ABSTRACT
Shiga toxin-producing Escherichia coli (STEC) is a foodborne pathogen, and serotype O157:H7 is typically associated with severe disease. Australia is unique in its STEC epidemiology, as severe cases are typically associated with non-O157 serogroups, and locally acquired O157 isolates are H-negative/nonmotile. The H-negative phenotype and reduced severity of disease compared to that associated with H7/motile strains are distinct features of Australian O157 strains, but the molecular mechanism behind this phenotype has not been reported. Accurate characterization of the H-negative phenotype is important in epidemiological surveillance of STEC. Serotyping is moving away from phenotype-based methods, as next generation sequencing allows rapid extrapolation of serotype through in silico detection of the O-antigen processing genes, wzx, wzy, wzm, and wzt, and the H-antigen gene, fliC. The detection and genotyping of fliC alone is unable to determine the motility of the strain. Typically, most Australian O157:H-negative strains carry an H7 genotype yet phenotypically are nonmotile; thus, many are mischaracterized as H7 strains by in silico serotyping tools. Comparative genomic analysis of flagellar genes between Australian and international isolates was performed and an insertion at nucleotide (nt) 125 in the flgF gene was identified in H-negative isolates. Chi-square results showed that this insertion was significantly associated with the H-negative phenotype (P < 0.0001). Phylogenetic analysis was also completed and showed that the Australian H-negative isolates with the insertion in flgF represent a clade within the O157 serogroup, distinct from O157:H7 serotypes. This study provides a genetic target for inferring the nonmotile phenotype of Australian O157 STEC, which increases the predictive value of in silico serotyping.
KEYWORDS: O157, STEC, epidemiology, whole-genome sequencing
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) is a foodborne pathogen affecting all six populated continents and is a serious public health concern (1). Infections can range from asymptomatic to causing life-threatening disease in humans, where children, the elderly, and those with otherwise compromised immune systems are at severe risk of disease. These strains of E. coli are distinct from commensal strains that inhabit the human gut due to the presence and expression of Shiga toxin (stx) genes and other potential virulence factors, which can be responsible for complications such as gastroenteritis, hemorrhagic colitis, and hemolytic uremic syndrome (HUS) (2). STEC-related HUS is a life-threatening condition that can affect adults and the elderly but is most common in children. In developed countries, STEC-related HUS is the leading cause of acute kidney failure in otherwise healthy children, and long-term complications commonly occur such as chronic renal insufficiency, neurological disorders, and cardiovascular diseases (3).
The O157:H7 serotype of STEC is typically associated with severe disease, and is responsible for the majority of outbreaks in the United States, Japan, and the United Kingdom (4). Australia is unique in its STEC epidemiology, however, as severe cases of STEC infection are typically associated with non-O157 serogroups, such as O111, and locally acquired O157 isolates are H-negative/nonmotile (5–7). Traditional methods of serotyping involve identifying the immunogenic structures, the lipopolysaccharide (O) antigen, and the flagellar (H) antigen phenotypically through antisera. With the introduction of next-generation sequencing technology, many laboratories are moving away from phenotype-based serotyping to extrapolation of serotype through in silico detection of O-antigen-processing and H-antigen-related genes (8). Detection and genotyping of the O-antigen-processing genes, wzx, wzy, wzm, and wzt, determines the serogroup of an isolate, while detection and genotyping of the fliC (flagellin) gene determines the H type (9). Currently, 53 different H types have been recognized in E. coli, and this is explained through the genotypic variation in the fliC gene (10); however, as ∼60 genes are involved in the expression of the flagellum in E. coli and other related species, detection and genotyping of the fliC gene alone cannot predict the motility of the strain (11). Therefore, many Australian O157:H-negative strains are mischaracterised through in silico serotyping as O157:H7, since they carry an intact fliC gene, yet are phenotypically nonmotile. The genetic mechanism(s) for the nonmotile phenotype have not been reported to date.
Epidemiological surveillance requires accurate molecular characterization of pathogens to effectively monitor trends in infections and to identify and track sources of outbreaks, so as to enable rapid public health response. Mischaracterization of O157:H-negative STEC strains as H7-positive will negatively impact surveillance of STEC, as the H-negative, nonmotile phenotype is a distinct feature of Australian O157 strains, which is also associated with reduced severity of disease compared to that associated with O157:H7 strains (6). To determine the genetic mechanism(s) for the H-negative phenotype in Australian O157 STEC strains, genomic comparisons between Australian and international O157 strains were performed. This study provides a potential genetic basis for the nonmotile phenotype of Australian O157 STEC to assist in the interpretation of in silico serotyping.
MATERIALS AND METHODS
Isolate selection.
All O157 isolates identified as stx1- or stx2-positive during routine clinical screening of referred feces by enrichment cultures and PCR from between 2007 and 2016 at the Queensland Health Forensic and Scientific Services (QHFSS) Public Health Microbiology reference laboratory were serotyped. Serotyping was performed by the Microbiological Diagnostic Unit (Melbourne, Australia). Patient travel history was noted if recorded. The data represent a total of 63 isolates, of which 13 isolates were initially chosen for whole-genome sequencing to search for motility-related mutations (see Table S1 in the supplemental material), and 50 further O157 isolates were examined for the specific flgF mutation by Sanger sequencing (see Table S2 in the supplemental material). These isolates represent all O157:H7 and H-negative STEC strains isolated from between 2007 and 2016 in Queensland, although only 2 representative isolates were included from a large previously reported outbreak (7). International STEC genomes were obtained from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/GenBank/) (see Table S3 in the supplemental material).
