Abstract
During maternal recognition of pregnancy (MRP), a conceptus-derived signal leads to the persistence of the corpus luteum and the maintenance of gestation. In the horse, the nature of this signal remains to be elucidated. Several studies have focused on the changes in gene expression during MRP, but little information exists at the protein level. The aim of this study was to identify the proteins at the embryo-maternal interface around signalling of MRP in the horse (day 13) by means of mass spectrometry. A distinct influence of pregnancy was established, with 119 proteins differentially expressed in the uterine fluid of pregnant mares compared to cyclic mares and with upregulation of several inhibitors of the prostaglandin synthesis during pregnancy. By creating an overview of the proteins at the embryo-maternal interface in the horse, this study provides a solid foundation for further targeted studies of proteins potentially involved in embryo-maternal interactions, MRP and pregnancy loss in the horse.
Introduction
Maternal recognition of pregnancy (MRP) covers the series of events leading to the persistence of the corpus luteum and a receptive uterine environment to support the maintenance of gestation1. In the cycling mare, pulsatile release of prostaglandin F2α (PGF2α) causes luteolysis, resulting in a decline in progesterone. This mechanism is inhibited during pregnancy by the presence of the conceptus2. In pigs, the conceptus derived signal which initiates MRP has been identified a long time ago as oestrogen3 and in ruminants as interferon tau4,5. However, the nature of this signal remains to be elucidated in the horse despite several decades of elaborate research on this topic6,7. Initial studies have focused on the identity of specific candidate signalling molecules and while the equine embryo produces substantial quantities of oestrogen as well as prostaglandins (PG) and limited amounts of interferons, no convincing evidence exists for their signalling role in MRP7. Potential embryonic signal targets involved in the luteostatic mechanism in the horse are prostaglandin-endoperoxide synthase 2 (PTGS2), an enzyme in the biosynthesis of PGF2α, and oxytocin, which stimulates endometrial PGF2α secretion through a positive feedback loop8. Both PTGS2 and oxytocin receptor expression (OXTR) are repressed during early pregnancy compared to cycling mares, with downregulation of PTGS2 at the RNA level and of OXTR at the protein level9–13.
During the last years, the topic of MRP in the horse has been broadened to all pathways involved in embryo-maternal communication around the timing of MRP. Signalling of MRP is a continuum of events, estimated to occur between days 12 and 14. Recipient mares can still get pregnant when an embryo is transferred to their uterus at day 12, but not at day 14 after ovulation14, while repression of PTGS2 occurs by day 13 of pregnancy11. By day 16, clear differences between pregnant and cyclic horses are observed. Transcriptomics of the equine endometrium and equine conceptuses have substantially contributed to the knowledge on pathways affected around the timing of MRP in the horse7,15–19. Technological advantages, including sequencing, favoured development of genomics and transcriptomics compared to proteomics20. However, mRNA abundances can only explain 40% of the variation in protein levels and the actual protein profile is influenced by post-transcriptional regulation mechanisms21. This appeals for complementing transcriptomics knowledge on MRP with quantitative proteomics. This can now be achieved through mass spectrometry (MS). Recent improvements in MS technologies, including data-independent-acquisition, allow reproducible label-free quantification of proteins in complex biological samples22.
Mass spectrometry of the embryo-maternal interface around MRP has been performed in several farm animals including pigs23,24, sheep25 and cattle26–28. In the horse, specific molecules with a potential role in MRP have been targeted by immunohistochemistry13,29–32 and global screening of uterine proteins has been performed in the context of endometritis33. However, the effect of pregnancy on the uterine secretome has not been assessed by means of high-throughput proteomics in the horse up to now. In a recent study, equine blastocysts were collected by uterine lavage on day 8 and an MS analysis was performed of the proteins secreted during culture of these embryos for 24 h and 48 h and of proteins present in the blastocoel fluid and the embryo capsule34. The authors detected prostaglandin F2 receptor inhibitor (PTGFRN) and a progesterone potentiating protein, FK506 binding protein 4 (FKBP4), in the blastocoel fluid, but it remained to be determined whether these proteins were actively secreted into the uterine lumen.
The aim of this study was to gain new insights into the embryo-maternal communication around the signalling of MRP in the horse. Since signalling of MRP is estimated to occur between Day 12 and Day 14, sampling was performed at Day 13 (±0.5 day). We hypothesize that high-throughput proteomics can provide complementary information to the transcriptomic reports. To this end, proteomics was performed by high definition data independent mass spectrometry (HDMSE) with ion mobility drift time-specific collision-energy35. In this way, proteins were identified and quantified in uterine fluid of pregnant and cyclic mares as well in the yolk sac fluid of the pregnant mares.
Results
Sampling
Only reproductively sound mares with negative bacteriology and cytology of the uterine fluid were used for the sampling. In two cycles, namely one pregnant (P) and one control cyclic (C) cycle, a double ovulation occurred. Response to hCG resulted in ovulation 24–36 h after administration. In four cycles, ovulation only occurred 3 days after hCG; once in a P cycle, where artificial insemination (AI) was performed at the same time and in this case, the mare was inseminated a second time 48 h after the first time and she ovulated the day after. In all other P cycles, ovulation occurred within 48 h after AI. In one mare, a line of fluid was noticed by ultrasound of the uterus 1 day after AI and she was treated by intramuscular administration of oxytocin.
Identification of proteins
The average protein concentration was similar in the uterine fluid (UF) of P (9.2 g/mL) and C (9.8 g/mL) mares, while the average protein concentration in the yolk sac (YS) was only 78 µg/mL.
For the first time, an overview was created of the proteins present in the UF and the YS at day 13 after ovulation in the horse. In the UF samples, a total of 10489 peptides were identified, accounting for 41% of all peptide like ions. Protein identification resulted in 1153 identifiable proteins (Supplementary file 1). After filtering and normalization, a total of 707 normalized proteins with at least two unique peptides were assessed for differential expression.
Differential expression of proteins was assessed for P versus C mares and pregnancy was associated with upregulation of 62 proteins (Table 1) and downregulation of 57 proteins (Table 2). For all proteins in this comparison, the log fold change, the adjusted p-value and the number of peptides are listed in Supplementary file 3.
Table 1.
Upregulated proteins in the uterine fluid of pregnant versus cyclic mares on day 13 after ovulation.
