individuals presented low CSF Aβ42 burden and some individuals with definitive diagnosis of AD showed high levels of CSF Aβ42 due to their lack of amyloid deposition (Fagan et al., 2006). The emergence of this situation posed challenges on discriminating individuals with AD from normal cognitive, and affected the diagnostic potential of these markers. To address this issue, a potential biomarker CSF T-tau/Aβ42 ratio demonstrated its predictive ability. It can be used to detect and measure the AD risk with cognitive decline in non-demented older adults, and individuals with higher ratio tend to have higher risk to develop AD (Fagan et al., 2007). Moreover, prior studies also showed that individuals with family history of AD had higher risk for AD than those without a family history. This indicates that the underlying genetic factors may play an important role in AD (Hohman et al., 2014).
The existing genome-wide association studies (GWAS) have analyzed Single Nucleotide Polymorphism (SNP) data and discovered a wide array of underlying genetic causes of AD and genetic associations with AD biomarkers as intermediate quantitative traits (QTs). For many conditions of complex diseases and traits, commonly used single marker analysis can identify a number of risk genetic loci, but these identified variants typically appear to explain only a modest portion of the theoretical estimates of genetic heritability (Goudey et al., 2013). One possible reason is that the univariate methods used in GWAS typically ignore the factor of underlying genetic interaction, which may contribute to the development of disease and trait variance.
Thus, one of several putative explanations for the “missing heritability” is that the trait variance can partially be explained by the SNP-SNP interaction effects in addition to their main effects (Becker et al., 2011). Therefore, genome-wide interaction studies (GWIS) have recently gained substantial attention (J. Li et al., 2015).
In this study, we performed both GWAS and GWIS in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. We used the AD-associated CSF T-tau/Aβ42 ratio as QT, and tested single-marker main effects and two-marker interactions at the genome-wide level.
2. Materials and Methods
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD).
We applied for and were granted permission to use data from the ADNI cohort (http://www.adni-info.org/) to conduct genetic association and interaction analyses.
2.1. Subjects
Participants are ADNI subjects (N=843) with CSF T-tau and Aβ42 measures and quality controlled genotyping data available at baseline. The sample included 199 cognitively normal (CN), 85 significant memory concern (SMC), 239 early mild cognitive impairment (EMCI), 207 late mild cognitive impairment (LMCI), and 113 AD participants. Table 1 shows selected participant characteristics at the baseline, which is the time point studied in this work.
Table 1.
The demographic and clinical characteristics of 843 ADNI participants at baseline studied in this work.
| CN (N=199) | SMC (N=85) | EMCI (N=239) | LMCI (N=207) | AD (N=113) | ||
|---|---|---|---|---|---|---|
| Age (years) | 74.4(7.79) | 72.0(5.48) | 71.4(7.30) | 72.4(7.61) | 75.2(8.19) | |
| Women | 96(48.2%) | 50(58.8%) | 102(43.6%) | 83(40.1%) | 45(39.8%) | |
| Education (years) | 16.21(2.82) | 16.00(2.79) | 16.16(2.80) | 16.38(2.53) | 16.40(2.56) | |
| APOE e4 allele present | 47(23.6%) | 31(36.5%) | 99(41.4%) | 112(54.1%) | 74(65.5%) | |
| CDR–SOB | 0.04(0.14) | 0.08(0.18) | 1.27(0.77) | 1.65(0.94) | 4.53(1.70) | |
| Mini mental status examination | 29.06(1.18) | 29.04(1.420) | 28.34(1.62) | 27.54(1.75) | 23.12(2.04) | |
| Logical memory immediate recall (WMS-R) | 14.42(3.00) | 14.44(3.34) | 11.09(2.68) | 7.18(3.06) | 4.15(2.70) | |
| Logical memory delayed recall (WMS-R) | 13.34(3.12) | 13.29(3.31) | 8.97(1.73) | 3.94(2.70) | 1.52(1.80) | |
| CSF T-tau (Total tau) | 69.76(31.76) | 66.95(31.68) | 77.66(46.97) | 98.22(52.50) | 126.26(54.47) | |
| CSF Aβ42 (amyloid-β42) | 198.09(52.87) | 201.42(49.43) | 184.50(51.41) | 162.92(52.80) | 136.90(36.25) | |
| Quantitative Traits (QTs) | T-tau/Aβ42 ratio | 0.40(0.27) | 0.37(0.24) | 0.50(0.45) | 0.70(0.47) | 0.98(0.49) |
Abbreviations: AD, Alzheimer’s disease; ADNI, Alzheimer’s Disease Neuroimaging Initiative; CDR–SOB, clinical dementia rating–sum of boxes; CN, cognitive normal; SMC, significant memory concern; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment; WMS-R, Wechsler Memory Scale-Revised. Data are number (%) or mean (s.d).
2.2. Quality control of genotyping data
The genotyping data of the ADNI-1, ADNI-GO and ADNI-2 cohorts were collected using either the Illumina 2.5M array (a byproduct of the ADNI whole genome sequencing sample) or the Illumina OmniQuad array (Saykin et al., 2010; Shen et al., 2010; Shen et al., 2014), and were downloaded from the LONI website (http://adni.loni.usc.edu). For the present analyses, we included SNPs that were present on both arrays.
