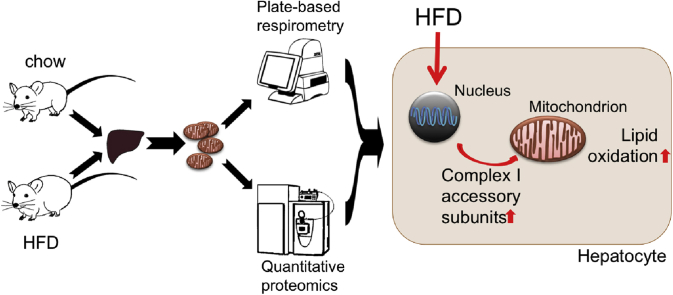
Abstract
Objective
Energy metabolism is challenged upon nutrient stress, eventually leading to a variety of metabolic diseases that represent a major global health burden.
Methods
Here, we combine quantitative mitochondrial respirometry (Seahorse technology) and proteomics (LC-MS/MS-based total protein approach) to understand how molecular changes translate to changes in mitochondrial energy transduction during diet-induced obesity (DIO) in the liver.
Results
The integrative analysis reveals that significantly increased palmitoyl-carnitine respiration is supported by an array of proteins enriching lipid metabolism pathways. Upstream of the respiratory chain, the increased capacity for ATP synthesis during DIO associates strongest to mitochondrial uptake of pyruvate, which is routed towards carboxylation. At the respiratory chain, robust increases of complex I are uncovered by cumulative analysis of single subunit concentrations. Specifically, nuclear-encoded accessory subunits, but not mitochondrial-encoded or core units, appear to be permissive for enhanced lipid oxidation.
Conclusion
Our integrative analysis, that we dubbed “respiromics”, represents an effective tool to link molecular changes to functional mechanisms in liver energy metabolism, and, more generally, can be applied for mitochondrial analysis in a variety of metabolic and mitochondrial disease models.
Keywords: Mitochondria, Respirometry, Proteomics, Mitochondrial pyruvate carrier, Liver disease, Bioenergetics, Obesity, Diabetes
Abbreviations: DIO, Diet induced obesity; ETC, Electron transport chain; HFD, High fat diet; TPA, Total protein approach
Graphical abstract
Highlights
-
•
Integrative analysis of respiration and quantitative proteomics of liver mitochondria in response to high-fat diets.
-
•
Evidence for mitochondrial pyruvate transport as important regulator of mitochondrial energy turnover.
-
•
High-confidence prediction of stress and disease-related networks facilitating lipid oxidation.
-
•
Evidence that nutritional stress impacts nuclear, but not mitochondrial, regulation of respiratory complex I.
1. Introduction
Mitochondria play a central role in energy metabolism as they convert nutrient to cellular energy. In response to physiological and environmental stress, mitochondria require functional adaptation to match increased ATP demand and to maintain metabolic homeostasis [1]. The complex adaptation of the organelle is integrated to assure cellular homeostasis and requires multiple adjustments of signaling pathways and structural proteins. In a homeostatic system, mitochondrial energy flux is balanced by two major processes: the reactions that produce proton motive force at the mitochondrial inner membrane and the reactions that consume proton motive force to fuel cellular functions [2]. The production of proton motive force depends on the capacity of the respiratory chain, substrate preferences, and routes of substrate supply; multiple distinct metabolic pathways regulate the latter. Proton motive force is consumed by ATP synthesis or dissipated as heat by the mitochondrial proton leak [3]. In most cell-types, electron flux is mainly controlled by ATP turnover, but feedback mechanisms also communicate upstream, regulating substrate supply as documented for the liver using top-down and elasticity control analyses of mitochondrial and cellular energy metabolism [4], [5], [6]. As cells are always exposed to fluctuations in energy supply and demand, intrinsic flexibility and allosteric control is partially supported by adjustments of protein concentrations. However, in particular chronic impairments of energy balance, such as oversupply (during overnutrition) or the lack of demand (e.g. the lack of exercise), may reach the limits of intrinsic flexibility and allosteric regulation, possibly even the limits of proteomic adaptation, thus establishing pathologies of the metabolic syndrome [7], [8].
The enormous complexity of mitochondrial regulation may only be captured using global molecular and functional analyses. Functional bioenergetic adjustments can be monitored overall as mitochondrial oxygen consumption, and partitioning oxygen consumption into functional modules may simplify the metabolic complexity. Respirometry enables determination of oxidative capacity and substrate preferences [9]. Using appropriate respiratory chain inhibitors and activators, oxygen consumption rates are instrumental to indirectly estimate consumers of proton motive force such as ATP synthase activity (state 3 respiration) and proton leak (state 4). In particular, state 4 respiration measurements bares pitfalls, as state 4 rates inevitably also depend on substrate oxidation capacity, thus only “estimate” mitochondrial proton conductance [10]. The ratio of state 3/state 4, termed respiratory control ratio (RCR), is considered a powerful, internally normalized parameter to detect defective mitochondrial properties [9]. Furthermore, RCRs may assist in clarifying whether changes of state 4 respiration are changes in proton leak (changing RCR) or in substrate oxidation capacity (maintaining RCR). Chemical uncouplers such as lipophilic acids (e.g. DNP, FCCP) are generally used to remove respiratory control and maximize substrate oxidation. However, the respiratory response is dose-sensitive and deleterious effects of chemical uncouplers could be overseen. Quantitative changes in ATP demand during physiological adaptation are probably best reflected in changes of state 3 respiration, which is balanced by substrate supply and ATP demand. Furthermore, state 3 rates are relatively robust by avoiding deleterious effects of non-physiological chemical inhibitors.
In the past, bulks of mitochondria were required to measure respiration in single-chambered Clark-type electrodes [11], hampering efforts to examine high sample numbers and various conditions in a timely manner. New technologies such as extracellular flux analyzers in the 24- or 96-well format, also dubbed “Seahorse technology,” referring to the initial vendor, have been developed to simultaneously analyze mitochondrial respiration with high sampling sizes in minor quantities [12]. The automated measurements not only enable screening efforts (e.g. for drugs) but also increase the statistical power to detect subtle bioenergetic adjustments, such as those found during physiological adaptation or multi-factorial metabolic diseases. Most of the molecular adjustments are controlled by systemic changes of gene programs rather than of single genes, and the complexity hampers our understanding on physiological regulation of mitochondrial energy metabolism. Thus, respiratory changes in response to (patho)physiological challenges cannot be attributed easily to single proteins or entire metabolic networks, while the bioenergetic impact of characterized proteins is assessable by mitochondrial respiration [e.g. [13], [14]]. To identify novel molecular players or signatures, global proteomics is required. Modern mass spectrometry and the latest approaches in label-free protein quantification enable quantitative proteomic analysis, up to the level of absolute protein concentrations [15]. Without a doubt, the progress in omics-analyses has promoted in-depth understanding of different types of physiological and pathogenic processes. However, the inherent problem of all omic datasets is the vast amount of information (transcripts, proteins, metabolites) that requires processing and filtering in order to reduce the number of protein candidates for further detailed analyses. How stress-related adjustments in the protein network impact cellular function still represents the ultimate question that drives therapeutic target identification for a variety of diseases. To the best of our knowledge, proteomic datasets have mostly been integrated only with other -omics data, such as transcriptomics or metabolomics, in order to characterize complex metabolic phenotypes [e.g. [16]]. It is clear, however, that the systematic, unbiased integration of proteomics with functional data would establish a powerful link between the molecular basis and specific cellular functions. Thus, the development of workflows to establish protein-function relationships for energy metabolism in complex systems such as intact mitochondria may benefit the understanding of metabolic and mitochondrial disease paradigms. Recently, proteomics was combined with enzyme kinetics to characterize tissue-specific types of the glycolytic and gluconeogenic pathways [17].