| Protein Symbol | Log FC | Adj. p-value | Gene Symbol | Gene Description |
|---|---|---|---|---|
| F7BAA0 | 2,27 | 0,03617 | GSTO1* | glutathione S-transferase omega 1 |
| F6Z0A9 | 2,02 | 0,01980 | RAC1* | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| F6VVU1; F6YMX5 | 1,96 | 0,02370 | MOB1A* | MOB kinase activator 1A |
| F6Y2H3; F6Y2V7 | 1,89 | 0,02907 | PEPD* | peptidase D |
| F7CCF5 | 1,81 | 0,01425 | LXN* | latexin |
| F7DIB3 | 1,80 | 0,03253 | SEC14L3* | SEC14 like lipid binding 3 |
| F6YAZ9; F7BYZ9 | 1,79 | 0,01633 | MYL12A* | myosin light chain 12A |
| F6RH25 | 1,55 | 0,01371 | DCPS* | decapping enzyme, scavenger |
| F6XV30 | 1,55 | 0,01980 | TBCA* | tubulin folding cofactor A |
| Q3S4D6 | 1,53 | 0,01915 | GM2A | GM2 ganglioside activator |
| F6XKI9 | 1,40 | 0,01142 | DNTTIP2* | deoxynucleotidyltransferase terminal interacting protein 2 |
| F6RTH0 | 1,39 | 0,01378 | TXNDC17* | thioredoxin domain containing 17 |
| F7CBN0 | 1,35 | 0,00336 | AKR1A1 | aldo-keto reductase family 1 member A1 |
| F6PWC8 | 1,25 | 0,00112 | PTGR1* | prostaglandin reductase 1 |
| F6XSN2 | 1,24 | 0,00106 | CCT7* | chaperonin containing TCP1 subunit 7 |
| F6XZQ1 | 1,24 | 0,00026 | CAPS* | calcyphosine |
| F7BAR2 | 1,23 | 0,02275 | TPMT | Thiopurine S-methyltransferase |
| F6W8C8 | 1,20 | 0,00004 | SERPINB6* | serpin family B member 6 |
| F6RGN2 | 1,19 | 0,04597 | FABP5* | fatty acid binding protein 5 |
| F6RMM1 | 1,17 | 0,00336 | SH3BGRL | SH3 domain binding glutamate rich protein like |
| F7CBR0; F7DZD2 | 1,11 | 0,00336 | LOC100050322 | Glutathione S-transferase |
| F7BHV8 | 1,11 | 0,00626 | TUBB4A* | tubulin beta 4A class IVa |
| Q8HZM6; F7A0T0 | 1,09 | 0,00106 | ANXA1 | Annexin A1 |
| F6XA04 | 1,06 | 0,00001 | YWHAE* | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon |
| F6XTY8 | 1,05 | 0,02235 | Unassigned | unassigned |
| F7D3E3 | 1,03 | 0,00004 | CMPK1* | cytidine/uridine monophosphate kinase 1 |
| F6W683 | 0,99 | 0,03617 | GMDS* | GDP-mannose 4,6-dehydratase |
| F7DB59 | 0,99 | 0,00106 | PAFAH1B3* | platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| F6SQ49 | 0,97 | 0,00112 | SMS* | spermine synthase |
| F6RL46 | 0,96 | 0,01211 | PGLS* | 6-phosphogluconolactonase |
| F6W9B1 | 0,93 | 0,01371 | ST13 | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| F7E0H3 | 0,91 | 0,01211 | TUBB* | tubulin beta class I |
| F6R8T8 | 0,90 | 0,01473 | ACY1 | aminoacylase 1 |
| F7D9J2 | 0,90 | 0,00053 | TKT* | transketolase |
| F6W039 | 0,85 | 0,00024 | ARHGDIA* | Rho GDP dissociation inhibitor alpha |
| F6ZHQ5 | 0,83 | 0,00336 | CLIC1 | chloride intracellular channel 1 |
| F7D1R1 | 0,82 | 0,01371 | PGK1 | Phosphoglycerate kinase 1 |
| F6TZS9 | 0,77 | 0,01633 | TPI1 | triosephosphate isomerase 1 |
| F7CIX6 | 0,76 | 0,00591 | ENO2* | enolase 2 |
| F6W3T1 | 0,73 | 0,00106 | LDHA | lactate dehydrogenase A |
| F7C5G3 | 0,73 | 0,01378 | PSMD11* | proteasome 26S subunit, non-ATPase 11 |
| F6ZE54 | 0,72 | 0,04589 | GPI | glucose-6-phosphate isomerase |
| F7BWW6 | 0,71 | 0,02824 | VCP* | valosin containing protein |
| F7CZS6 | 0,71 | 0,01052 | MDH1 | malate dehydrogenase 1 |
| F6PJY2 | 0,71 | 0,04256 | LZTFL1* | leucine zipper transcription factor like 1 |
| F7DMY1 | 0,70 | 0,04029 | CBFB* | core-binding factor beta subunit |
| F6UJ33 | 0,69 | 0,02943 | PFN1 | profilin |
| F6VSN2 | 0,69 | 0,00056 | GSTP1* | glutathione S-transferase pi 1 |
| F7DXG8 | 0,69 | 0,01915 | CFL1* | cofilin 1 |
| F7APS1; F6ZWS7 | 0,68 | 0,00106 | CSTB; LOC100050835* | Cystatin B |
| F7BE95; F6U2P8 | 0,67 | 0,03059 | UBE2V1* | ubiquitin conjugating enzyme E2 V1 |
| F7ALV0 | 0,67 | 0,00336 | TXN | Thioredoxin |
| F7BPT4 | 0,61 | 0,00336 | EZR* | Ezrin |
| F6S5E7 | 0,59 | 0,01211 | TARS* | threonyl-tRNA synthetase |
| F6QXW2 | 0,58 | 0,01378 | PEBP1* | phosphatidylethanolamine binding protein 1 |
| F6XLG0; F7DY67 | 0,57 | 0,04597 | PNP; LOC100058767 | Purine nucleoside phosphorylase |
| F7DZV9 | 0,57 | 0,00961 | YWHAB* | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta |
| F7CI32; F7ASU6; F7DKR3 | 0,51 | 0,03433 | SELENBP1* | selenium binding protein 1 |
| F7B5P1 | 0,49 | 0,03253 | CNDP2* | CNDP dipeptidase 2 (metallopeptidase M20 family) |
| F6YZ13 | 0,48 | 0,03604 | S100A13* | S100 calcium binding protein A13 |
| F6ZEV8 | 0,46 | 0,01618 | DBI* | diazepam binding inhibitor, acyl-CoA binding protein |
| F6X6A6; F6XKX6; F6Z5Z4 | 0,45 | 0,00423 | LOC100052020; LOC100054282 | Peptidyl-prolyl cis-trans isomerase |
Table 2.
Downregulated proteins in the uterine fluid of pregnant versus cyclic mares on day 13 after ovulation.
| Protein ID | Log FC | adj. P-Value | Gene Symbol | Gene Description |
|---|---|---|---|---|
| F6USV6 | −0,44 | 0,04220 | NOL11* | nucleolar protein 11 |
| F7BF31 | −0,52 | 0,01207 | SPI2* | alpha-1-antitrypsin |
| F6WZW6 | −0,57 | 0,01980 | PSMA1 | proteasome subunit alpha 1 |
| F6YLA3 | −0,62 | 0,00626 | TXNRD1* | thioredoxin reductase 1 |
| F6YVT0 | −0,71 | 0,01004 | RASGRP4* | RAS guanyl releasing protein 4 |
| F6ZFH9 | −0,72 | 0,02902 | YWHAG* | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma |
| F7AED2 | −0,72 | 0,01528 | LOC100050100* | alpha-1-acid glycoprotein 2-like |
| F7CZW9 | −0,73 | 0,02003 | SERPING1* | serpin family G member 1 |
| F6T7X3 | −0,75 | 0,00626 | LOC100065767 | membrane primary amine oxidase |
| F6RRV1 | −0,76 | 0,00368 | FETUB* | fetuin B |
| F7BKK5 | −0,76 | 0,04705 | GSTM3* | glutathione S-transferase mu 3 |
| F6PQ46 | −0,78 | 0,01052 | CP* | ceruloplasmin |
| F6R942; F6RI47 | −0,79 | 0,00336 | A2M* | Alpha-2-macroglobulin |
| F6SJ41 | −0,82 | 0,03640 | PFN2 | profilin 2 |
| F6XWM5 | −0,82 | 0,00041 | HP* | haptoglobin |
| F6RMD0 | −0,87 | 0,00516 | CFB* | complement factor B |
| F6ZD04 | −0,89 | 0,01443 | PYGB | glycogen phosphorylase B |
| P69905 | −0,89 | 0,01010 | HBA1 | hemoglobin subunit alpha 1 |
| F7AJP3 | −0,90 | 0,02643 | CHI3L1* | chitinase 3 like 1 |
| F6VTZ7 | −0,91 | 0,00072 | CFAP58* | cilia and flagella associated protein 58 |
| F6RDD3; F6VE37 | −0,91 | 0,00072 | HBB | hemoglobin subunit beta |
| F7BFJ1 | −0,92 | 0,01242 | F2 | coagulation factor II, thrombin |
| F6RZ27 | −0,93 | 0,00336 | APOA4* | apolipoprotein A4 |
| F6WMT7; F7C7Y1 | −0,93 | 0,00053 | KRT71; KRT73* | keratin 71; keratin 73 |
| F6QS41; F7BQS9 | −0,95 | 0,00336 | MROH2A* | maestro heat like repeat family member 2A |
| F6Z2L5 | −0,96 | 0,00005 | APOA1* | apolipoprotein A1 |
| F6XM13 | −0,98 | 0,01371 | APOD* | apolipoprotein D |
| F6XLB1 | −0,99 | 0,00056 | LTF | Lactotransferrin |
| F6XRU1; F6YAV2 | −1,01 | 0,00217 | SERPINB11* | serpin family B member 11 |
| F7DTV1 | −1,04 | 0,01115 | PON1* | paraoxonase 1 |
| F6SGV0 | −1,07 | 0,00338 | TTF2* | transcription termination factor 2 |
| Q29482 | −1,07 | 0,01298 | CLU | clusterin |
| F6SRP7 | −1,10 | 0,01010 | CAP1 | adenylate cyclase associated protein 1 |
| F6QX36 | −1,13 | 0,00119 | ITIH1* | inter-alpha-trypsin inhibitor heavy chain 1 |
| F6TJX5 | −1,16 | 0,03059 | TPP1* | tripeptidyl peptidase 1 |
| F7BZ41 | −1,19 | 0,02235 | CTSL* | cathepsin L |
| F7BNQ2 | −1,20 | 0,00217 | C4BPA* | complement component 4 binding protein alpha |
| F7AMJ7 | −1,20 | 0,02095 | STK38* | serine/threonine kinase 38 |
| F6RM73 | −1,22 | 0,00366 | APOA2 | Apolipoprotein A-II |
| F6YNT8 | −1,27 | 0,02370 | PEBP4* | phosphatidylethanolamine binding protein 4 |
| F6VUW2 | −1,30 | 0,00626 | CTSS* | cathepsin S |
| F6TE92 | −1,33 | 0,02043 | AGL | amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| F7C0E6 | −1,36 | 0,00259 | PLS1* | plastin 1 |
| F6X5J6 | −1,36 | 0,02043 | ADSL | adenylosuccinate lyase |
| F7BCH1 | −1,37 | 0,00000 | INHBA | Inhibin beta A chain |
| F6PUX2 | −1,41 | 0,00026 | MSN* | moesin |
| F7DXH4 | −1,44 | 0,00106 | VIPAS39* | VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| P01008 | −1,54 | 0,00178 | SERPINC1 | serpin family C member 1 |
| F7CWC8 | −1,57 | 0,00119 | unassigned | Amine oxidase [flavin-containing] |
| F7CWT0 | −1,58 | 0,00056 | P19* | P19 lipocalin |
| F6WRK2 | −1,59 | 0,00199 | MANBA* | mannosidase beta |
| F7BLE3 | −1,69 | 0,01851 | unassigned | unassigned |
| F6QYS3 | −1,75 | 0,01765 | ECM1* | extracellular matrix protein 1 |
| F6R8P9; F6RM27 | −2,13 | 0,00112 | TTLL7* | tubulin tyrosine ligase like 7 |
| F6SJN4 | −2,22 | 0,00556 | UBOX5* | U-box domain containing 5 |
| F7CHR8 | −2,23 | 0,01530 | CCDC36* | coiled-coil domain containing 36 |
| F6VST0; F6W6H2 | −3,34 | 0,00004 | NEFL* | neurofilament, light polypeptide |
In the YS samples, a total of 6500 peptide ions were identified, representing 51% of all peptide like ions and resulting in 903 identifiable proteins (Supplementary file 2). For the YS proteins, the primary goal was identification, rather than quantification, as different nature of the fluids impedes assessment of differential expression of proteins in YS versus UF.