Quality control (QC) was performed using the PLINK software (version 1.90) (Purcell et al., 2007). SNPs were removed from the analysis if any of the following criteria were not satisfied: (1) SNPs on chromosome 1–22; (2) call rate per SNP ≥ 95%; (3) minor allele frequency ≥ 5% (1,845,510 SNPs were removed based on Criteria 1, 2 and 3); and (4) Hardy-Weinberg equilibrium (HWE) test of p ≥ 10−6 using CN subjects only (198 SNPs were removed). Participants not meeting any of the following criteria were removed from further analyses: (1) call rate per participant ≥ 90% (none); (2) sex check (1 participant was removed); and (3) identity check for related pairs (8 sibling pairs and one triplet were identified with PI_HAT ≥ 0.25, 9 participants (one from each pair or triplet) were randomly selected and included in this study).
Population stratification analysis was performed using EIGENSTRAT (Price et al., 2006), and confirmed using STRUCTURE (Pritchard et al., 2000). It yielded 89 participants who did not cluster with the remaining subjects and with the CEU HapMap samples who are primarily of European ancestry (non-Hispanic Caucasians). These 89 participants were excluded from the analysis. Among the remaining 1,079 subjects, only 843 subjects have both genotyping data and phenotypes (T-tau and Aβ42) after quality control (QC), and thus the other 236 participants were excluded.
After QC, 843 subjects and 563,980 SNPs remained for the subsequent genome-wide association and interaction analyses.
2.3. CSF T-tau/Aβ42 biomarker
In our study, the CSF levels of T-tau and Aβ42 at baseline were used. The methods for CSF acquisition and biomarker measurement have been reported previously (Hampel et al., 2010; Jagust et al., 2009; Shaw et al., 2009). For this analyses, the Aβ42 and T-tau data were log-transformed to better approximate normality in distribution (Dickerson et al., 2013), and the values greater or smaller than 4 SDs (standard deviation) from the mean value of Aβ42 and T-tau were regarded as extreme outliers and excluded from the analyses. After QC, 843 valid CSF samples remained.
2.4. Method of GWAS and GWIS
GWAS was used to evaluate the SNPs main effects at the genome-wide level. Our study performed a genotypic model based GWAS using PLINK 1.90 to detect the association between SNPs and the T-tau/Aβ42 ratio with age, gender and clinical diagnosis (five values (1–5) indicating CN, SMC, EMCI, LMCI and AD respectively) as covariates. Manhattan plots and quantile-quantile (Q-Q) plots were generated using Haploview (http://www.broad.mit.edu/mpg/haploview/) and R (http://www.r-project.org) respectively.
For GWIS detecting the SNP-SNP interactions, software tool INTERSNP (Herold et al., 2009) was used for two-marker analysis. The input files of the tool were PLINK genotype files. Firstly, single-marker test was performed for GWAS as previously described. The SNPs that met the threshold (uncorrected p-value ≤ 0.01) were included in the subsequent interaction analysis. Linear regression model was used for an additive interaction test (full model including both additive and dominance effects plus interaction term versus reduced model that does not contain interaction terms) on all possible SNP pairs among the previous SNPs selected in first step. We detected the epistasis interactions with the T-tau/Aβ42 ratio as QT while controlling for covariates including the baseline age, gender, and clinical diagnosis. There were about 22 million unique SNP pairs to be examined, and the Bonferroni corrected p-value < 0.05 was used as the statistical significance threshold.
2.5. Post hoc analysis
We performed a hierarchical linear regression among the significant interactions, used IBM SPSS 20 to estimate the amount of variance (R2) on the T-tau/Aβ42 level accounted for by these interaction terms. We first included the same set of covariates (age, gender, and diagnosis) in the linear model, and then included apolipoprotein E (APOE) status, the best-known AD risk gene (Akiyama et al., 1993), and two main effects of SNPs from the significant pair. Finally, we included the SNP-SNP interaction term and computed additional variance explained by interaction term. The difference in R2 for the significant model was calculated in SPSS as ΔR2 = R2full (full model with interaction term) – R2domain (reduced model without interaction term).
3. Results
3.1. Genome-wide association study results
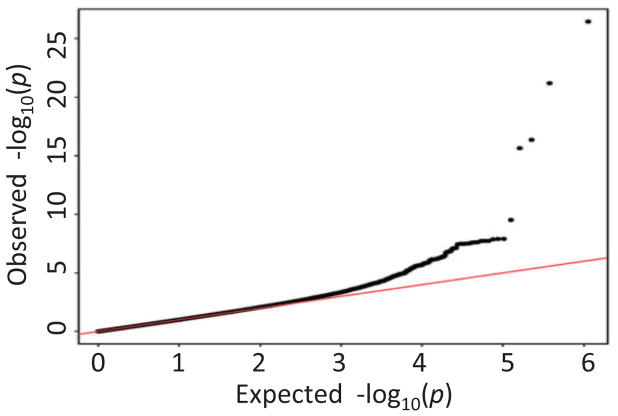
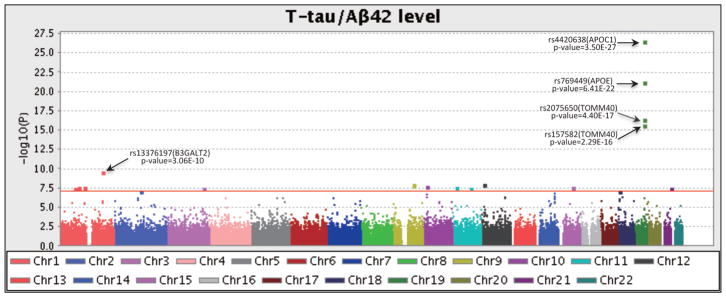
The demographic and clinical characteristics of the 843 participants at baseline were shown in Table 1. Q-Q plot (Fig. 1) shows five evident outliers at the high end of the range, and indicates no evidence of spurious inflation. The Manhattan plot in Fig. 2 shows the same five outliers with high significance, and there are a few other significant hits shown above the red line (Bonferroni-corrected threshold p-value = 0.05).