In this study, we combine LC-MS/MS-based total protein approach (TPA) with multi-well respiratory flux analysis of isolated mitochondria to link absolute protein concentrations with changes in mitochondrial energy transduction. This integrative analysis aims to identify direct functional and mechanistic interactions to improve our mechanistic understanding how molecular key players and networks impact mitochondrial bioenergetics, and how cellular energy metabolism adapts to stress situations. As proof-of-principle for this integrative workflow, that we dub “respiromics,” we focus on nutrient-stressed energy metabolism in the obese mouse liver, a model of broad medical interest to elucidate how Western lifestyle and excessive food intake promote progression from non-alcoholic fatty liver to hepatosteatosis [18] and the progression from high blood glucose to diabetes.
Instead of analyzing isolated datasets of mitochondrial respiration rates and of protein lists, we enhance the mechanistic picture using global correlation analysis. The integrative analysis highlights the mitochondrial pyruvate carrier (MPC) as prime element controlling respiration in the liver. Furthermore, we show that mitochondrial lipid oxidation of DIO mice is specifically enhanced by an array of (partially novel) disease-related proteins controlling catabolism of fatty acids and amino acids. The TPA also enabled quantitative summation and quantification of multimeric protein complexes. Herein, we found that respiratory complex I, in particular, is elevated by the sum of nuclear-encoded accessory subunits of complex I, rather than by mitochondrial encoded or core subunits, and associates to enhanced lipid oxidation. This proof-of-principle study shows that “respiromic” analyses are capable of revealing functional sets of stress-induced and disease-related proteins that deserve further investigation as biomarkers or therapeutic targets.
2. Materials and methods
2.1. Animals
C57/Bl6J mice were housed at room temperature (23 °C) in a 12 h-light/dark cycle. Starting at the age of six weeks, the mice had ad libitum access to either chow diet (CHOW: Altromin 1310, 14 kcal% fat, 59 kcal% carbohydrate, 27 kcal% protein) or high fat diet (HFD: Surwit Diet D12331, 58 kcal% fat, 25.5 kcal% carbohydrate, 16.4 kcal% protein, Research diets) for 15.5 weeks. Animal maintenance and experimental procedures were approved by the guidelines of the Institutional Animal Care and Committee of the Helmholtz Center Munich.
2.2. Mitochondrial isolation
Livers were dissected and immediately placed in ice-cold isolation buffer (STE; 250 mM Sucrose, 5 mM TRIS, 2 mM EGTA, pH adjusted to 7.4 at 4 °C) for the isolation of mitochondria by differential centrifugation at 4 °C. The livers were washed in STE buffer, cut to small pieces, washed several times, and homogenized in STE containing 0.5% fatty acid-free BSA (Sigma No: A3803-50G) using a glass dounce homogenizer. After centrifugation at 800g for 10 min, the supernatant was filtered and transferred to a fresh tube. The suspension was centrifuged at 8000g for 10 min for sedimentation of the mitochondrial fraction. Excessive residual fat was wiped off the tube walls, and the pellet was resuspended in fresh STE. During the final centrifugation step, the initial speed of 2000g was increased after 5 min to 4000g for another 5 min. The final pellet was resuspended in a minimal amount of STE buffer and the protein content was quantified using Biuret reagent.
2.3. Mitochondrial respiration
Mitochondrial respiration rates were measured in a XF96 extracellular flux analyzer (Seahorse Bioscience, Agilent). The measuring buffer (MAS) consisted of 70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA, 0.5% fatty acid-free BSA and the pH was adjusted to 7.2 at 37 °C. Mitochondria, respiratory chain substrates, and inhibitors were added as follows: 2.5 μg mitochondria per well were incubated with 10 mM succinate and 2 μM rotenone; 5 μg mitochondria per well were incubated with 10 mM pyruvate and 3 mM malate, and 10 μg mitochondria per well were incubated with 50 μM palmitoyl-carnitine and 3 mM malate. After two initial measurement cycles, ADP was injected at 4 mM (final concentration) and a single measurement cycle recorded. Subsequent injection of 2.5 μg/ml oligomycin shifted mitochondria to state 4 (proton leak respiration), injection of 4 μM FCCP induced maximal substrate oxidation, and 4 μM antimycin A blocked mitochondrial respiration for baseline correction. Every condition per animal was measured in multiple wells and averaged.
2.4. Complex I activity assay
Complex I activity was measured with a “Complex I Enzyme Activity Microplate Assay Kit (Colorimetric)” (ab109721) from Abcam according to the manufacturer's instructions.
2.5. Protein digestion and peptide fractionation
Sample preparation for mass spectrometry was performed by denaturing liver mitochondria with 2% SDS and 100 mM DTT. Protein lysates and extracts containing 100 μg of total protein were processed in the 30 k filtration units (Microcon, Millipore) centrifuged at 10,000 ×g using the MED-FASP protocol [21]. Endoproteinase Lys-C and trypsin were used for sequential digestion of proteins. The enzyme to protein ratios were 1/50. Duplicates of each sample were prepared and analyzed. Protein and peptide concentrations were assayed using the ‘WF’-assay [22].
2.6. LC-MS/MS analysis
Aliquots containing ∼4 μg peptide were separated on a reverse phase column (20 cm × 75 μm inner diameter) packed with 1.8 μm C18 particles (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) using a 3 h acetonitrile gradient in 0.1% formic acid at a flow rate of 250 nl/min. The LC was coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Germany) via a nanoelectrospray source (Proxeon Biosystems). The Q Exactive HF was operated in data dependent mode with survey scans of 300–1650 m/z acquired at a resolution of 60,000. Up to the top 15 most abundant isotope patterns with charge m/z ≥ 2 from the survey scan were selected with an isolation window of 1.4 Th and fragmented by HCD with normalized collision energies of 25. The maximum ion injection times for the survey scan and the MS/MS scans were 20 ms and 60 ms, respectively. The ion target value for MS1 and MS2 scan modes were set to 3 × 106 and 1 × 105, respectively. The dynamic exclusion was 30 s. The raw data have been deposited to the ProteomeXchange Consortium [PMID:24727771] via the PRIDE partner repository with the dataset identifier PXD002289 (Reviewer account details: Username: reviewer14529@ebi.ac.uk, Password: dl0046sZ).