Gene Ontology enrichment and pathway analysis
Categorization in the Gene Ontology (GO) terms ‘molecular function’, ‘biological process’ and ‘cellular component’ is provided for all quantified proteins in the comparison of P versus C in Supplementary file 3.
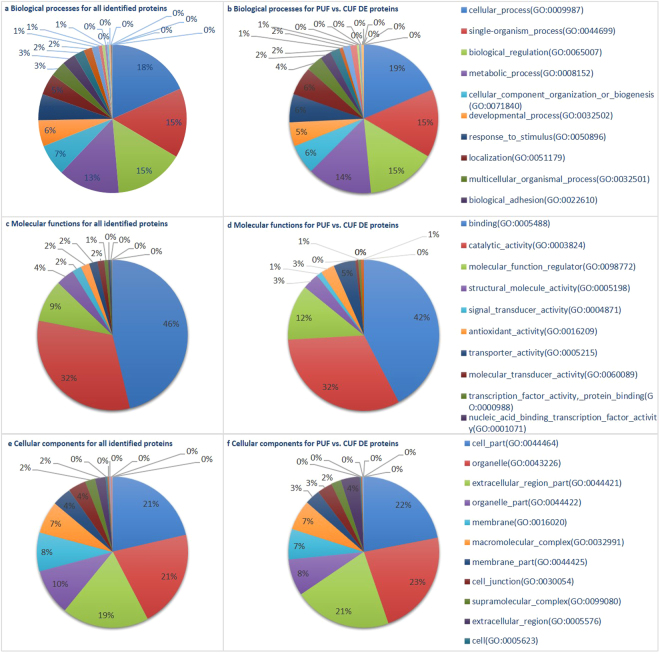
Figure 1 summarizes the GO categories in which the differentially expressed proteins are involved. The main category to which most proteins are assigned is ‘cellular process (GO:0009987)’ for the biological processes and ‘binding (GO:0005488)’ for the molecular functions. This coincides with the results in porcine uterine fluid24, but these are also the major categories when all proteins are taken into account. Overall, the differences in categorization between the groups are small.
Figure 1.
Categorization in Gene Ontology terms of all identified proteins in the uterine fluid (UF) and of differentially expressed (DE) proteins in the uterine fluid of pregnant (P) versus cyclic (C) mares. The main GO biological processes (a,b), molecular functions (c,d) and structural components (e,f) are represented for all quantified proteins in the equine uterine fluid (a,c,e) as well as for proteins found to be differentially expressed in the uterine fluid of pregnant versus cyclic mares (b,d,f).
Gene Ontology (GO) enrichment revealed no statistical overrepresentation when a Bonferroni correction for multiple testing was used (FDR < 0.05). No up- or downregulated KEGG pathways were detected either at a Benjamini-Hochberg corrected p-value of 0.05.
Embryo-maternal interaction
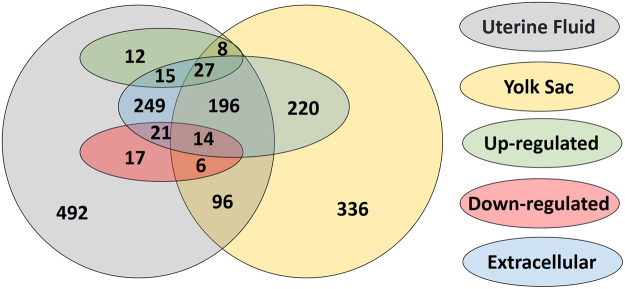
Comparison of the proteins identified in the UF of the P mares and in the YS of the corresponding embryo revealed 347 common proteins, 806 proteins which were only detected in the UF and 556 proteins which were only found in the YS. Figure 2 represents an overview of these UF specific proteins, YS specific proteins and common proteins, with specific display of the proteins up- and downregulated during pregnancy and of the proteins categorized in the extracellular space.
Figure 2.
Proteins identified in the uterine fluid and the yolk sac fluid. The number of common proteins as well as the number of proteins specific for the uterine fluid or the yolk sac fluid are displayed. The proteins which were found to be upregulated or downregulated in the uterine fluid of pregnant mares compared to cyclic mares are depicted separately. Proteins categorized in the extracellular space are also indicated.
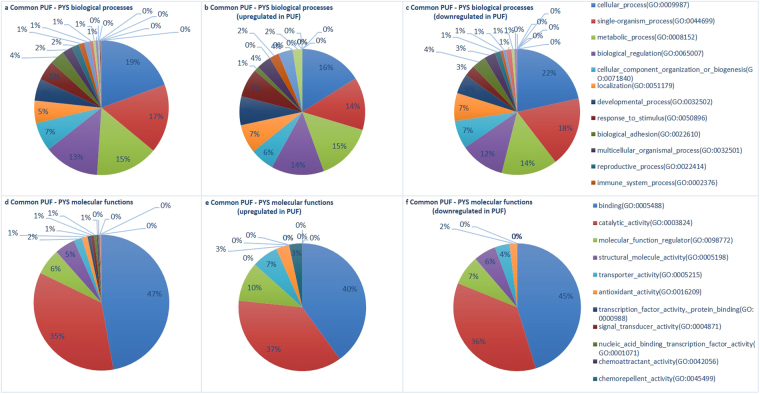
A list of the 347 common proteins is provided in Supplementary file 4, including the functions in which these proteins are involved. Figure 3 summarizes the GOs in which these common proteins were involved. Similar to the results for the UF in Fig. 1, the main GO categories in which the common proteins are involved are also ‘cellular process (GO:0009987)’ and ‘binding (GO:0005488)’ and differences in categorization are small. Common proteins in YS and UF which were also upregulated in P versus C, showed a higher representation in the biological processes ‘developmental process (GO:0032502)’ and ‘response to stimulus (GO:0050896)’. Molecular functions in which these proteins were more involved are ‘transporter activity (GO:0005215)’ and ‘transcription factor activity - protein binding (GO:0000988)’, while the common proteins which were downregulated in P versus C were rather represented in ‘structural molecule activity (GO:0005198)’.
Figure 3.
Categorization in Gene Ontology terms of common proteins in the uterine fluid of pregnant mares (PUF) and the yolk sac fluid (PYS) of the corresponding embryos. The main GO biological processes (a–c) and molecular functions (d–f) are represented for all common proteins in equine uterine fluid and yolk sac fluid (a,d), as well as for the subset of common proteins which were found to be upregulated (b,e) or downregulated (c,f) in the uterine fluid of pregnant mares compared to cyclic mares.