Fig. 1.
Quantile–quantile (Q–Q) plot of the observed −log10(p-values) from the GWAS of T-tau/Aβ42 level versus those expected under the null hypothesis.
Fig. 2.
Manhattan plot of the observed −log10(p-values) from the GWAS CSF. More than 560,000 SNPs were tested for association to T-tau/Aβ42 level under an genotypic model with age, gender and diagnosis as covariates. Genome-wide associations identified 22 significant SNPs (Bonferroni-corrected threshold is p-value < 0.05 and represented by the red line), and the top 4 significant SNPs were on chromosome 19 within the APOE and its neighboring regions.
In single-marker analyses, 24 SNPs exhibited genome-wide significance to the T-tau/Aβ42 ratio (Table 2). As expected, the most significant loci were identified on chromosome 19, including rs4420638 (p=3.50E-27) from the APOC1 region, rs769449 (p=6.41E-22) within the APOE region, and rs2075650 (p=4.40E-17) and rs157582 (p=2.29E-16) within the TOMM40 region. Other SNPs identified in this study are shown in Table 2. Table 2 also shows the variances explained by each identified SNP after controlling two sets of covariates: (1) age, gender, and diagnosis; and (2) age, gender, diagnosis, and the APOE e4 status. The main effects of rs4420638, rs769449, rs2075650, and rs157582 account for 10%, 9.1%, 7.1%, and 6.8% of phenotypic variance respectively while controlling for age, gender and diagnosis, but account for ≤0.1% variance after removing the APOE e4 status. The most significant AD-risk factor APOE e4 SNP (rs429358) accounts for 12.9% variance (Table 3). The total amount of additional variance explained by 24 identified SNPs is 11.3% after accounting for age, gender, diagnosis and APOE e4 status, and up to 41.6% while including all factors (age, gender, diagnosis, APOE and 24 SNPs).
Table 2.
Identified significant genome-wide association loci with quantitative trait T-tau/Aβ42 in this study.
| NO. | CHR | rs_No. | Gene | Single-marker p_value |
Corrected p-value |
R Square | No. | CHR | rs_No. | Gene | Single-Marker p_Value |
Corrected p-value |
R Square | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Modela | Modelb | Modela | Modelb | ||||||||||||
| 1 | 19 | rs4420638 | APOC1 | 3.50E-27 | 1.97E-21 | 0.100 | 0.001 | 13 | 1 | rs10127852 | LPAR3 | 2.46E-08 | 0.0137 | 0.034 | 0.023 |
| 2 | 19 | rs769449 | APOE | 6.41E-22 | 3.62E-16 | 0.091 | 0.0 | 14 | 15 | rs9806191 | DAPK2 | 2.56E-08 | 0.0143 | 0.034 | 0.023 |
| 3 | 19 | rs2075650 | TOMM40 | 4.40E-17 | 2.48E-11 | 0.071 | 0.0 | 15 | 11 | rs7129826 | DBX1 | 2.99E-08 | 0.0167 | 0.033 | 0.023 |
| 4 | 19 | rs157582 | TOMM40 | 2.29E-16 | 1.25E-10 | 0.068 | 0.001 | 16 | 21 | rs11910985 | S100B | 3.17E-08 | 0.0177 | 0.033 | 0.023 |
| 5 | 1 | rs13376197 | B3GALT2 | 3.04E-10 | 0.00017 | 0.042 | 0.031 | 17 | 21 | rs1981331 | S100B | 3.19E-08 | 0.0178 | 0.033 | 0.023 |
| 6 | 12 | rs3020811 | LRP6 | 1.24E-08 | 0.0069 | 0.030 | 0.026 | 18 | 3 | rs9872004 | AADACL1 | 3.26E-08 | 0.0181 | 0.033 | 0.023 |
| 7 | 9 | rs2280302 | FBP1 | 1.28E-08 | 0.0071 | 0.035 | 0.026 | 19 | 1 | rs17027633 | ATP5F1 | 3.26E-08 | 0.0182 | 0.033 | 0.023 |
| 8 | 9 | rs2280301 | FBP1 | 1.37E-08 | 0.0076 | 0.035 | 0.026 | 20 | 1 | rs6662771 | SGIP1 | 3.44E-08 | 0.0192 | 0.033 | 0.023 |
| 9 | 10 | rs12265790 | ITGA8 | 1.85E-08 | 0.0103 | 0.034 | 0.024 | 21 | 11 | rs12797204 | DLG2 | 3.71E-08 | 0.0206 | 0.033 | 0.025 |
| 10 | 10 | rs7896076 | ITGA8 | 1.87E-08 | 0.0104 | 0.034 | 0.024 | 22 | 18 | rs6506440 | ARHGAP28 | 7.51E-08 | 0.0414 | 0.032 | 0.026 |
| 11 | 10 | rs11253637 | ITGA8 | 1.91E-08 | 0.0107 | 0.034 | 0.024 | 23 | 2 | rs6541929 | CNTNAP5 | 7.64E-08 | 0.0421 | 0.032 | 0.027 |
| 12 | 1 | rs1539581 | ATP5F1 | 2.34E-08 | 0.0130 | 0.034 | 0.024 | 24 | 2 | rs17267326 | CNTNAP5 | 8.23E-08 | 0.0453 | 0.031 | 0.028 |
Model: Percent of additional variance in T-tau/Aβ42 level explained by the main effect of SNPs after accounting for age, gender, and diagnosis.