2.7. Data analysis
The MS data was analyzed using the software environment MaxQuant [23] version 1.2.6.20. Proteins were identified by searching of MS and MS/MS data against UniProtKB/Swiss-Prot database (May 2013) containing 50,807 sequences. The FDR was derived by analyzing the decoy database. Carbamidomethylation of cysteines was set as a fixed modification. The initial allowed mass deviation of the precursor ion was up to 6 ppm, and for the fragment masses, it was up to 20 ppm. The maximum false peptide discovery rate was specified as 0.01. Protein concentrations were calculated on the basis of spectral protein intensity using the Total Protein Approach (TPA) [15] using following equation:
where MS_signal and Total MS_signal refer to total MS1 signal intensity of the protein i and the total protein MS1 signal.
2.8. Computational filtering for mitochondrial proteins and molecular mass
In order to exclude confounding effects of non-mitochondrial proteins in some analyses, the mitochondrial proteome was computationally filtered for proteins located in mitochondria using the MitoCarta 2.0, a compendium of proteins that are located in mitochondria [24]. The experimental proteome and the Mitocarta collection were compared based on gene-ID (Entrez-ID, retrieved from UniProt [25], April 2016) and gene symbol (including synonyms). Discrepancies of annotations were manually curated by comparing the gene descriptions. The total mass of mitochondrial and non-mitochondrial proteins in the suspension after LC-MS/MS analysis was calculated based on protein masses retrieved from UniProt (March 2016) and the protein concentrations calculated by TPA.
2.9. Gene Set Enrichment Analysis, construction of heatmaps, and network analysis
Gene Set Enrichment Analysis was performed with GSEA 2.2.1 [26] with the gene set versions 5.1 of KEGG and GO databases (c2.cp.kegg.v5.1.symbols.gmt, c5.all.v5.1.symbols.gmt). The input for the main paper was filtered for mitochondrial proteins ahead of enrichment analysis, GO terms for cellular compartment were not included but can be generated from the original proteomic data deposited at the ProteomeXchange Consortium. Gene Symbols were used as Chip platform, and minimum size for gene sets was set to 5. For all other parameters, the default settings were used.
Enrichment analysis for KEGG pathways and GO terms irrespective of group affiliation or protein concentration was performed with Enrichr [27] with the databases of April 2016.
Enrichment for disease genes was performed using the “Gene set → diseases” Data mining tool by Fontaine and Andrade-Navarro [28]. The heat map of the 30 top up- and down-regulated proteins was generated with GenePattern [29] using default settings. Protein–protein networks were created using String-db using default settings [30].
2.10. Correlation analysis
State 2, 3, 4, and 3u respiration rates fueled by palmitoyl-carnitine/malate, pyruvate/malate, or succinate were correlated with every single protein of the mitochondrial proteome and tested for Spearman correlation using JMP 12, as not all protein concentrations follow Gaussian distribution. Respiration rates and complex I activity were correlated with total respiratory complex concentrations by Pearson correlation and linear regression analysis was performed using GraphPad Prism 6.
2.11. Statistical analysis
Statistical analysis was performed using GraphPad Prism 6, JMP 12, and ‘Perseus’ software (http://www.biochem.mpg.de/5111810/perseus). Respiratory data and proteomic data, including summed up concentrations of respiratory complexes, were tested for statistical significance by t-test. Significance of enrichment was calculated by GSEA 2.2.1 [26] or Enrichr [27]. Changes in stoichiometry of respiratory complexes were tested for significance by 2-way-ANOVA with Sidak correction for multiple comparisons.
3. Results
3.1. Obesity increases mitochondrial lipid oxidation in liver mitochondria
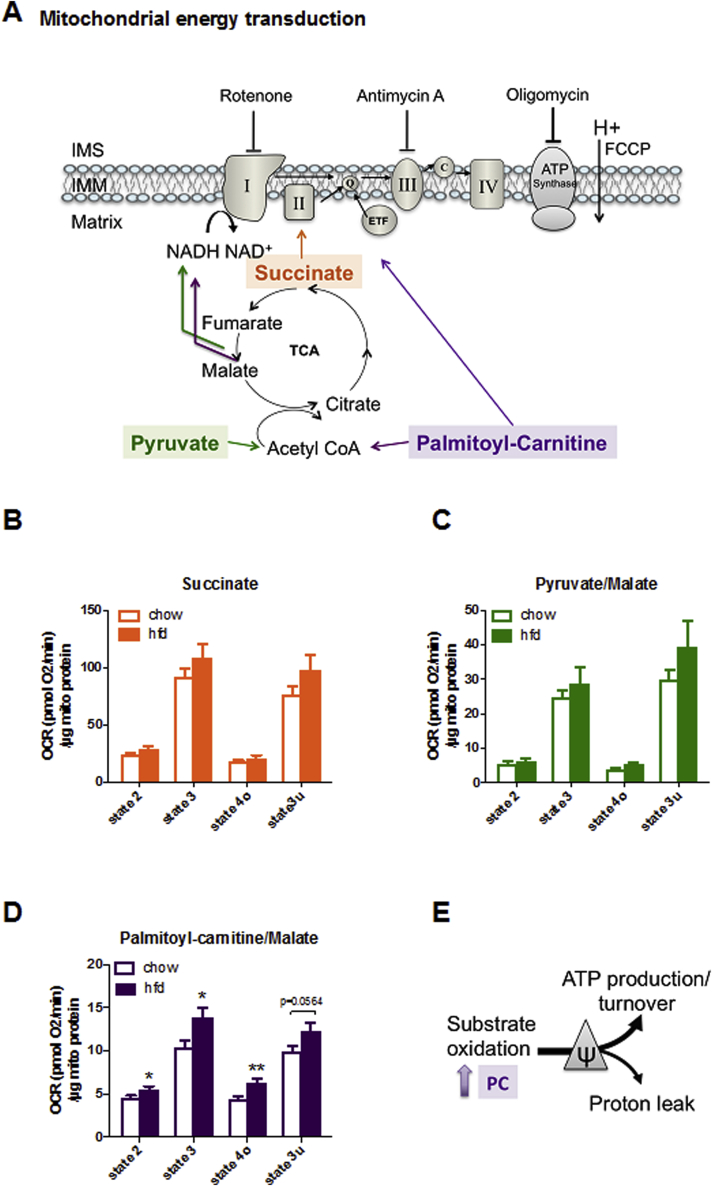
High-fat diet feeding for four months induced obesity in mice (40.1 g ± 1.0 g body mass, about 26% higher than chow-fed controls (31.8 g ± 0.3 g; p < 0.0001; n = 8)). Mitochondrial respiration was tested for differences in substrate preferences and oxidation rates (depicted in Figure 1A). Pyruvate and palmitoyl-carnitine (PC) are catabolized to acetyl-CoA but cannot independently drive tricarboxylic acid (TCA) cycle flux, thus requiring malate as source of oxaloacetate to spark the TCA cycle. Initially, mitochondria were incubated with their respective substrate only (state 2). Then, ADP was injected to induce phosphorylation (state 3), followed by oligomycin to inhibit the ATP synthase (state 4) and the chemical uncoupler FCCP to induce maximal respiration (state 3u). While neither pyruvate/malate nor succinate-fueled respiration was significantly affected (Figure 1B, C), PC respiration rates were increased in response to HFD (Figure 1D). The increased proton leak respiration (state 4) with PC is not due to increased proton conductance but a result of increased substrate (PC) oxidation. This is corroborated by unchanged RCR (Suppl. Fig. 1) and strong trends towards elevated state 3u respiration (p = 0.0564). Notably, the mitochondrial adjustments at the functional level still appear subtle, hampering efforts to identify molecular mechanisms. Collectively, we demonstrate that liver mitochondria of HFD-fed mice increase the capacity of ATP production through increased lipid oxidation with minor effects on mitochondrial proton leak (Figure 1E).