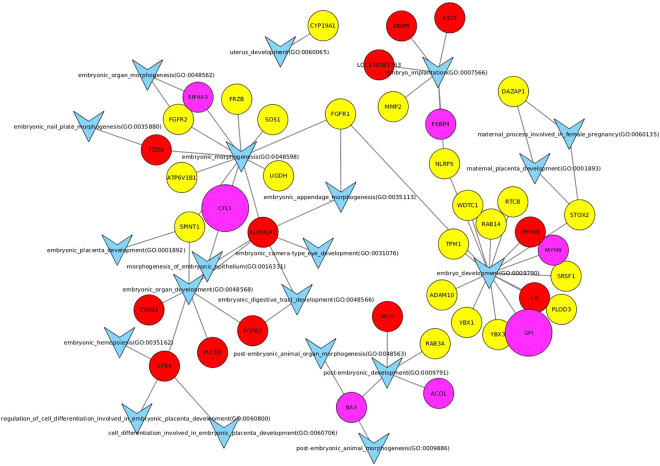
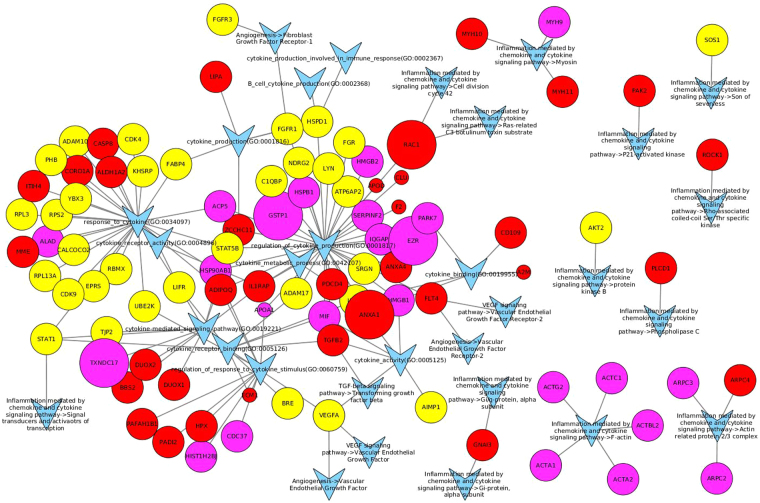
The embryo-maternal interaction was further visualized by Cytoscape 3.3.0 in Fig. 4. The most prominent GO terms in this network are ‘embryo development’ (GO:0009790) and ‘embryo morphogenesis’ (GO:0048598), with a main contribution of proteins originating from the yolk sac, and embryo implantation (GO:0007566) with the involvement of both uterine and embryonic proteins. In Fig. 5, the contribution of growth factors and cytokines in equine embryo-maternal signalling is visualized. The most extensive networks with various proteins found in the yolk sac fluid and/or the uterine fluid of pregnant mares are involved in ‘regulation of cytokine production’ (GO:0001817), ‘response to cytokine’ (GO:0034097) and the downstream GO’s ‘cytokine receptor binding‘ (GO:0005126), ‘cytokine mediated signalling pathway’ (GO:0019221) and ‘regulation of response to cytokine stimulus’ (GO:0060759).
Figure 4.
Involvement of proteins found in the yolk sac fluid and the uterine fluid of pregnant mares in GO terms and pathways representing embryo-maternal interaction. All GO terms and pathways that include ‘embryo’, ‘maternal’ or ‘uterus’ in their description were selected, together with all identified proteins in either the yolk sac or uterine fluid of pregnant horses belonging to these GO terms or pathways. These GO terms, pathways and proteins were then visualized using Cytoscape 3.3.0. Proteins found only in the uterine fluid are represented as red circles, proteins found only in the yolk sac as yellow circles and proteins found in both as purple circles. Proteins significantly (FDR corrected p-value < 0.05) up- or downregulated in the uterine fluid of pregnant mares are respectively larger and smaller circles (size not scaled with magnitude of up- or downregulation). GO terms and pathways are represented as a blue ‘V’, with lines indicating whether a GO term or pathway is associated with a protein.
Figure 5.
Involvement of proteins found in the yolk sac fluid and the uterine fluid of pregnant mares in GO terms and pathways representing embryo-maternal interaction. All GO terms and pathways that include ‘cytokine’ or ‘growth factor’ in their description were selected, together with all identified proteins in either the yolk sac or uterine fluid belonging to these GO terms or pathways. These GO terms, pathways and proteins were then visualized using Cytoscape 3.3.0. Proteins found only in the uterine fluid are represented as red circles, proteins found only in the yolk sac as yellow circles and proteins found in both as purple circles. Proteins significantly (FDR corrected p-value < 0.05) up- or downregulated in the uterine fluid of pregnant mares are respectively larger and smaller circles (size not scaled with magnitude of up- or downregulation). GO terms and pathways are represented as a blue ‘V’, with lines indicating whether a GO term or pathway is associated with a protein.
Discussion
Maternal recognition of pregnancy is an intriguing subject in the horse and extensive research on the molecular processes involved has been performed in the field of transcriptomics7,18. However, information on the downstream translation to proteins is scarce. In this study, quantitative proteomics of the uterine luminal fluid assessing the effect of pregnancy was performed for the first time in the horse. At the same time, proteins in the embryonic yolk sac fluid were mapped to provide insight into the embryo-maternal interaction.
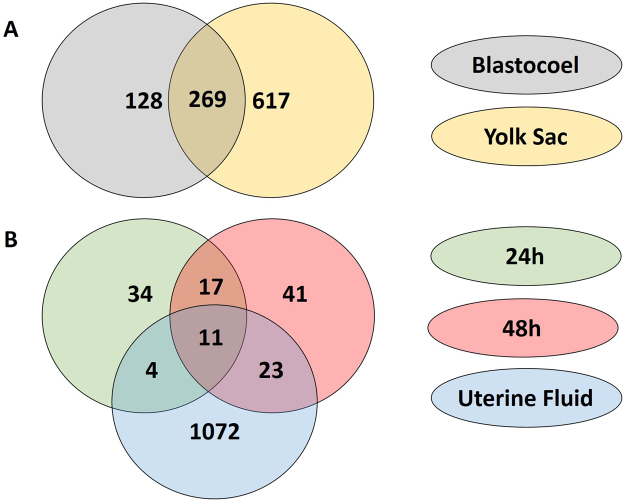
With 119 proteins differentially expressed in the uterine fluid of P versus C mares, a distinct influence of pregnancy was established. In general, a function of more than 40% of the differentially expressed proteins in the UF was categorized as ‘binding (GO:0005488)’, coinciding with the findings in pigs and cattle, where the majority of proteins were also allocated to molecular binding24,28 (Fig. 1). ‘Binding’ also represents the main category to which the common proteins in UF and YS were allocated, with subtly higher representation of proteins upregulated during pregnancy in categories linked to embryo-maternal interaction, namely ‘developmental process (GO:0032502)’, ‘response to stimulus (GO:0050896)’, ‘transporter activity (GO:0005215)’ and ‘transcription factor activity - protein binding (GO:0000988)’ at the expense of the more general GO term ‘structural molecule activity (GO:0005198)’ (Fig. 3). Cellular component categorization allocated 45% of the identified UF proteins to the extracellular space (Fig. 2). This coincides with the findings of Swegen et al.34, who specifically targeted secreted proteins by analysing embryo-conditioned medium. This supports the fact the proteins detected in our study mainly represent the proteins secreted in the uterine fluid rather than endometrial cells shed in the uterine lumen. This also accounts for the proteins which were found to be differentially expressed during pregnancy. Sixty four % of these proteins were categorized in the extracellular space; the other may have originated from occasional shedding of embryonic and endometrial cells into the uterine lumen. Figure 2 represents an overview of all UF specific, YS specific and common proteins, including their differential expression in P vs C and their allocation to the extracellular space. Interestingly, the majority of proteins commonly found in UF and YS are indeed present in the extracellular space. These represent candidate proteins involved in embryo-maternal interaction and signalling. In general, our results greatly coincided with the findings of Swegen et al.34 who worked with day 8 blastocysts to examine proteins present in and secreted by early equine embryos. Figure 6 shows the number of proteins which were commonly found in the blastocoel fluid and the YS and those found to be secreted in embryo-conditioned medium at 24 h and 48 h and in the UF in our study. More than two third of the proteins reported in the blastocoel fluid were also detected in the YS and more than one third of the proteins found to be secreted after 48 h of embryo culture were also detected in the UF. Overlap of the results validates our independent findings on the one hand and indicates conserved expression of several proteins throughout development on the other hand.
Figure 6.
Comparison of proteins detected in the uterine fluid (UF) and the yolk sac (YS) with the proteins reported by Swegen et al.34. Figure 6A shows the number of proteins which were found in the blastocoel fluid by Swegen et al.34 and the YS in our study and Fig. 6B illustrates the proteins those to be secreted in embryo-conditioned medium at 24 h and 48 h by Swegen et al.34 and in the UF in our study. Numbers are based on the reported gene symbols.