Model: Percent of additional variance in T-tau/Aβ42 level explained by the main effect of SNPs after accounting for age, gender, diagnosis, and the APOE
Table 3.
Seven significant SNP-SNP interaction pairs identified in the GWIS of the T-tau/Aβ42 ratio.
| NO. | SNP1×SNP2 | GENE | CHR | Main Effect | Interaction | Explained Variance (R Square) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| p-value | p-value | Corrected p-value | Age+Gendera | Dxb | APOEc | SNP1+SNP2d | SNP1* SNP2e | ||||
| 1 | rs1514061× | PLXNA4* | 7 | 3.41E-06 | 8.35E-10 | 0.0181 | 0.01 | 0.164 | 0.129 | 0.015 | 0.051 |
| rs6467419 | PLXNA4* | 7 | 0.00167353 | ||||||||
|
| |||||||||||
| 2 | rs1514061× | PLXNA4* | 7 | 3.41E-06 | 1.97E-10 | 0.0043 | 0.01 | 0.164 | 0.129 | 0.019 | 0.047 |
| rs4453471 | CDH13 | 16 | 0.00377003 | ||||||||
|
| |||||||||||
| 3 | rs7303599× | ADIPOR2* | 12 | 7.91E-05 | 5.76E-11 | 0.0013 | 0.01 | 0.164 | 0.129 | 0.023 | 0.042 |
| rs7146454 | ADSSL1* | 14 | 0.00733134 | ||||||||
|
| |||||||||||
| 4 | rs7303599× | ADIPOR2* | 12 | 7.91E-05 | 6.91E-10 | 0.0150 | 0.01 | 0.164 | 0.129 | 0.021 | 0.041 |
| rs167396 | GSN* | 9 | 0.00583631 | ||||||||
|
| |||||||||||
| 5 | rs1482548× | INHBA* | 7 | 0.007007 | 2.95E-10 | 0.0064 | 0.01 | 0.164 | 0.129 | 0.015 | 0.038 |
| rs12894119 | NIN* | 14 | 0.00867969 | ||||||||
|
| |||||||||||
| 6 | rs9550406× | MTUS2* | 13 | 0.00543688 | 1.49E-09 | 0.0324 | 0.01 | 0.164 | 0.129 | 0.010 | 0.034 |
| rs6471951 | RLBP1L1 | 8 | 0.00604041 | ||||||||
|
| |||||||||||
| 7 | rs211953× | CXADR | 21 | 0.000478713 | 3.03E-10 | 0.0066 | 0.01 | 0.164 | 0.129 | 0.024 | 0.031 |
| rs4881147 | PITRM1* | 10 | 0.0015086 | ||||||||
The Bonferroni corrected p-values (< 0.05) and R2 of the SNP*SNP interaction term are shown in bold.
Age+Gender: Percent of variance in T-tau/Aβ42 level explained by age, gender.
Dx: Percent of variance in T-tau/Aβ42 level explained by diagnosis after accounting for age, gender.
APOE: Percent of additional variance in T-tau/Aβ42 level explained by the APOE genotype after accounting for age, gender and diagnosis.
SNP1+SNP2: Percent of additional variance in T-tau/Aβ42 level explained by the combined main effect of SNP1 and SNP2 after accounting for age, gender, diagnosis, and the APOE genotype.
SNP1*SNP2: Percent of additional variance in T-tau/Aβ42 level explained by the interaction effect of SNP1 and SNP2 after accounting for age, gender, diagnosis, APOE genotype, SNP1 and SNP2.
Nearest gene proximal to the SNP.
3.2. Genome-wide interaction study results
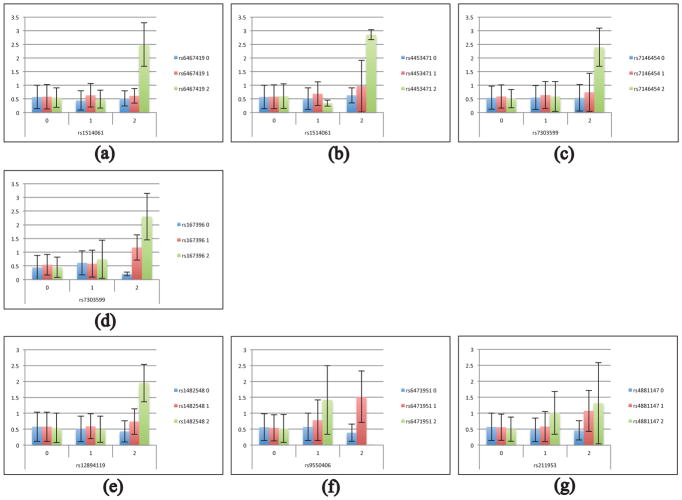
Our two-marker interaction model considered age, gender, and clinical diagnosis as covariates. 307 pairs of SNPs showed statistically significant interaction effects on the T-tau/Aβ42 level (Bonferroni-corrected p-value<0.05). Only 7 pairs passed the cell-size criterion: all the cell sizes in 3-by-3 contingency table are required to be either more than 5 or equal to 0. The results of two-marker interaction were: rs1514061 (PLXNA4*) - rs6467419 (PLXNA4*), rs1514061 (PLXNA4*) - rs4453471 (CDH13), rs7303599 (ADIPOR2*) - rs7146454 (ADSSL1*), rs7303599 (ADIPOR2*) - rs167396 (GSN*), rs1482548 (INHBA*) - rs12894119 (NIN*), rs9550406 (MTUS2*) - rs6471951 (RLBP1L1), rs211953 (CXADR) - rs4881147 (PITRM1*), where * indicating nearest gene proximal to the SNP. Details are available in Table 3.