Figure 1.
Respirometry of isolated liver mitochondria from chow vs HFD fed mice. (A) The scheme depicts distinct substrate entry points into the respiratory chain. State 2 (substrate only), state 3 (phosphorylating respiration in presence of ADP), state 4o (ATP synthase inhibited by oligomycin), and state 3u (uncoupled respiration) were corrected for non-mitochondrial oxygen consumption using antimycin A. Respiration rates with (B) pyruvate/malate, (C) palmitoyl-carnitine/malate, and (D) succinate were normalized to mitochondrial protein concentrations. (E) Mitochondrial energy transduction is controlled by substrates entering the electron transport chain (ETC) produce proton motive force (ψ) which can be used for either ATP production or proton leak activity. In HFD liver mitochondria, lipid oxidation is enhanced. Data are expressed as mean ± SEM; n = 8 animals per group, *p < 0.05, **p < 0.01 by t-test comparing HFD vs chow.
3.2. Quantitative liver mitoproteomics
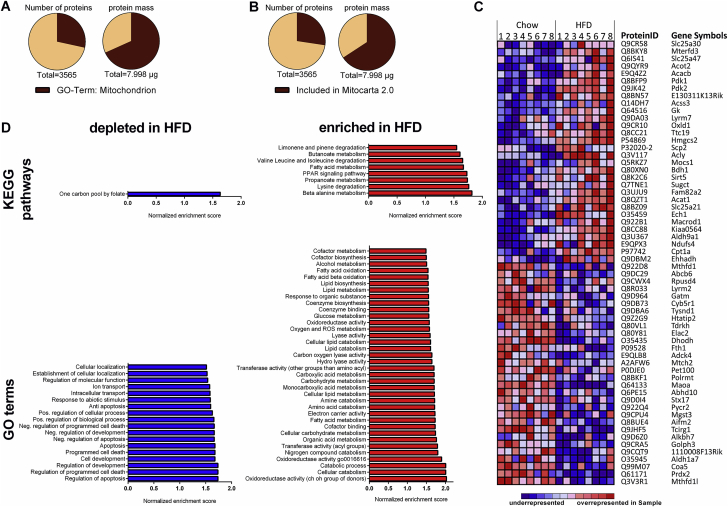
Mitochondrial suspensions of livers from chow- and HFD-fed mice were analyzed with LC-MS/MS, and protein concentrations were calculated using the total protein approach (TPA) method. 3565 proteins were identified, encoded by 3529 different genes (Suppl. Table 1). Classifying proteins with either the Gene Ontology (GO)-term ‘Mitochondrion’ (Figure 2A) or with the Mitocarta 2.0 database [24] (Figure 2B) uncovered about three-fold higher diversity of unique non-mitochondrial proteins than of mitochondrial proteins. However, the TPA method enabled the calculation of cumulative protein mass, verifying the enrichment of absolute mitochondrial protein content by the isolation procedure (Figure 2A, B). Similar degrees of mitochondrial enrichment are seen in the sum of individual protein concentrations (Suppl. Fig. 2). In order to exclude non-mitochondrial proteins in the following enrichment analyses geared towards mechanistic mitochondrial adjustments, the protein list was filtered using the Mitocarta 2.0 database (Suppl. Table 2). 976 proteins were confirmed as mitochondrial, of which 161 were significantly altered by HFD feeding (p < 0.05). The heat map (Figure 2C) illustrates the top 30 significantly induced (upper panel) and reduced (lower panel) protein concentrations in response to HFD. Gene Set Enrichment Analysis using GSEA v2.2.1 [7], [26] was performed on all mitochondrial proteins using KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO databases (based on database version 5.1 in GSEA v2.2.1). The stress-induced recruitment of metabolic pathways, in particular related to lipid metabolism, is substantiated by enrichment of eight KEGG pathways and 38 GO terms for molecular function and biological processes in HFD mitochondria (p < 0.05) (Figure 2D). In contrast, one KEGG pathway and 18 GO terms were significantly depleted (p < 0.05), but these were mostly related to processes such as apoptosis, development, localization, and proliferation, that are not directly linked with metabolism (Figure 2D).
Figure 2.
HFD-induced changes of the liver mitochondrial proteome. The numeric and mass proportion of mitochondrial protein within the isolated suspension was summed using mitochondrial annotation (A) of the GO database or (B) of the Mitocarta 2.0 database. Specific protein masses were assessed by multiplying protein concentrations × respective molecular masses retrieved from the UniProt database. (C and D) Gene Set Enrichment Analysis [26] was performed with the mitochondrial protein subset defined by Mitocarta 2.0. (C) The heat map illustrates top 30 significantly up- and downregulated proteins of the HFD group sorted by fold change. For each protein, the relative concentration compared to its mean concentration is color-coded on a scale from minimum (blue) to maximum (red) concentration for each sample. (D) KEGG pathways (upper panels) and GO-terms (lower panels) of significantly depleted (blue, left panels) or enriched (red, right panels) in response to HFD feeding.