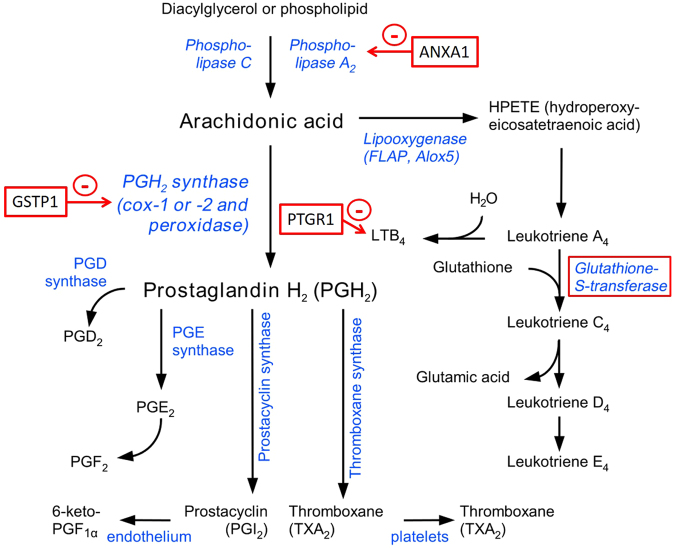
In the context of MRP, prostaglandin synthesis is of special interest. For three proteins involved in this pathway, namely prostaglandin reductase 1 (PTGR1), glutathione transferase 1 (GSTP1) and annexin A1 (ANXA1), significantly higher amounts were detected in the uterine fluid of pregnant mares compared with cyclic mares. Apart from acting on 15-oxo-PGE1, 15-oxo-PGE2 and 15-oxo-PGE2-alpha as 15-oxo-prostaglandin 13-reductase, PTGR1 catalyzes leukotriene B4 into its biologically less active metabolite, being the key step in the metabolic inactivation of leukotriene B4, as depicted in Fig. 7.
Figure 7.
Inhibitors of prostaglandin synthesis in uterine fluid of pregnant mares. Eicosanoid pathway, adapted from Wikipedia. Proteins found to be upregulated in the uterine fluid of pregnant mares are marked in red.
While glutathione transferases are also generally involved in the biosynthesis of prostaglandins and leukotrienes, as well as progesterone and testosterone36, a specific anti-inflammatory effect of GSTP1 by reduction of PTGS2, formerly known as cyclooxygenase-2 (COX-2), has been described37. Furthermore, transport of GSTP1 across the plasma membrane was demonstrated37. Based upon these observations with recombinant human GSTP1 in mice and the high homology of equine GSTP1 with other species, equine GSTP1 in uterine fluid might cross the plasma membrane and target intracellular PTGS2. Interestingly, GSTP1 was also detected in the YS of the equine conceptuses. In this regard, the pregnancy associated upregulation of GSTP1 observed in the equine uterine fluid could be involved in the luteostatic mechanism by inhibiting PTGS2. Further research is needed to examine this hypothesis, as this is the first report on the presence of GSTP1 in equine uterine fluid.
Another anti-inflammatory factor with an inhibitory effect on prostaglandin synthesis, more specifically on phospholipase A2, is annexin A1 (ANXA1)38,39. Annexin A1 was upregulated in the uterine fluid of the pregnant mares when compared to the cyclic mares and this association of annexins with pregnancy coincides with literature. An increase in ANXA1 was also reported in the uterine luminal fluid of pregnant ewes from day 10 to day 1225. Several annexins have been linked to embryo-maternal interaction. Annexin 4 (ANXA4) was found to increase over time from day 10 to day 13 in both cyclic and pregnant pigs24, and we previously reported greater quantities of ANXA4 in the oviductal fluid of pregnant mares, when compared to cyclic mares40. In our study, we detected annexin 1, 2, 3, 4, 5, 7 8 and 11 in the UF, while ANXA2 and ANXA5 were also found in the YS. Swegen et al.34 also reported the presence of ANXA2 in both the equine blastocoel fluid and the embryo-conditioned medium after 48 h. The only annexin found to be upregulated during pregnancy was ANXA1. Annexin 1 is an inhibitor of phospholipase A2, a rate-limiting enzyme which liberates arachidonic acid for the synthesis of prostaglandins and for which a lower enzyme activity of phospholipase A2 has been demonstrated in pregnant mares compared to cyclic mares on day 1441. In our study, phospholipase A2 group IIA (PLA2G2A) tended to be downregulated in the uterine fluid of pregnant mares with a logFC of −1.31 compared to the cyclic condition, but it was not significant at a 0.05 FDR. Both PLA2G2A and phospholipase A2 group VII (PLA2G7) were detected in the YS; the latter was also found in the equine embryo-conditioned medium after 48 h34. Overall, our data suggest pregnancy associated interference with the luteolytic eicosanoid pathway with upregulation of inhibitory factors at different levels of the prostaglandin synthesis pathway.
A close interaction between prostaglandins and oxytocin has been described in the context of MRP in the horse with downregulation of the oxytocin receptor protein in the pregnant endometrium on day 1413. In the present study, the presence of oxytocin in the uterine luminal fluid was examined, but it was not detected. However, this does not mean it was not present in the original samples; the collection method might have retained some peptides and missing values are intrinsic to mass spectrometry42,43. Phosphoinositide phospholipase C (PLCD1), involved in the oxytocin receptor signalling pathway, was detected, but not significantly affected by pregnancy44. The reduced expression of OXTR during pregnancy has been hypothesized to be induced by an observed decrease in the gene expression of oestrogen receptor 1 (ESR1) in the pregnant equine endometrium15. Several proteins related to ESR1 were also found to be affected in the uterine fluid. Surprisingly, pregnant mares showed a strong upregulation of deoxynucleotidyltransferase terminal interacting protein 2 (DNTTIP2), previously known as oestrogen receptor binding protein (ERBP). Binding of DNTTIP2 to ESR1 enhances its transcription45. Upregulation of DNTTIP2, which would lead to increased transcription of ESR1 in pregnant mares is contradictory to findings in literature and further targeted research is required to clarify this aspect. Downstream of the ESR1, the influence of oestrogen on the ezrin–radixin–moesin (ERM) family of actin-binding proteins has been studied, mainly in the context of breast cancer46,47. The distribution pattern of ERM-proteins in the blastocyst and the uterus has been linked to the implantation potential in mice. Protein analysis of uterine fluid has demonstrated the presence of ezrin (EZR) and moesin (MSN) in cattle26,27 and pigs23. In the horse, upregulation of EZR and downregulation of MSN was detected, while an inverse association with pregnancy was noted in cattle26,27. We detected both EZR and MSN in the YS and they were also found in the blastocoel fluid34.
Apart from the specific interest in proteins involved in prostaglandin synthesis, we also aimed to create a general overview of the proteins present at the embryo-maternal interface and potentially involved in signalling. Supplementary File 4 presents all proteins which were commonly found in the UF of P mares and in the YS and the functions of each protein are included. To visualize their role in embryo-maternal interaction and signalling, the proteins involved in GO terms including ‘embryo’, ‘maternal’ or ‘uterus’ are depicted in Fig. 4 and those linked to GO terms ‘growth factor’ and ‘cytokine’ in Fig. 5. The origin of the proteins can be distinguished in red (UF), yellow (YS) and pink (UF and YS) and up- and downregulation during pregnancy is represented by enlargement or shrinkage of the protein respectively. Interestingly, most proteins which were found to be upregulated during pregnancy were detected both in UF and in YS, while downregulation during pregnancy generally coincided with absence of these proteins in the YS, indicating a potentially important role of the embryo in the production of these proteins during pregnancy. Several common proteins were found at the embryo-maternal interface during MRP in cattle, including aconitase 1 (ACO1), which was specifically detected in the uterine fluid of pregnant and not in cyclic heifers, as well as glucose-6-phospate isomerase (GPI), which has been detected in the uterine fluid of both pregnant and cyclic heifers and for which an embryonic source has been presumed based on transcriptomics27. In our study, both proteins were found in the YS and the UF of P mares, with significant upregulation of GPI in P versus C. Two other proteins which were commonly found in UF and YS, namely FK506 binding protein 4 (FKBP4) (Fig. 4) and heat shock protein 90 (HSP90AB1) (Fig. 5), have been elaborately discussed by Swegen et al.34 concerning their progesterone supportive role. Co-operation of both factors is necessary for activation of the progesterone receptor48, FKBP4 has shown to be crucial for uterine receptivity and implantation in mice49 and FKBP4 deficit has been associated with pregnancy loss in human50. While FKBP4 was detected in equine blastocoel fluid and speculated to be involved in signalling, it was not detected in the embryo-conditioned medium34. Interestingly, we did find both FKBP4 and HSP90AB1, not only in YS, but also in UF, even though their presence was not affected by pregnancy. While further confirmation of the role of specific proteins is required, the overview created in this study can be used as a basis for further targeted studies in the horse.