3.3. Post hoc analysis
Table 3 also shows the two-marker interaction results of post hoc analysis on T-tau/Aβ42 level. Age, gender, and diagnosis were first included in the model and accounted for 17.4% of variance on the T-tau/Aβ42 level. APOE status was then accounted for an additional 12.9% of variance. For each interaction, we ran a hierarchical linear regression model. We first added in the genetic main effects, and then the genetic interaction term to determine the variance associated with the interaction term alone. For rs1514061 (PLXNA4*) - rs6467419 (PLXNA4*), the SNPs’ main effects accounted for 1.5% of variance, and the interaction term accounted for 5.1% of variance (6.6% combined). For rs1514061 (PLXNA4*) - rs4453471 (CDH13), the main effects accounted for 1.9% of variance, and the interaction accounted for 4.7% of variance (6.6% combined). For rs7303599 (ADIPOR2*) - rs7146454 (ADSSL1*), the main effects accounted for 2.3% of variance, and the interaction term accounted for 4.2% of variance (6.5% combined). For rs7303599 (ADIPOR2*) - rs167396 (GSN*), the main effects accounted for 2.1% of variance, and the interaction term accounted for 4.1% of variance (6.2% combined). For rs1482548 (INHBA*) - rs12894119 (NIN*), the main effects accounted for 1.5% of variance, and the interaction term accounted for 3.8% of variance (5.3% combined). For rs9550406 (MTUS2*) - rs6471951 (RLBP1L1), the main effects accounted for 1.0% of variance, and the interaction accounted for 3.4% of variance (4.4% combined). For rs211953 (CXADR) - rs4881147 (PITRM1*), the main effects accounted for 2.4% of variance, and the interaction term accounted for 3.1% of variance (5.5% combined).
4. Discussion
In this work, we performed GWAS and GWIS of the CSF biomarker T-tau/Aβ42 ratio, using a sample of 843 subjects from the ADNI database. To our knowledge, this genome-wide study on detecting two-marker interaction is the first GWIS on the quantitative trait of the T-tau/Aβ42 level.
In single-marker analysis, we identified the SNPs in APOE, APOC1 and TOMM40 genes (Fig. 2), which showed high-level genome-wide significant associations to the T-tau/Aβ42 ratio. We also revealed 20 additional significant loci, within or proximal to LRP6, S100B, DLG2, CNTNAP5, B3GALT2, FBP1, ITGA8, ATP5F1, LPAR3, DAPK2, DBX1, AADACL1, SGIP1, and ARHGAP28 genes (Table 2). The previously reported AD risk genes APOE, APOC1, and TOMM40 were replicated in our GWAS (Supplementary Table s4). In addition, the S100B, CNTNAP5, LRP6, and DLG2 genes were also reported to have pathological relevances in AD. S100B shows a pathological relevance for degeneration of the central nervous system in AD (Petzold et al., 2003), and overexpression of S100B in the neuritic plaques of AD is related to the degree of neuritic pathology in Aβ plaques (Peskind et al., 2001). CNTNAP5 encodes the protein belonging to the neurexin family functioning in the central nervous system as cell adhesion molecules and receptors, and has been implicated as a risk factor for posterior cortical atrophy variant of AD (Schott et al., 2016). Neuronal LRP6 mediated Wnt signaling has an impact on synaptic function and cognition, and genetic variants in the LRP6 gene have been linked to AD risk (Liu et al., 2014). A proteomics study showed AD-dependent changes in the DLG2 level in the hippocampus, and DLG2 exhibits an early-up, late-down expression pattern during AD pathology (Hondius et al., 2016). Our exploratory GWAS nominates the others novel loci, such as B3GALT2, FBP1, ITGA8, ATP5F1, LPAR3, DAPK2, DBX1, AADACL1, SGIP1 and ARHGAP28, meeting the genome-wide significance. These potential T-tau/Aβ42 related quantitative trait loci (QTLs) warrant further investigation.
SNP-SNP interaction studies may explain part of the “missing heritability”. The recent studies (Shen et al., 2014) in ADNI cohort demonstrated “case-control” studies for testing epistasis interaction. In this study, we preformed two-marker interaction analyses using the T-tau/Aβ42 ratio as quantitative trait for increasing statistical power and reducing required sample sizes. Our method revealed 7 pairs of SNPs within or proximal to 11 genes meeting the criterion of the cell size either more than 5 or equal to 0 and a Bonferroni corrected threshold (corrected p-value ≤ 0.05). As we expected, the significant variants in these pairs all have marginal dominance effects, but their interactions can explain a relatively high-level variances of the T-tau/Aβ42 ratio (Table 3), and high-level AD risk. The bar charts of the QT measures across SNP-by-SNP genotype combinations are shown in Fig. 3.
Fig. 3.
Seven SNP pairs with significant genome-wide interaction effects on the T-tau/Aβ42 ratio: The mean T-tau/Aβ42 ratio is plotted against each pairwise genotype combination and the error bar indicates the standard deviation.
In previous studies, PLXNA4 has been reported to be associated with precise positioning of OPCs (oligodendrocyte precursor cells) in developing cerebral cortex. Then it has also suggested that PLXNA4 does not influence APP processing or Aβ production but its isoform differentially affects tau protein phosphorylation (Jun et al., 2014) involved in AD pathogenesis, leading to neurofibrillary tangle formation and neuronal death (Wang et al., 2016). CDH13 gene has been linked to brain function or neuropsychiatric disorders, affecting morphometry of the temporal lobes (a typical AD biomarker) (Kohannim et al., 2012). With these observations, the identified PLXNA4-PLXNA4 and PLXNA4-CDH13 interactions may have a potential on contributing to the tau pathway instead of Aβ.