3.3. Integration of respiratory and proteomic data
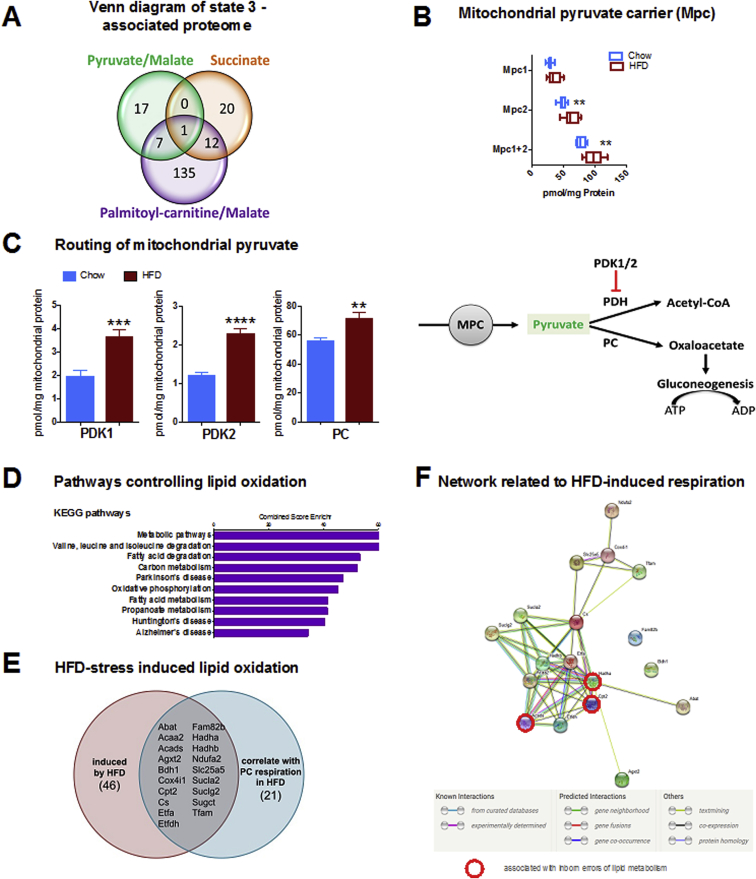
Next, we thought to investigate how changes in mitochondrial oxidative phosphorylation associate with and are controlled by specific mitochondrial proteins. Thus, we integrated substrate-specific respiration rates with absolute concentrations of all identified mitochondrial proteins (n = 976). These were tested against their respective respiration rates by correlation analysis, considering Spearman correlation coefficients as significant with p < 0.05 (Suppl. Table 3, Suppl. Fig. 3A). Given the absence of changes in proton conductance, we focused on physiological meaningful ATP production/turnover rates, represented by correlations with state 3 respiration. The integrative analysis associated 192 of 976 mitochondrial proteins to ATP production/turnover (Figure 3A, Suppl. Table 3), which were depicted in substrate-specific subsets (Figure 3A, Suppl. Fig. 3B). Proteins correlating substrate-specifically represent potential factors in the control of specific substrate supply (pyruvate, succinate or PC), while proteins correlating with two substrate oxidation rates may control common pathways (e.g. both pyruvate and PC supply NADH for oxidation). The control of common pathways would also apply to proteins correlating with all three substrate oxidation rates. However, the correlation with respiration irrespective of substrate would rather suggest that the candidate protein controls or is controlled by ATP demand. Strikingly, only the mitochondrial pyruvate carrier subunit 1 (MPC1) overlaps with state 3 rates of all substrates. Notably, the influence of HFD stress on mitochondrial pyruvate transport is also recovered in the proteomic approach by significant induction of MPC2, resulting in the significantly elevated sum of MPC subunits (Figure 3B). While the stoichiometry of the heterodimeric MPC is not clarified yet [31], [32], the proteomic recovery suggests a 1:2 ratio (MPC1/2). Given the strong correlation of MPC1 with all substrates, mitochondrial pyruvate transport appears to crucial for balancing the production and consumption of proton motive force. The strong correlation between MPC1 and state 3 oxidation of all substrates supports a model of ATP demand controlling mitochondrial pyruvate influx. The proteomic data suggest reduced pyruvate decarboxylation by elevated pyruvate dehydrogenase kinase (PDK1 and PDK2) levels (Figure 3C) and increased pyruvate carboxylation by increased pyruvate carboxylase (PC) concentration. This would be in line with the model, as pyruvate would be rerouted into ATP-demanding anabolic pathways, such as hepatic gluconeogenesis, a typical driver for pathologically high blood glucose levels. The energetic significance of the MPC-dependent funneling of pyruvate is coherent with recent results reporting the requirement of MPC for liver gluconeogenesis [19], [20]. Noticeably, from the cellular bioenergetics point of view, gluconeogenesis assists in dissipating energy of the nutrient-overloaded hepatic system, despite all negative effects of gluconeogenesis in the sequelae of metabolic disease.
Figure 3.
Correlation analysis of mitochondrial respiration and proteome. (A) 976 mitochondrial proteins (as annotated in the Mitocarta 2.0) were correlated with state 3 respiration for different substrates. The numbers in the Venn-diagram depict significantly correlating proteins (Spearman ρ ≥ 0.5, p < 0.05). The single protein correlating with all substrates is MPC1. (B) Absolute protein concentrations of Mpc1, Mpc2, and Mpc1 + Mpc2. (C) Routing of mitochondrial pyruvate during HFD-feeding based on proteomic data. Enhanced imported pyruvate is not decarboxylated, as increased levels of pyruvate dehydrogenase kinases PDK1 and PDK2 inhibit pyruvate dehydrogenase (PDH). Increased pyruvate carboxylase (PC) levels support enhanced pyruvate carboxylation, forming oxaloacetate for gluconeogenesis, a highly ATP-demanding process driving respiration. (D) Enrichment analysis for KEGG pathways was performed for proteins significantly correlating with state 3 palmitoyl-carnitine (PC)/malate respiration. The top 10 significantly enriched KEGG pathways are displayed with their respective combined score calculated by Enrichr. (E) Prediction of proteins controlling lipid respiration in response to nutrient stress. Protein levels significantly induced by HFD (p < 0.05) were filtered specifically for correlation with HFD-induced PC/malate state 3 respiration. (F) The functional link between HFD-stress induced proteins is strongly supported by the string-db network [30]. Acads, Cpt2, Hadha (indicated by red circle) were previously identified as disease genes for inborn errors of lipid metabolism [28]. All data represent n = 8 animals. Boxplots indicate 25–75 percentiles, with the vertical line indicating the median and whiskers from minimum to maximum (B) or as mean ± standard error of mean (C). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 in a t-test comparing chow vs HFD.
Concerning increased lipid supply during HFD, enhanced lipid oxidation requires a complex protein network. An array of 155 proteins correlated with lipid-dependent ATP production (Figure 3A, Suppl. Fig. 3C). Their link to lipid oxidation is confirmed by KEGG pathway enrichment, elucidating “fatty acid degradation” and “fatty acid metabolism”, alongside with “valine, leucine and isoleucine degradation”, “propanoate metabolism” and “oxidative phosphorylation” (Figure 3D). Similarly, GO term enrichment analysis revealed enrichment for “fatty acid beta oxidation”, “(respiratory) electron transport chain” and “NADH dehydrogenase (activity)” in this set of proteins (see Suppl. Fig. 3D). Notably, strong correlations of proteins with state 3 respiration were not necessarily paralleled with significant protein concentration changes, therefore masking their significance in mitochondrial bioenergetics without integrating functional respirometry. To further filter for nutrient stress-related key proteins from the 155-protein list, we considered only proteins that are significantly elevated by HFD and that correlate with PC respiration only under HFD conditions (Suppl. Table 4). Of 46 proteins specifically associating with PC/malate fueled state 3 respiration in HFD, 19 were significantly increased by HFD feeding, i.e. represent potential HFD stress-induced proteins (Figure 3E). Functional interaction between 16 out of 19 proteins is supported by string-db analysis (Figure 3F) [30], including Acads, Cpt2, and Hadha, which are all implicated in inborn errors of lipid metabolism according to disease gene enrichment analysis and/or are demonstrated as regulators of beta oxidation [28], [33], [34]. Thus, integrating functional bioenergetics and proteomics appears instrumental to effectively and potently filter for disease-relevant proteins.