In addition to the role in prostaglandin and progesterone metabolism, involvement in proteolysis and lipid metabolism was also prominent in the commonly detected proteins in our study and the one of Swegen et al.34, also coinciding with previous findings on transcriptomics around MRP15. Several cathepsins (G, D, L and S) were detected in the UF with downregulation of cathepsin L (CTSL) and S during pregnancy. Pregnancy associated downregulation of CTSL1 was also found at the transcriptome level15. Considering lipid metabolism, we detected differential expression of lipocalin (P19), apolipoprotein A1 (APOA1) and apolipoprotein D (APOD). These proteins are important transporters of essential lipids to the developing conceptus. Retinol binding protein (RBP), which also belongs to the lipocalin family, and APOA1 have been detected in uterine fluid of pregnant and cyclic pigs, cattle and sheep23–25,27,28, with increasing amounts between day 10 and day 13 in both pregnant and cyclic pigs24. In the horse, lipocalin (P19), apolipoprotein A1 (APOA1) and apolipoprotein D (APOD) were all downregulated in the uterine fluid of the pregnant mares. Pregnancy associated upregulation of APOA1 was reported at the transcriptome level15 and presence of P19 and APOA1 in the yolk sac fluid illustrates their role in the embryo-maternal dialogue. Therefore, lower amounts in the uterine fluid during pregnancy rather indicate the transport and binding to the conceptus. Lipocalin P19 or uterocalin is a progesterone induced protein, which is abundantly present in the equine uterine secretions during dioestrus and early pregnancy51,52. While the early developing equine conceptus moves around the uterus, it entirely depends upon the uterine secretions for its nutrition and P19 can function as a carrier for essential lipids and amino acids53. Coinciding with our findings, P19 has been detected in the trophoblast and the yolk sac fluid of the equine embryo51,52 and it is one of the most abundant proteins in the embryonic capsule54–56. Therefore, the lower amount of P19 in P versus C is probably due to binding of substantial quantities to the embryo.
While a novel and informative overview is created, it has to be borne in mind that no statistically significant results were obtained at the level of molecular functions, biological processes and pathways. Differential expression of individual proteins was observed between the different UF conditions, and these proteins were categorized in GO terms, but statistical analysis showed no significant overrepresentation of any of the GO terms or KEGG pathways. Furthermore, it should be noted that MS intrinsically suffers from missing values and conclusions based on the absence of proteins cannot be made42,43. However, the field of proteomics has greatly evolved in recent years, providing the possibility for statistically robust quantitative comparison of individual protein levels in complex biological samples, like uterine fluid22. HDMSE specifically has been shown to provide good proteome coverage and reproducibility35. At the same time, however, analysis of GO terms and pathways for proteomics is still in its infancy57,58. As many of the here described bioinformatics approaches for proteomic analysis were originally developed for genomics, a similar but more matured field, their performance can be expected to show a similar growth as that of the genomic approaches. Moreover, the similarity between these fields potentially allows an integrated approach in which results from several omics studies can be combined.
In conclusion, proteins present in the equine uterine and embryonic yolk sac fluid around the signalling of MRP at day 13 were identified and quantified at large scale for the first time in the horse. We detected upregulation of several inhibitors of prostaglandin synthesis, including PTGR1, GSTP1 and ANXA1, in the uterine fluid of pregnant mares. Overall, an overview was created of the proteins playing a role at the embryo-maternal interface in the horse. This study provides a solid foundation for further targeted studies of proteins potentially involved in embryo-maternal interactions, maternal recognition of pregnancy and pregnancy loss in the horse.
Methods
Sampling
All animal handlings were approved by the Ethical Committee of the Faculty of Veterinary Medicine (EC2013/118) of Ghent University. All methods were performed in accordance with the relevant guidelines and regulations. A switch back design was followed with 5 mares undergoing two different types of cycles: a pregnant cycle (P) and a cyclic control cycle (C). In this way, the samples were paired using the same mare as its own control for pregnancy and the experimental unit was the mare. The order of P and C cycles was randomly altered for the different mares. No resting cycles were included. During the breeding season, five reproductively sound Warmblood mares between 4 and 13 years old were monitored by transrectal ultrasound. Reproductive soundness was confirmed by negative cytology and bacteriology. Mares displaying uterine oedema and a follicle exceeding 35 mm received 1500 IU hCG intravenously and were either inseminated the next day with fresh semen of the same stallion (P) or left unbred (C). Ovulation was evaluated twice daily by ultrasound. In both groups, sampling was performed 13 days after detection of ovulation. To recover undiluted uterine fluid in order to avoid negative effects of excessive Ringer’s salts on MS59, intra-uterine application of a tampon (OB Mini; Johnson & Johnson, Beerse, Belgium) was performed based upon the method described by Wolf et al.33. A double gloved technique was used to avoid vaginal contamination. The tampon was left in the uterus during 10 minutes and upon removal it was placed in a Falcon tube at 4 °C until further processing. Subsequently, the mare’s uterus was flushed with sterile Ringer’s solution by means of a modified endotracheal tube to recover the embryo (P).
To process the uterine fluid, 1 mL of sterile water (B60, Biosolve, Valkenswaard, The Netherlands) was infused on top of the tampon and the tampon was attached in the upper part of the Falcon tube by fixing the cord with the cap. Subsequently, the Falcon tube was centrifuged for 20 minutes at 1000 × g at 4 °C. The supernatant was collected and stored in a Protein LoBind Eppendorf tube (Eppendorf AG, Hamburg, Germany) at −80 °C. Meanwhile, the embryo was isolated in a petri dish and the yolk sac fluid was collected by aspiration with a 21 G needle and stored in a Protein LoBind Eppendorf at −80 °C.
A total of 15 samples were collected, consisting of uterine fluid (UF) (n = 10) from five biological replicates coinciding with the five mares (1–5) for the P and C treatment cycles, as well as yolk sac fluid (YS) (n = 5) from the P cycles.
Sample preparation for mass spectrometry analysis
After thawing, protein concentration in each sample was determined using the Coomassie (Bradford) Protein Assay Kit (Thermo Fisher Scientific, San José, CA, USA) according to the manufacturer’s instructions. Further processing was performed for 10 µg protein of each uterine fluid sample and for 500 ng protein of the yolk sac samples. Samples were dissolved in 20 μL 0.5 M triethylammonium bicarbonate (TEABC; Sigma-Aldrich, St. Louis, MO, USA). Two µl of reducing agent (10 µM DTT; Invitrogen, Merelbeke, Belgium) were added followed by incubation for 1 h at 60 °C. Subsequently, 1 µl of alkylizing agent (200 mM methyl methanethiosulfonate (MMTS) in isopropanol; Sigma-Aldrich, St. Louis, MO, USA) was added and samples were incubated for 10 min at room temperature. Digestion was performed overnight at 37 °C with trypsin lys C (1:20, trypsin:protein w/w, Promega, Leiden, The Netherlands) in TEABC buffer with 1 mM CACL(2) and 5% acetonitrile (Biosolve, Valkenswaard, The Netherlands). Samples were vacuum-dried and stored at −20 °C until analysis.
Data acquisition by HDMSE analysis
The peptides were separated using a nanoscale UPLC system (nanoAcquityUPLC, Waters, Milford, USA) coupled to a Synapt G2-Si mass spectrometer (Waters). Peptides were first trapped in 0.1% formic acid on a 180 µm × 20 mm C18 Trap column. Separation was performed on a HSS C18 1.8 m, 100 m × 250 mm analytical column at a flow rate of 300 nL/min and a temperature of 45 °C. As mobile phase A a 0.1% formic acid with 4% DMSO in water solution was used and 80% ACN containing 0.1% formic acid constituted mobile phase B. Peptides were separated for 60 min at 1–40% solvent B and for 1 min 40–85% solvent B. Seven minutes of rinsing (85% solvent B) re-equilibrated the column to the initial conditions. Eluted peptides were analysed in positive mode ESI-MS using High Definition MSE (HDMSE) with a collision energy look up table as described in22. The spectral acquisition time of low and elevated energy scans was 0.6 s over an m/z range of 50–2000. [Glu1]-Fibrinopeptide B was used for post-acquisition lock mass correction. All UF samples were analysed in the same run; three technical replicates (R1–R3) were run for each sample and four quality controls (QC) were included in which all samples were pooled.
Identification and quantification of peptides and proteins
All data were processed in Progenesis QIP (Progenesis QIP 2.0, Nonlinear Dynamics, Waters), including normalization and quality control. A database with UniProt IDs was created by conversion of Ensembl gene identifiers for Equus caballus (n = 22295) to Uniprot IDs using http://www.uniprot.org/uploadlists/ and including common contaminants (http://www.thegpm.org/crap/). As only secreted proteins are expected to be found, it can be argued that this database should be limited to only these secreted proteins. However, there is much debate on the accuracy of FDR calculations with such limited databases60–62 and as such a cautious approach was taken in which all proteins were assessed. Using Progenesis QIP, peptides were identified against this database with a FDR of 4%63 and allowing maximum one miscleavage. Protein quantification was based on the Hi-3 method64, which uses the average of the three most intense peptides of each protein for its quantification. Resulting normalized abundances for each protein, as well as unique peptide counts were further used for analysis of differential expression.
Analysis of differential expression
Analysis of differential expression was performed for the UF samples. Only normalized abundancies of proteins with at least two unique peptides (n = 707) were included in the analysis. Pairwise comparisons of differential expression were made for P versus C with the individual horses as a blocking factor, using R Bioconductor limma package65 and a FDR of 0.05.