ADIPOR2 (Adiponectin Receptor 2) is a protein coding gene, and adiponectin is the most abundant adipokine secreted from adipose tissue. Globular adiponectin has been reported to induce a pro-inflammatory response in human astrocytic cells (Chan et al., 2012; Wan et al., 2014). Aβ caused neuroinflammation plays a critical role in the development of neurodegenerative disorder in AD pathogenesis. ADSSL1 is an Aβ toxicity modifier gene, and also an intracellular protein responsible for catalyzing the first step of de novo biosynthesis of AMP. Its genetic variation has been shown to affect AD neuropathology and episodic memory (Rosenthal et al., 2012). GSN (Gelsolin) is a protein coding gene, and Gelsolin is one of the most abundant actin–binding proteins. Gelsolin binds to Aβ protein, inhibits its fibrillization, solubilizes preformed Aβ fibrils, and helps in its clearance from the brain (Yang et al., 2014). It is involved in several pathological processes, including AD (Deng et al., 2015). With these observations, the identified ADIPOR2-ADSSL1 and ADIPOR2-GSN interactions could be related to the Aβ pathway.
The protein encoded by the INHBA gene has been linked to neuroprotection via preventing neurons from mitochondrial dysfunction, a major cause of excitotoxicity. The corresponding process, providing protection against ischemic brain damage, could be altered in AD, or aging-related neurodegenerative conditions (Lau et al., 2015). The NIN gene encodes ninein (GSK3B interacting protein), and variants of GSK3B have been shown to be linked with AD and interacted with the APOE genotype (Izzo et al., 2013). The MTUS2 gene encodes microtubule associated tumor suppressor candidate 2, also known as cardiac zipper protein or CAZIP. CAZIP has been shown to play a role in the development and function of the heart and nervous system in vertebrates (Du Puy et al., 2009). PITRM1 is responsible for significant Aβ degradation, and the impairment of its activity results in Aβ accumulation (Brunetti et al., 2016). The possible mechanisms behind INHBA-NIN, MTUS2-RLBP1L1, and CXADR-PITRM interactions warrant further investigation.
In summary, some of the genes identified in our GWAS and GWIS have shown interesting associations with tauopathies and/or amyloid pathology related to AD from prior knowledge of current literatures, such as APOE, APOC1, TOMM40, LRP6, S100B, DLG2, CNTNAP5, PLXNA4, CDH13, ADIPOR2, ADSSL1, GSN, and PITRM genes (see Supplementary Tables s4a and s4b). Supplementary Tables s4a and s4b showed that these 13 genes were reported in previous genomic, cell culture, mouse model and biomarker studies, and shown to be significantly associated to CSF Aβ42, CSF T-tau or other AD endo-phenotypes. However, in this work, only APOE, APOC1, TOMM40 genes showed significant associations to the CSF Aβ42, T-tau and T-tau/Aβ42 levels, other genes were not identified by GWAS and GWIS of Aβ42 alone or T-tau alone (Supplementary Table s2 and Table s3). This indicates that, when CSF Aβ42 alone and T-tau alone show less power for detecting the risk variants, the T-tau/Aβ42 ratio has the potential to serve as a more powerful quantitative trait to identify significant variants. In addition, our study also revealed numerous SNPs and SNP-SNP pairs that had not yet been associated with AD pathology, which warrant further investigation or replication in future studies.
5. Conclusions
Aimed at studying a major AD biomarker as phenotype, we performed GWAS and GWIS to detect the main genetic effects as well as SNP-SNP interaction effects on the CSF T-tau/Aβ42 ratio. The single-marker analysis replicated the APOE, APOC1 and TOMM40 genes, which are previously confirmed AD risk genes. We also identified 14 additional loci within or proximate to LRP6, S100B, DLG2, CNTNAP5, B3GALT2, FBP1, ITGA8, ATP5F1, LPAR3, DAPK2, DBX1, AADACL1, SGIP1, and ARHGAP28. The two-marker interaction analysis identified a number of novel interaction findings, which showed strong associations with the T-tau/Aβ42 ratio. These were interactions between PLXNA4 and PLXNA4, between PLXNA4 and CDH13, between ADIPOR2 and ADSSL1, between ADIPOR2 and GSN, between INHBA and NIN, between MTUS2 and RLBP1L1, and between CXADR and PITRM1. The effects of SNP-SNP interactions showed high-level statistical significance, while the corresponding single-marker effects were marginal. SNP-SNP interaction effects may help address part of “miss heritability”.
Our genome-wide association study and interaction study have the following strengths. (1) Continuous quantitative trait T-tau/Aβ42 can not only gain higher statistical power, but also contribute to detecting potential risk variants related to T-tau and/or Aβ42 at the same time. (2) Five values (1–5) indicating CN, SMC, EMCI, LMCI and AD respectively, provide a rank ordered spectrum of the AD progression. (3) In this study, both GWAS and GWIS consider age, gender, and clinical diagnosis as covariates. In the post hoc linear regression analysis we included confounding factors APOE ε4 allele (rs429358) on top of the above three covariates, and so provided more accurate estimate of the interaction effects on CSF T-tau/Aβ42 ratio.