3.4. Complex I concentration controls mitochondrial lipid oxidation in the liver
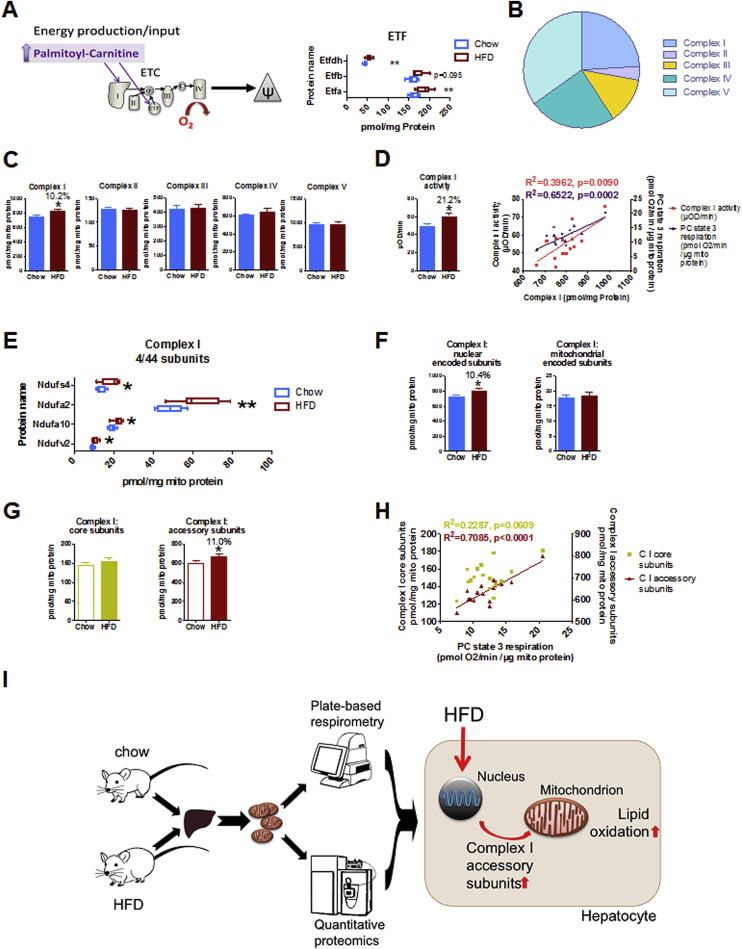
Next, we analyzed in depth the relation between proteomic changes of the electron transport chain and HFD-induced lipid respiration, based on evidence from correlation and enrichment analyses (Figure 3A, Suppl. Table 3, Suppl. Fig. 3C). We find that all ETF (electron transfer flavoprotein) subunits are increased by HFD feeding, thus contributing to the increased lipid oxidation in HFD (Figure 4A). During lipid oxidation, however, energy from lipid oxidation is also donated to complex I via NADH [35]. Looking into the relative proportions of respiratory complexes, the TPA approach revealed high abundance of respiratory complexes I, IV and V in liver mitochondria (Figure 4B). However, the TPA approach also enabled determination of quantitative changes of entire respiratory complexes by summing up absolute subunit concentrations. This analysis enabled the discovery of systemic functional changes that may be blurred by analyzing subtle changes in single proteins (Figure 4C). Strikingly, HFD feeding significantly increased the cumulative concentration of complex I (about 10%) while other respiratory complexes were unchanged. Consistently with PC/malate-fueled state 3 respiration, complex I activity was also increased and correlated significantly with complex I concentrations (Figure 4D), thus demonstrating that the cumulative complex I concentration is functionally relevant. Not surprisingly, pyruvate respiration rates also correlate with complex I content (Suppl. Fig. 4A), as decarboxylation of pyruvate and catabolism of acetyl-CoA in the TCA cycle fully donates pyruvate's electrons to NADH and eventually complex I. However, pyruvate respiration was not significantly changed in response to HFD (Figure 1B). Of the remaining respiratory complexes, only complex IV shows association with PC and pyruvate respiration (Suppl. Fig. 4B–E), but it was not significantly elevated in HFD (Figure 4C). Thus, we conclude that at least the HFD-induced lipid oxidation is facilitated through increased complex I content. Notably, HFD-induced stress imposed only mild impacts on single subunit concentrations of complex I (Suppl. Fig. 5A, B), similar to the remaining respiratory complexes (Suppl. Fig. 5C–F). Pertaining to absolute subunit concentrations, we observed the induction of Ndufs4, Ndufa2, Ndufab10, as well as Ndufv2, which is the only complex I core subunit (Figure 4E).
Figure 4.
HFD-induced lipid oxidation is facilitated by increased complex I content and activity, regulated by nuclear accessory subunits. (A) Scheme showing palmitoyl-carnitine entry into the electron transport chain (ETC). Electron transfer flavoprotein (ETF) subunits are increased in HFD-liver mitochondria. (B) The pie chart shows relative proportion of each complex of the ETC. Single subunit concentrations were summed up for each respiratory complex. (C) Total cumulative concentration of respective ETC complex subunits, depicted as group mean values. Asterisk indicates significantly increased complex I concentration in HFD. (D) Respiratory complex I activity by colorimetric assay (bar chart). Cumulated complex I concentrations correlate significantly with complex I activity (red) and PC-state 3 respiration (purple). (E) The absolute concentrations of complex I subunits that are significantly changed by HFD (see Fig. S5 for the complete list of absolute subunit concentrations). (F) The additive concentration of nuclear-encoded complex I subunits (left panel) is increased by HFD, while mitochondrial-encoded subunits are not (right panel). (G) The cumulative concentrations of complex I core and accessory subunits were correlated with (H) PC-state 3 respiration. (I) The integrative analysis of respirometry and proteomics supports model of bioenergetic adaptation in response to HFD that permits increasing lipid oxidation by upregulation of nuclear-encoded accessory complex I subunits. All data represent n = 8 animals. Boxplots indicate 25–75 percentiles, with the vertical line indicating the median and whiskers from minimum to maximum (A, E) or as mean ± standard error of mean. *p < 0.05, **p < 0.01 comparing chow vs HFD by t-test (A, C, D, E, F, G). (D, H) R2 and p-value of Pearson-correlation are given, linear regression is shown where Pearson-correlation is significant (p < 0.05).
3.5. Obesity impacts nuclear- but not mitochondrial-encoded subunit concentrations and functional capacity of complex I
Next, we focused on mammalian respiratory complex I, which consists of seven mitochondrial-encoded subunits, while the residual 37 are nuclear-encoded [36], [37]. Summing up mitochondrial- and nuclear-encoded subunit concentrations separately demonstrated that the HFD-induced elevation of entire complex I component concentration is impacted by nuclear-encoded subunits, but not mitochondrial-encoded subunits (Figure 4F). Importantly, the amount of nuclear-encoded but not mitochondrial-encoded subunits correlated strongly with PC and pyruvate-fueled state 3 respiration (Suppl. Fig. 6A). The impact of nuclear-encoded subunits on mitochondrial respiration was also observed through correlations with complex IV (Suppl. Fig. 6C), where nuclear-control of activity is well established [38]. We found no associations regarding nuclear-mitochondrial imbalance on complex III and V (Suppl. Fig. 6B, D). The 14 core subunits of complex I are highly conserved and build the catalytic part of complex I, while the 30 accessory or “supernumerary” subunits are under suspect to control assembly, regulation, stability, or protection against oxidative stress [39]. Interestingly, only the sum of accessory complex I subunits, but not of core subunits, was increased upon HFD feeding (Figure 4G) and correlates with PC/malate fueled state 3 respiration (Figure 4H). Notably, the regulation and function of complex I during obesity is important for some metabolic disease drugs, such as metformin, which is currently in the light of structural analyses concerning complex I [40]. Taken together, these integrative analyses reveal that the increase of nuclear-encoded accessory complex I subunits may be permissive, if not required, for increased lipid oxidation in liver mitochondria of HFD fed mice (Figure 4I). The plasticity of liver mitochondrial respiration per se appears to be mainly permitted by nuclear regulation of complex I and IV subunits.