Gene Ontology enrichment and pathway analysis
Gene Ontology (GO) terms (molecular functions, biological processes and cellular locations) were downloaded for each protein with the PANTHER Classification System66. For the pair-wise comparison of P and C, a statistical overrepresentation test against all quantified proteins was performed for all significantly up- and downregulated (FDR < 0.05) proteins. These tests were done for all primary GO classes: molecular functions, biological processes and cellular components. Pathways were analysed with Bioconductor’s67 GAGE package68. LogFC values of all quantified proteins were used as input against Equus caballus background reference pathways from KEGG.
Proteins involved in the embryo-maternal interaction were visualized using Cytoscape 3.3.0. To visualize embryo-maternal signalling, all GO terms and pathways that include ‘cytokine’ or ‘growth factor’ in their description were selected, together with all identified proteins in either the yolk sac or uterine fluid belonging to these GO terms or pathways. The connection between these GO terms, pathways and proteins was then visualized using Cytoscape 3.3.0. The same methodology was used to create a network based on GO terms and pathways including ‘embryo’, ‘maternal’ or ‘uterus’ in their description.
Data availability
All data are available in the Supplementary files.
Electronic supplementary material
Acknowledgements
The authors would like to thank Petra Van Damme and Isabel Lemahieu for their excellent technical assistance. Katrien Smits works as a postdoctoral researcher for the Research Foundation Flanders (grant FWO13/PDO/08). This research was funded by the Research Foundation Flanders (FWO project GO35511N) and by Ghent University (GOA project 01G01112).
Author Contributions
K.S. performed the experiments and wrote the manuscript. S.W. provided the bioinformatics analysis. K.V.S. was responsible for the mass spectrometry. M.V.D.V., V.D.L., C.V., K.R. and J.G. contributed to the sampling of the mares. F.V.N., L.P., D.D. and A.V.S. participated in the study design. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23537-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Short R. Implantation and the maternal recognition of pregnancy. Foetal Autonomy. 1969;2:31. [Google Scholar]
- 2.Kindahl H, Knudsen O, Madej A, Edqvist LE. Progesterone, prostaglandin F-2 alpha, PMSG and oestrone sulphate during early pregnancy in the mare. J Reprod Fertil Suppl. 1982;32:353–359. [PubMed] [Google Scholar]
- 3.Bazer FW, Thatcher W. Theory of maternal recognition of pregnancy in swine based on estrogen controlled endocrine versus exocrine secretion of prostaglandin F 2α by the uterine endometrium. Prostaglandins. 1977;14:397–401. doi: 10.1016/0090-6980(77)90185-X. [DOI] [PubMed] [Google Scholar]
- 4.Lamming GE, et al. Local action of trophoblast interferons in suppression of the development of oxytocin and oestradiol receptors in ovine endometrium. J Reprod Fertil. 1995;105:165–175. doi: 10.1530/jrf.0.1050165. [DOI] [PubMed] [Google Scholar]
- 5.Lamming GE, Mann GE. Control of endometrial oxytocin receptors and prostaglandin F2 alpha production in cows by progesterone and oestradiol. J Reprod Fertil. 1995;103:69–73. doi: 10.1530/jrf.0.1030069. [DOI] [PubMed] [Google Scholar]
- 6.Allen WR. Fetomaternal interactions and influences during equine pregnancy. Reproduction. 2001;121:513–527. doi: 10.1530/rep.0.1210513. [DOI] [PubMed] [Google Scholar]
- 7.Klein C, Troedsson MH. Maternal recognition of pregnancy in the horse: a mystery still to be solved. Reprod Fertil Dev. 2011;23:952–963. doi: 10.1071/RD10294. [DOI] [PubMed] [Google Scholar]
- 8.Vanderwall DK, Silvia WJ, Fitzgerald BP. Concentrations of oxytocin in the intercavernous sinus of mares during luteolysis: temporal relationship with concentrations of 13, 14-dihydro-15-keto-prostaglandin F2 alpha. J Reprod Fertil. 1998;112:337–346. doi: 10.1530/jrf.0.1120337. [DOI] [PubMed] [Google Scholar]
- 9.Goff AK, Pontbriand D, Sirois J. Oxytocin stimulation of plasma 15-keto-13, 14-dihydro prostaglandin F-2 alpha during the oestrous cycle and early pregnancy in the mare. J Reprod Fertil Suppl. 1987;35:253–260. [PubMed] [Google Scholar]
- 10.Starbuck GR, Stout TA, Lamming GE, Allen WR, Flint AP. Endometrial oxytocin receptor and uterine prostaglandin secretion in mares during the oestrous cycle and early pregnancy. J Reprod Fertil. 1998;113:173–179. doi: 10.1530/jrf.0.1130173. [DOI] [PubMed] [Google Scholar]
- 11.Boerboom D, et al. Expression of key prostaglandin synthases in equine endometrium during late diestrus and early pregnancy. Biol Reprod. 2004;70:391–399. doi: 10.1095/biolreprod.103.020800. [DOI] [PubMed] [Google Scholar]
- 12.Ealy AD, Eroh ML, Sharp DC., III. Prostaglandin H synthase Type 2 is differentially expressed in endometrium based on pregnancy status in pony mares and responds to oxytocin and conceptus secretions in explant culture. Anim Reprod Sci. 2010;117:99–105. doi: 10.1016/j.anireprosci.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 13.de Ruijter-Villani M, van Tol HT, Stout TA. Effect of pregnancy on endometrial expression of luteolytic pathway components in the mare. Reprod Fertil Dev. 2015;27:834–845. doi: 10.1071/RD13381. [DOI] [PubMed] [Google Scholar]
- 14.Wilsher S, Clutton-Brock A, Allen WR. Successful transfer of day 10 horse embryos: influence of donor-recipient asynchrony on embryo development. Reproduction. 2010;139:575–585. doi: 10.1530/REP-09-0306. [DOI] [PubMed] [Google Scholar]
- 15.Klein C, Scoggin KE, Ealy AD, Troedsson MH. Transcriptional profiling of equine endometrium during the time of maternal recognition of pregnancy. Biol Reprod. 2010;83:102–113. doi: 10.1095/biolreprod.109.081612. [DOI] [PubMed] [Google Scholar]
- 16.Klein C, Troedsson MH. Transcriptional profiling of equine conceptuses reveals new aspects of embryo-maternal communication in the horse. Biol Reprod. 2011;84:872–885. doi: 10.1095/biolreprod.110.088732. [DOI] [PubMed] [Google Scholar]
- 17.Merkl M, et al. Microarray analysis of equine endometrium at days 8 and 12 of pregnancy. Biol Reprod. 2010;83:874–886. doi: 10.1095/biolreprod.110.085233. [DOI] [PubMed] [Google Scholar]
- 18.Bauersachs S, Wolf E. Transcriptome analyses of bovine, porcine and equine endometrium during the pre-implantation phase. Anim Reprod Sci. 2012;134:84–94. doi: 10.1016/j.anireprosci.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Klein C. Novel equine conceptus?endometrial interactions on Day 16 of pregnancy based on RNA sequencing. Reprod Fertil Dev. 2015 doi: 10.1071/RD14489. [DOI] [PubMed] [Google Scholar]
- 20.Wright PC, Noirel J, Ow SY, Fazeli A. A review of current proteomics technologies with a survey on their widespread use in reproductive biology investigations. Theriogenology. 2012;77:738–765 e752. doi: 10.1016/j.theriogenology.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature reviews. Genetics. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Distler U, Kuharev J, Tenzer S. Biomedical applications of ion mobility-enhanced data-independent acquisition-based label-free quantitative proteomics. Expert review of proteomics. 2014;11:675–684. doi: 10.1586/14789450.2014.971114. [DOI] [PubMed] [Google Scholar]
- 23.Kayser JP, Kim JG, Cerny RL, Vallet JL. Global characterization of porcine intrauterine proteins during early pregnancy. Reproduction. 2006;131:379–388. doi: 10.1530/rep.1.00882. [DOI] [PubMed] [Google Scholar]
- 24.Jalali BM, Bogacki M, Dietrich M, Likszo P, Wasielak M. Proteomic analysis of porcine endometrial tissue during peri-implantation period reveals altered protein abundance. Journal of proteomics. 2015;125:76–88. doi: 10.1016/j.jprot.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Brooks K, Burns GW, Moraes JG, Spencer TE. Analysis of the Uterine Epithelial and Conceptus Transcriptome and Luminal Fluid Proteome During the Peri-Implantation Period of Pregnancy in Sheep. Biol Reprod. 2016;95(88):1–17. doi: 10.1095/biolreprod.116.141945. [DOI] [PubMed] [Google Scholar]
- 26.Munoz M, et al. Proteome of the early embryo-maternal dialogue in the cattle uterus. J Proteome Res. 2012;11:751–766. doi: 10.1021/pr200969a. [DOI] [PubMed] [Google Scholar]