The limitations of our study are as follows: (1) We examined 22 million SNP-SNP pairs and conducted an exhaust test among the SNPs. More effective and efficient strategies remain to be developed. (2) To control for potential false positives of the GWIS findings, we used two methods. One is the Bonferroni method, which corrects for multiple comparison by using a threshold of α/n and is well-known to be a conservative approach. Another method used in the work is the cell-size criterion, which excludes rare genotype combinations to avoid potential false positives. In this work, we set α=0.05 and n=21,717,345 for Bonferroni correction; and for the cell-size criterion, all the cell sizes in the 3-by-3 contingency table are required to be either more than 5 or equal to 0. There are 307 pairs SNPs passed the first threshold, and subsequently only 7 pairs among 307 pairs passed the second threshold for further study. Although these are relatively stringent criteria for controlling false positives, future replication studies are required to confirm the identified interaction signals. (3) We performed the GWAS and GWIS of CSF T-tau/Aβ42 ratio using data-driven method. Future studies could utilize prior biological knowledge, such as biological networks, pathways databases, special tissues and other functional annotation data, to enhance statistical power and improve biological interpretability. (4) Future studies are necessary to replicate and validate the findings in independent datasets, and to uncover potential mechanisms underlying tagged by the identified SNPs and genes in our study.
Supplementary Material
Acknowledgments
Supported in part by grants from National Natural Science Foundation of China (81301297), National Key Scientific Instrument and Equipment Development Projects of China (2012YQ04014001, 2012YQ04014010), and Fundamental Research Funds for the Central Universities (HEUCF160412); and by NIH R01 EB022574, R01 LM011360, U01 AG024904, RC2 AG036535, R01 AG19771, P30 AG10133, UL1 TR001108, NSF IIS-1117335, DOD W81XWH-14-2-0151, and NCAA 14132004 at IU.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Disclosure Statement
The authors have no actual or potential conflicts of interest including any financial, personal, or other relationships with other people or organizations that could inappropriately influence (bias) our work.
References
- Akiyama H, Ikeda K, Kondo H, Kato M, McGeer PL. Microglia express the type 2 plasminogen activator inhibitor in the brain of control subjects and patients with Alzheimer’s disease. Neurosci Lett. 1993;164(1–2):233–235. doi: 10.1016/0304-3940(93)90899-v. [DOI] [PubMed] [Google Scholar]
- Becker T, Herold C, Meesters C, Mattheisen M, Baur MP. Significance levels in genome-wide interaction analysis (GWIA) Ann Hum Genet. 2011;75(1):29–35. doi: 10.1111/j.1469-1809.2010.00610.x. [DOI] [PubMed] [Google Scholar]
- Blennow K, Hampel H. CSF markers for incipient Alzheimer’s disease. Lancet Neurol. 2003;2(10):605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- Brunetti D, Torsvik J, Dallabona C, Teixeira P, Sztromwasser P, Fernandez-Vizarra E, … Bindoff LA. Defective PITRM1 mitochondrial peptidase is associated with Abeta amyloidotic neurodegeneration. EMBO Mol Med. 2016;8(3):176–190. doi: 10.15252/emmm.201505894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KH, Lam KS, Cheng OY, Kwan JS, Ho PW, Cheng KK, … Xu A. Adiponectin is protective against oxidative stress induced cytotoxicity in amyloid-beta neurotoxicity. PLoS One. 2012;7(12):e52354. doi: 10.1371/journal.pone.0052354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R, Hao J, Han W, Ni Y, Huang X, Hu Q. Gelsolin regulates proliferation, apoptosis, migration and invasion in human oral carcinoma cells. Oncol Lett. 2015;9(5):2129–2134. doi: 10.3892/ol.2015.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Wolk DA Alzheimer’s Disease Neuroimaging I. Biomarker-based prediction of progression in MCI: Comparison of AD signature and hippocampal volume with spinal fluid amyloid-beta and tau. Front Aging Neurosci. 2013;5:55. doi: 10.3389/fnagi.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Puy L, Beqqali A, Monshouwer-Kloots J, Haagsman HP, Roelen BA, Passier R. CAZIP, a novel protein expressed in the developing heart and nervous system. Dev Dyn. 2009;238(11):2903–2911. doi: 10.1002/dvdy.22107. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, … Holtzman DM. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59(3):512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007;64(3):343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- Goudey B, Rawlinson D, Wang Q, Shi F, Ferra H, Campbell RM, … Kowalczyk A. GWIS--model-free, fast and exhaustive search for epistatic interactions in case-control GWAS. BMC Genomics. 2013;14(Suppl 3):S10. doi: 10.1186/1471-2164-14-S3-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel H, Blennow K, Shaw LM, Hoessler YC, Zetterberg H, Trojanowski JQ. Total and phosphorylated tau protein as biological markers of Alzheimer’s disease. Exp Gerontol. 2010;45(1):30–40. doi: 10.1016/j.exger.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold C, Steffens M, Brockschmidt FF, Baur MP, Becker T. INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics. 2009;25(24):3275–3281. doi: 10.1093/bioinformatics/btp596. [DOI] [PubMed] [Google Scholar]