4. Conclusions
Novel technologies such as multi-well, plate-based respirometry and new analytic developments in mass-spectrometric analysis provide powerful tools to increase our mechanistic understanding of physiological functions and disease patterns. Here, we demonstrate that the combination of these technologies and the integrative analysis uncovers direct links between mitochondrial function and its molecular basis, with the potential to establish causal relationships for mechanistic discovery in disease-mechanisms. Importantly, measuring absolute values for respiration and protein concentrations is required to establish a robust association between oxidative function and the mitoproteome. The absolute protein concentration output of the TPA approach enables to overcome subtle single protein changes by summation to functional protein modules, thereby enhancing molecular insights into mechanisms. The respiratory complexes provide the ideal proof-of-principle for this workflow as they are directly related to mitochondrial oxygen consumption and assembled of multiple subunits. We substantiate in physiologically-challenged liver mitochondria that this integrative methodological approach improves the linkage map between cellular energy metabolism and the molecular protein adjustments in disease models. We cannot formally exclude an impact of blood cell mitochondria, as the mice were not perfused prior to liver dissection. However, due to vigorous washing steps, the proportion should be small and differential influence of immune cells was not observed when inspecting inflammation marker of the background proteome. The integrative analysis reveals that the mitochondrial pyruvate carrier associates strongest to mitochondrial energy turnover in the liver, and that elevated mitochondrial lipid oxidation is specifically associated, and possibly facilitated, by nuclear-encoded complex I subunits.
First, we confirmed that increased systemic lipid metabolism of HFD feeding and obesity is reflected by the increase of PC-fueled respiratory capacity at the level of isolated liver mitochondria [41], [42]. The increased respiratory rates in the mouse liver are in accordance with data on pre-pathological changes of the human liver before accumulating dysfunctional properties at later stages [18]. Analyzing the mouse liver mitoproteome, we show that HFD induces numerous quantitative changes, but the molecular adjustments appear rather subtle when inspecting single protein concentrations, hampering attempts to identify distinct molecular signature changes for mitochondrial respiration. Therefore, we hypothesized that functionally-linked protein subsets are responsible for robust regulation of hepatocyte energy metabolism during overnutrition, prior to chronic stress-induced dysfunction that disturbs protein-function relationships. Indeed, KEGG/GO term-enrichment analyses cluster the majority of upregulated proteins to metabolism, in particular fatty acid oxidation, including carnitine palmitoyltransferase 1 (Cpt1, Figure 2C), which is the rate limiting enzyme for the long chain fatty acid transport into mitochondria and mitochondrial beta oxidation [43]. While most of the enrichment data appear to be in accordance with the functional respirometric data, uncertainty remained as to which proteomic changes control distinct parts of the oxidative phosphorylation machinery.
While KEGG pathways and GO terms aim to partition the proteome into functional subsets using a broad collection of various parameters, we attempted to build specific functional links to directly associate proteins or multimeric complex concentrations with the functional readout “mitochondrial respiration.” Herein, we deliver lists of single protein concentrations correlating with substrate-specific or –unspecific mitochondrial respiration. Strikingly, MPC1 correlates strongest with ATP-linked state 3 respiration, irrespective of substrate. Therefore, mitochondrial pyruvate transport appears as master regulator of mitochondrial energy turnover in the liver, possibly not only controlling substrate supply, but is also being controlled by ATP demand. The model relating MPC to ATP demand is further supported by proteomic data that suggest funneling of pyruvate into gluconeogenesis, a highly energy demanding process. Similar observations were made in the liver of the MPC knockout mouse [19], [20]. Besides the basic understanding of mitochondrial energy metabolism upon physiological challenge, the integrative approach may also assist to detect disease-related mechanisms. By systemically filtering the mitoproteome for proteins related to HFD-induced lipid oxidation, string database analysis connected a single network, which included genes causing inborn errors of lipid oxidation (as depicted in Figure 3E). The integrative analysis further suggested the involvement of multiple respiratory complex subunits in the response to HFD, particularly complex I subunits. Taking advantage of absolute quantitation by LC-MS/MS-based TPA, total subunit concentrations of respective respiratory complexes were instrumental to substantiate the claim of specific complex I adjustments by HFD feeding, providing compelling evidence for the relation between mitochondrial lipid oxidation and nuclear complex I subunits. Thus, HFD-induced mitochondrial adjustments are controlled via nucleus-mitochondrion communication rather than being mitochondrion-autonomous, affecting nuclear transcription and translation. The imbalance in mito-nuclear regulation during HFD feeding may foster cellular stress and disease as shown for mismatching mitochondrial and nuclear genotypes [44]. In the case of aging muscle, nuclear-mitochondrial communication was reported to be mediated by declining NAD+ in the nucleus, reversible, and PGC-1alpha/beta-independent [45]. In further expansion to this study, the integration of global nuclear gene regulation may be feasible to understand which factors wire mitochondrial biology to nuclear control.
Taken together, the integration of mitochondrial functional analysis and quantitative proteomic analysis is suitable for revealing molecular signatures controlling energetic adjustments in diet-induced obesity, providing a tool to explore proteins that impact mitochondrial respiration and substrate preferences in disease, physiological and environmental challenges.
Author contributions
E.W. performed and analyzed all mitochondrial bioenergetics and bioinformatics experiments; J. W. performed and analyzed proteomics, TPA calculations, and gave advice on experimental design and the manuscript; M.J. conceptualized and supervised the research plan, and assisted in mitochondrial bioenergetics experiments; E.W. and M.J. interpreted all data and wrote the manuscript.
Competing financial interest
The authors declare no competing financial interests.