- 27.Forde N, Bazer FW, Spencer TE, Lonergan P. ‘Conceptualizing’ the Endometrium: Identification of Conceptus-Derived Proteins During Early Pregnancy in Cattle. Biol Reprod. 2015;92(156):1–13. doi: 10.1095/biolreprod.115.129296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forde, N. et al. Proteomic analysis of uterine fluid during the pre-implantation period of pregnancy in cattle. Reproduction, 10.1530/REP-13-0010(2014). [DOI] [PubMed]
- 29.Tachibana Y, et al. Expression of endometrial immune-related genes possibly functioning during early pregnancy in the mare. J Reprod Dev. 2013;59:85–91. doi: 10.1262/jrd.2012-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein C, Scoggin KE, Troedsson MH. The expression of interferon-stimulated gene 15 in equine endometrium. Reprod Domest Anim. 2011;46:692–698. doi: 10.1111/j.1439-0531.2010.01731.x. [DOI] [PubMed] [Google Scholar]
- 31.Hartt LS, et al. Temporal and spatial associations of oestrogen receptor alpha and progesterone receptor in the endometrium of cyclic and early pregnant mares. Reproduction. 2005;130:241–250. doi: 10.1530/rep.1.00596. [DOI] [PubMed] [Google Scholar]
- 32.Watson, E. D., Buckingham, J., Bjorksten, T. & Nikolakopoulos, E. Immunolocalization of oxytocin and neurophysin in the mare uterus. J Reprod Fertil Suppl 289–296 (2000). [PubMed]
- 33.Wolf CA, Maslchitzky E, Gregory RM, Jobim MI, Mattos RC. Effect of corticotherapy on proteomics of endometrial fluid from mares susceptible to persistent postbreeding endometritis. Theriogenology. 2012;77:1351–1359. doi: 10.1016/j.theriogenology.2011.10.042. [DOI] [PubMed] [Google Scholar]
- 34.Swegen, A. et al. From Peptide Masses to Pregnancy Maintenance: A Comprehensive Proteomic Analysis of The Early Equine Embryo Secretome, Blastocoel Fluid, and Capsule. Proteomics17, 10.1002/pmic.201600433 (2017). [DOI] [PubMed]
- 35.Distler U, et al. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nature methods. 2014;11:167–170. doi: 10.1038/nmeth.2767. [DOI] [PubMed] [Google Scholar]
- 36.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annual review of pharmacology and toxicology. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 37.Luo L, et al. Recombinant protein glutathione S-transferases P1 attenuates inflammation in mice. Molecular immunology. 2009;46:848–857. doi: 10.1016/j.molimm.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Wallner BP, et al. Cloning and expression of human lipocortin, a phospholipase A2 inhibitor with potential anti-inflammatory activity. Nature. 1986;320:77–81. doi: 10.1038/320077a0. [DOI] [PubMed] [Google Scholar]
- 39.Huang KS, et al. Two human 35 kd inhibitors of phospholipase A2 are related to substrates of pp60v-src and of the epidermal growth factor receptor/kinase. Cell. 1986;46:191–199. doi: 10.1016/0092-8674(86)90736-1. [DOI] [PubMed] [Google Scholar]
- 40.Smits K, et al. The Equine Embryo Influences Immune-Related Gene Expression in the Oviduct. Biol Reprod. 2016;94:36. doi: 10.1095/biolreprod.115.136432. [DOI] [PubMed] [Google Scholar]
- 41.Ababneh MM, Troedsson MH. Endometrial phospholipase A2 activity during the oestrous cycle and early pregnancy in mares. Reprod Domest Anim. 2013;48:46–52. doi: 10.1111/j.1439-0531.2012.02023.x. [DOI] [PubMed] [Google Scholar]
- 42.Albrecht D, Kniemeyer O, Brakhage AA, Guthke R. Missing values in gel-based proteomics. Proteomics. 2010;10:1202–1211. doi: 10.1002/pmic.200800576. [DOI] [PubMed] [Google Scholar]
- 43.Webb-Robertson BJ, et al. Review, evaluation, and discussion of the challenges of missing value imputation for mass spectrometry-based label-free global proteomics. J Proteome Res. 2015;14:1993–2001. doi: 10.1021/pr501138h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park ES, et al. Phospholipase C-delta1 and oxytocin receptor signalling: evidence of its role as an effector. Biochem J. 1998;331(Pt 1):283–289. doi: 10.1042/bj3310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bu H, et al. ERBP, a novel estrogen receptor binding protein enhancing the activity of estrogen receptor. Biochem Biophys Res Commun. 2004;317:54–59. doi: 10.1016/j.bbrc.2004.02.179. [DOI] [PubMed] [Google Scholar]
- 46.Montt-Guevara MM, et al. Androgens Regulate T47D Cells Motility and Invasion through Actin Cytoskeleton Remodeling. Frontiers in endocrinology. 2016;7:136. doi: 10.3389/fendo.2016.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song J, et al. Expression and clinicopathological significance of oestrogen-responsive ezrin-radixin-moesin-binding phosphoprotein 50 in breast cancer. Histopathology. 2007;51:40–53. doi: 10.1111/j.1365-2559.2007.02730.x. [DOI] [PubMed] [Google Scholar]
- 48.Tranguch S, Smith DF, Dey SK. Progesterone receptor requires a co-chaperone for signalling in uterine biology and implantation. Reprod Biomed Online. 2006;13:651–660. doi: 10.1016/S1472-6483(10)60655-4. [DOI] [PubMed] [Google Scholar]
- 49.Tranguch S, et al. Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14326–14331. doi: 10.1073/pnas.0505775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen HY, et al. Expression of FK506-binding protein 52 (FKBP52) in chorionic villi with early recurrent spontaneous abortion. J Matern Fetal Neonatal Med. 2015;28:1165–1169. doi: 10.3109/14767058.2014.947572. [DOI] [PubMed] [Google Scholar]
- 51.Crossett B, Allen WR, Stewart F. A 19 kDa protein secreted by the endometrium of the mare is a novel member of the lipocalin family. Biochem J. 1996;320(Pt 1):137–143. doi: 10.1042/bj3200137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ellenberger, C. et al. Immunolocalisation of the uterine secretory proteins uterocalin, uteroferrin and uteroglobin in the mare’s uterus and placenta throughout pregnancy. Theriogenology70, 746–757, 10.1016/j.theriogenology.2008.04.050 S0093-691X(08)00290-2 [pii] (2008). [DOI] [PubMed]
- 53.Suire S, Stewart F, Beauchamp J, Kennedy MW. Uterocalin, a lipocalin provisioning the preattachment equine conceptus: fatty acid and retinol binding properties, and structural characterization. Biochem J. 2001;356:369–376. doi: 10.1042/bj3560369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinn BA, Hayes MA, Waelchli RO, Kennedy MW, Betteridge KJ. Changes in major proteins in the embryonic capsule during immobilization (fixation) of the conceptus in the third week of pregnancy in the mare. Reproduction. 2007;134:161–170. doi: 10.1530/REP-06-0241. [DOI] [PubMed] [Google Scholar]
- 55.Stewart F, Charleston B, Crossett B, Barker PJ, Allen WR. A novel uterine protein that associates with the embryonic capsule in equids. J Reprod Fertil. 1995;105:65–70. doi: 10.1530/jrf.0.1050065. [DOI] [PubMed] [Google Scholar]
- 56.Smits, K. et al. Influence of the uterine environment on the development of in vitro-produced equine embryos. Reproduction143, 173–181, 10.1530/REP-11-0217 REP-11-0217 [pii] (2012). [DOI] [PubMed]
- 57.Schmidt A, Forne I, Imhof A. Bioinformatic analysis of proteomics data. BMC systems biology. 2014;8(Suppl 2):S3. doi: 10.1186/1752-0509-8-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaudel M, Sickmann A, Martens L. Introduction to opportunities and pitfalls in functional mass spectrometry based proteomics. Biochim Biophys Acta. 2014;1844:12–20. doi: 10.1016/j.bbapap.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 59.Piwowar AM, Lockyer NP, Vickerman JC. Salt effects on ion formation in desorption mass spectrometry: an investigation into the role of alkali chlorides on peak suppression in time-of-flight-secondary ion mass spectrometry. Anal Chem. 2009;81:1040–1048. doi: 10.1021/ac8020888. [DOI] [PubMed] [Google Scholar]
- 60.Noble WS. Mass spectrometrists should search only for peptides they care about. Nature methods. 2015;12:605–608. doi: 10.1038/nmeth.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noble WS, Keich U. Mass spectrometrists should search for all peptides, but assess only the ones they care about Reply. Nature methods. 2017;14:644–644. doi: 10.1038/nmeth.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sticker A, Martens L, Clement L. Mass spectrometrists should search for all peptides, but assess only the ones they care about. Nature methods. 2017;14:643–644. doi: 10.1038/nmeth.4338. [DOI] [PubMed] [Google Scholar]
- 63.Li GZ, et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 64.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nature protocols. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nature methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the Supplementary files.