- Hohman TJ, Koran ME, Thornton-Wells TA Alzheimer’s Disease Neuroimaging I. Genetic modification of the relationship between phosphorylated tau and neurodegeneration. Alzheimers Dement. 2014;10(6):637–645. e631. doi: 10.1016/j.jalz.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondius DC, van Nierop P, Li KW, Hoozemans JJ, van der Schors RC, van Haastert ES, … Smit AB. Profiling the human hippocampal proteome at all pathologic stages of Alzheimer’s disease. Alzheimers Dement. 2016;12(6):654–668. doi: 10.1016/j.jalz.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Izzo G, Forlenza OV, Santos B, Bertolucci PH, Ojopi EB, Gattaz WF, Kerr DS. Single-nucleotide polymorphisms of GSK3B, GAB2 and SORL1 in late-onset Alzheimer’s disease: interactions with the APOE genotype. Clinics (Sao Paulo) 2013;68(2):277–280. doi: 10.6061/clinics/2013(02)RC01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, Reiman EM … Alzheimer’s Disease Neuroimaging, I. Relationships between biomarkers in aging and dementia. Neurology. 2009;73(15):1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Asai H, Zeldich E, Drapeau E, Chen C, Chung J, … Farrer LA. PLXNA4 is associated with Alzheimer disease and modulates tau phosphorylation. Ann Neurol. 2014;76(3):379–392. doi: 10.1002/ana.24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabogo D, Rauw G, Amritraj A, Baker G, Kar S. ss-amyloid-related peptides potentiate K+-evoked glutamate release from adult rat hippocampal slices. Neurobiol Aging. 2010;31(7):1164–1172. doi: 10.1016/j.neurobiolaging.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Kohannim O, Hibar DP, Stein JL, Jahanshad N, Hua X, Rajagopalan P … Alzheimer’s Disease Neuroimaging, I. Discovery and Replication of Gene Influences on Brain Structure Using LASSO Regression. Front Neurosci. 2012;6:115. doi: 10.3389/fnins.2012.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau D, Bengtson CP, Buchthal B, Bading H. BDNF Reduces Toxic Extrasynaptic NMDA Receptor Signaling via Synaptic NMDA Receptors and Nuclear-Calcium-Induced Transcription of inhba/Activin A. Cell Rep. 2015;12(8):1353–1366. doi: 10.1016/j.celrep.2015.07.038. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang Q, Chen F, Yan J, Kim S, Wang L, … Shen L. Genetic Interactions Explain Variance in Cingulate Amyloid Burden: An AV-45 PET Genome-Wide Association and Interaction Study in the ADNI Cohort. Biomed Res Int. 2015;2015:647389. doi: 10.1155/2015/647389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Parrado AR, Samtani MN, Narayan VA Alzheimer’s Disease Neuroimaging I. Variations in the FRA10AC1 Fragile Site and 15q21 Are Associated with Cerebrospinal Fluid Abeta1–42 Level. PLoS One. 2015;10(8):e0134000. doi: 10.1371/journal.pone.0134000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Tsai CW, Deak F, Rogers J, Penuliar M, Sung YM, … Bu G. Deficiency in LRP6-mediated Wnt signaling contributes to synaptic abnormalities and amyloid pathology in Alzheimer’s disease. Neuron. 2014;84(1):63–77. doi: 10.1016/j.neuron.2014.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaetova-Ladinska EB, Abdel-All Z, Mugica ES, Li M, Craggs LJ, Oakley AE, … Kalaria RN. Tau proteins in the temporal and frontal cortices in patients with vascular dementia. J Neuropathol Exp Neurol. 2015;74(2):148–157. doi: 10.1097/NEN.0000000000000157. [DOI] [PubMed] [Google Scholar]
- Pan C, Korff A, Galasko D, Ginghina C, Peskind E, Li G, … Zhang J. Diagnostic Values of Cerebrospinal Fluid T-Tau and Abeta(4)(2) using Meso Scale Discovery Assays for Alzheimer’s Disease. J Alzheimers Dis. 2015;45(3):709–719. doi: 10.3233/JAD-143099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskind ER, Griffin WS, Akama KT, Raskind MA, Van Eldik LJ. Cerebrospinal fluid S100B is elevated in the earlier stages of Alzheimer’s disease. Neurochem Int. 2001;39(5–6):409–413. doi: 10.1016/s0197-0186(01)00048-1. [DOI] [PubMed] [Google Scholar]
- Petzold A, Keir G, Lim D, Smith M, Thompson EJ. Cerebrospinal fluid (CSF) and serum S100B: release and wash-out pattern. Brain Res Bull. 2003;61(3):281–285. doi: 10.1016/s0361-9230(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, … Sellers TA. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45(4):362–370. 370e361–362. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, … Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal SL, Wang X, Demirci FY, Barmada MM, Ganguli M, Lopez OL, Kamboh MI. Beta-amyloid toxicity modifier genes and the risk of Alzheimer’s disease. Am J Neurodegener Dis. 2012;1(2):191–198. [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, … Weiner MW. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott JM, Crutch SJ, Carrasquillo MM, Uphill J, Shakespeare TJ, Ryan NS, … Mead S. Genetic risk factors for the posterior cortical atrophy variant of Alzheimer’s disease. Alzheimers Dement. 2016 doi: 10.1016/j.jalz.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC … Alzheimer’s Disease Neuroimaging, I. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65(4):403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD … Alzheimer’s Disease Neuroimaging, I. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53(3):1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM … Alzheimer’s Disease Neuroimaging, I. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8(2):183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Mah D, Simtchouk S, Klegeris A, Little JP. Globular adiponectin induces a pro-inflammatory response in human astrocytic cells. Biochem Biophys Res Commun. 2014;446(1):37–42. doi: 10.1016/j.bbrc.2014.02.077. [DOI] [PubMed] [Google Scholar]
- Wang H, Sun FR, Tan L, Wang HF, Zhang W, Wang ZX, … Tan L. Association study of the PLXNA4 gene with the risk of Alzheimer’s disease. Ann Transl Med. 2016;4(6):108. doi: 10.21037/atm.2016.03.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chauhan A, Mehta S, Mehta P, Gu F, Chauhan V. Trichostatin A increases the levels of plasma gelsolin and amyloid beta-protein in a transgenic mouse model of Alzheimer’s disease. Life Sci. 2014;99(1–2):31–36. doi: 10.1016/j.lfs.2014.01.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.