Acknowledgements
We thank Ms Katharina Zettl for excellent technical assistance, Ms Silke Morin and Dr. Susanne Keipert for editing of the manuscript. This work was supported by funding in part to M.J. from the German Diabetes Center (DZD) and in part to J.W. from the Max-Planck-Society (MPS) for the Advancement of Science and the German Research Foundation (DFG/Gottfried Wilhelm Leibniz Prize).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.01.002.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Brand M.D. The efficiency and plasticity of mitochondrial energy transduction. Biochemical Society Transactions. 2005;33:897–904. doi: 10.1042/BST0330897. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 3.Jastroch M., Divakaruni A.S., Mookerjee S., Treberg J.R., Brand M.D. Mitochondrial proton and electron leaks. Essays in Biochemistry. 2010;47:53–67. doi: 10.1042/bse0470053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soboll S., Oh M.-H., Brown G.C. Control of oxidative phosphorylation, gluconeogenesis, ureagenesis and ATP turnover in isolated perfused rat liver analyzed by top-down metabolic control analysis. European Journal of Biochemistry. 1998;254:194–201. doi: 10.1046/j.1432-1327.1998.2540194.x. [DOI] [PubMed] [Google Scholar]
- 5.Rolfe D.F.S., Brand M.D. Proton leak and control of oxidative phosphorylation in perfused, resting rat skeletal muscle. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1996;1276:45–50. doi: 10.1016/0005-2728(96)00029-1. [DOI] [PubMed] [Google Scholar]
- 6.Brown G.C., Lakin-Thomas P.L., Brand M.D. Control of respiration and oxidative phosphorylation in isolated rat liver cells. European Journal of Biochemistry. 1990;192:355–362. doi: 10.1111/j.1432-1033.1990.tb19234.x. [DOI] [PubMed] [Google Scholar]
- 7.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J. PGC-1alpha-responsive genes involved in oxidative phosphoryylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 8.Morino K., Petersen K.F., Dufour S., Befroy D., Frattini J., Shatzkes N. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. Journal of Clinical Investigation. 2005;115:3587–3593. doi: 10.1172/JCI25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochemical Journal. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Divakaruni A.S., Brand M.D. The regulation and physiology of mitochondrial proton leak. Physiology. 2011;26:192–205. doi: 10.1152/physiol.00046.2010. [DOI] [PubMed] [Google Scholar]
- 11.Clark L.C.J. Monitor and control of blood and tissue oxygen tensions. ASAIO Journal. 1956;2:41–48. [Google Scholar]
- 12.Rogers G.W., Brand M.D., Petrosyan S., Ashok D., Elorza A.A., Ferrick D.A. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faubert B., Vincent E.E., Griss T., Samborska B., Izreig S., Svensson R.U. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1α. Proceedings of the National Academy of Sciences. 2014;111:2554–2559. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie X., Li M., Lu B., Zhang Y., Lan L., Chen L. Down-regulating overexpressed human lon in cervical cancer suppresses cell proliferation and bioenergetics. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wisniewski J.R., Rakus D. Multi-enzyme digestion FASP and the ‘Total Protein Approach’-based absolute quantification of the Escherichia coli proteome. Journal of Proteomics. 2014;109:322–331. doi: 10.1016/j.jprot.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y., Williams Evan G., Dubuis S., Mottis A., Jovaisaite V., Houten Sander M. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiśniewski J.R., Gizak A., Rakus D. Integrating proteomics and enzyme kinetics reveals tissue-specific types of the glycolytic and gluconeogenic pathways. Journal of Proteome Research. 2015;14:3263–3273. doi: 10.1021/acs.jproteome.5b00276. [DOI] [PubMed] [Google Scholar]
- 18.Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metabolism. 2015;21:739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 19.McCommis Kyle S., Chen Z., Fu X., McDonald William G., Colca Jerry R., Kletzien Rolf F. Loss of mitochondrial pyruvate carrier 2 in the liver leads to defects in gluconeogenesis and compensation via pyruvate-alanine cycling. Cell Metabolism. 2015;22:682–694. doi: 10.1016/j.cmet.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray Lawrence R., Sultana Mst R., Rauckhorst Adam J., Oonthonpan L., Tompkins Sean C., Sharma A. Hepatic mitochondrial pyruvate carrier 1 is required for efficient regulation of gluconeogenesis and whole-body glucose homeostasis. Cell Metabolism. 2015;22:669–681. doi: 10.1016/j.cmet.2015.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wisniewski J.R., Mann M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Analytical Chemistry. 2012;84:2631–2637. doi: 10.1021/ac300006b. [DOI] [PubMed] [Google Scholar]
- 22.Wisniewski J.R., Gaugaz F.Z. Fast and sensitive total protein and Peptide assays for proteomic analysis. Analytical Chemistry. 2015;87:4110–4116. doi: 10.1021/ac504689z. [DOI] [PubMed] [Google Scholar]
- 23.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 24.Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Research. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium T.U. UniProt: a hub for protein information. Nucleic Acids Research. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrade-Navarro M.A., Fontaine J.F. Gene set to diseases: disease enrichment analysis on human gene sets with literature data. Genomics and Computational Biology. 2016;2:e33. [Google Scholar]
- 29.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nature Genetics. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 30.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bricker D.K., Taylor E.B., Schell J.C., Orsak T., Boutron A., Chen Y.-C. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herzig S., Raemy E., Montessuit S., Veuthey J.-L., Zamboni N., Westermann B. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 33.Rinaldo P., Matern D., Bennett M.J. Fatty acid oxidation disorders. Annual Review of Physiology. 2002;64:477–502. doi: 10.1146/annurev.physiol.64.082201.154705. [DOI] [PubMed] [Google Scholar]
- 34.Rector R.S., Payne R.M., Ibdah J.A. Mitochondrial trifunctional protein defects: clinical implications and therapeutic approaches. Advanced Drug Delivery Reviews. 2008;60:1488–1496. doi: 10.1016/j.addr.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oelkrug R., Kutschke M., Meyer C.W., Heldmaier G., Jastroch M. Uncoupling protein 1 decreases superoxide production in brown adipose tissue mitochondria. Journal of Biological Chemistry. 2010;285:21961–21968. doi: 10.1074/jbc.M110.122861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirst J., Carroll J., Fearnley I.M., Shannon R.J., Walker J.E. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 37.Balsa E., Marco R., Perales-Clemente E., Szklarczyk R., Calvo E., Landázuri Manuel O. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metabolism. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Poyton R.O., McEwen J.E. Crosstalk between nuclear and mitochondrial genomes. Annual Review of Biochemistry. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 39.Vinothkumar K.R., Zhu J., Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bridges Hannah R., Jones Andrew J.Y., Pollak Michael N., Hirst J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochemical Journal. 2014;462:475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marvyn P.M., Bradley R.M., Mardian E.B., Marks K.A., Duncan R.E. Data on oxygen consumption rate, respiratory exchange ratio, and movement in C57BL/6J female mice on the third day of consuming a high-fat diet. Data Brief. 2016;7:472–475. doi: 10.1016/j.dib.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin T.L., Alquier T., Asakura K., Furukawa N., Preitner F., Kahn B.B. Diet-induced obesity alters AMP kinase activity in hypothalamus and skeletal muscle. Journal of Biological Chemistry. 2006;281:18933–18941. doi: 10.1074/jbc.M512831200. [DOI] [PubMed] [Google Scholar]
- 43.Orellana-Gavaldà J.M., Herrero L., Malandrino M.I., Pañeda A., Sol Rodríguez-Peña M., Petry H. Molecular therapy for obesity and diabetes based on a long-term increase in hepatic fatty-acid oxidation. Hepatology. 2011;53:821–832. doi: 10.1002/hep.24140. [DOI] [PubMed] [Google Scholar]
- 44.Latorre-Pellicer A., Moreno-Loshuertos R., Lechuga-Vieco A.V., Sanchez-Cabo F., Torroja C., Acin-Perez R. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016;535:561–565. doi: 10.1038/nature18618. [DOI] [PubMed] [Google Scholar]
- 45.Gomes Ana P., Price Nathan L., Ling Alvin J.Y., Moslehi Javid J., Montgomery M.K., Rajman L. Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.