Whole-genome sequencing.
DNA extraction was performed on the QIAsymphony SP, using the DSP DNA minikit (Qiagen, Germany), following the Tissue HC 200 V DSP protocol. Extracted DNA was quantitated on a plate reader using the Quant-IT kit (Thermo Fisher Scientific).
Library preparation of DNA samples was performed by using the Nextera XT library preparation kit (Illumina). After library preparation, sequencing was performed using the NextSeq 500 Mid Output V2 kit (Illumina) on the NextSeq 500 sequencer (Illumina, CA, USA).
Sequence reads for the isolates were trimmed with Trimmomatic v0.36 and quality checked by FastQC v0.11.5 (12, 13). De novo assemblies were generated with the SPAdes assembler, and assemblies were annotated using the RAST (Rapid Annotations using Subsystems Technology) server (14, 15).
Comparative analysis of the flagellar genes was done to determine whether consistent differences in sequence between the O157 motile and nonmotile genomes exist within the flagellar genes, operon sequences, and promoter regions. Using Geneious R7 (Biomatters, New Zealand) (16) and CLC Genomics Workbench 7 (Qiagen, Germany), reference genes from E. coli strains EDL933 and Sakai (obtained from NCBI) and promoter sequences from E. coli strain K-12 (obtained from RegulonDB; http://regulondb.ccg.unam.mx/) (17) and cross-referenced in BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) (18) against the EDL933/Sakai genomes were aligned manually to the sequenced genomes.
In silico serotyping.
Genome assemblies from the 13 whole-genome sequenced isolates were submitted to SerotypeFinder (https://cge.cbs.dtu.dk/services/SerotypeFinder/) in FASTA file format, using a threshold of 85% identity (ID) and a minimum length of 60% (9).
Sanger Sequencing.
A total of 50 O157 STEC isolates were sequenced from DNA prepared as crude Tris-EDTA buffer boils. The primers used for amplifying (forward primer, 5′-ACAACGAAGGTCTGGCATCC-3′; reverse primer, 5′-CCGTGGCTTTCACCAGTTTC-3′) and sequencing (forward primer, 5′-GACCAGATCCTCAACACGCT-3′; reverse primer, 5′-CCTGAATGCTGCCATTACGC-3′) the flgF gene region were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (19) and quality checked with NetPrimer (Premier Biosoft, Palo Alto, CA). Amplification primers were used to PCR amplify the flgF gene region, following the AmpliTaq Gold Hot Start PCR protocol (Applied Biosystems). PCR products were treated with Antarctic phosphatase (New England BioLabs, MA) and exonuclease I (Thermo Fisher Scientific, MA, USA), and then sequenced with the ABI 3130 genetic analyzer (Applied Biosystems, CA).
The consensus sequences were obtained by aligning both forward- and reverse-sequenced strands using ChromasPro software (Technelysium, Australia) and further analyzed using Geneious R7.
Phylogenomic analysis.
Core single nucleotide polymorphism (SNP) analysis was performed using Snippy (v3.0) (https://github.com/tseemann/snippy), with the Sakai genome as a reference. Snippy results for the analyzed genomes were aligned as core SNP alignments and used to generate a phylogenomic tree using Geneious R7. The phylogenomic tree was constructed with RAxML, based on 36 genomes obtained from NCBI GenBank (n = 23) and sequenced in this study (n = 13), which included 35 STEC O157:H7 and H-negative strains and an outgroup O55:H7 strain. Interactive tree of life (iTOL) v4.0.2 was used for visualization of the phylogenomic tree (https://itol.embl.de/) (20).
Statistics.
Chi-square tests were used to determine if genetic differences identified between O157:H-negative and H7 isolates were significant. A P value of < 0.05 was considered to represent a significant difference between the two groups.
Accession number(s).
The whole-genome sequence (WGS) data from the 13 isolates sequenced in this study have been deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under the BioProject accession number PRJEB23370.
RESULTS
Identification of flagellar gene sequence differences in Australian O157:H-negative isolates by comparative genomics.
During the period from 2007 to 2016, there were a total of 213 STEC isolates grown from all STEC cases notified by the Queensland Public Health Microbiology reference laboratory. Of these, 91 were serogroup O157 (43%), and 85 were phenotypically serotyped as O157:H-negative (93%). Thirteen of these O157 isolates (one H7 strain, M74957, and 12 H-negative strains, M7371, M76137, M75212, M78680, M79992, M76796, M75061, M74378, M71373, M78862, M76948, and M77548) were whole-genome sequenced and analyzed for genetic differences that could explain the H-negative phenotype. SerotypeFinder predicted the serotype for all 13 sequenced isolates as O157:H7, despite 12/13 isolates phenotypically serotyping as O157:H-negative.
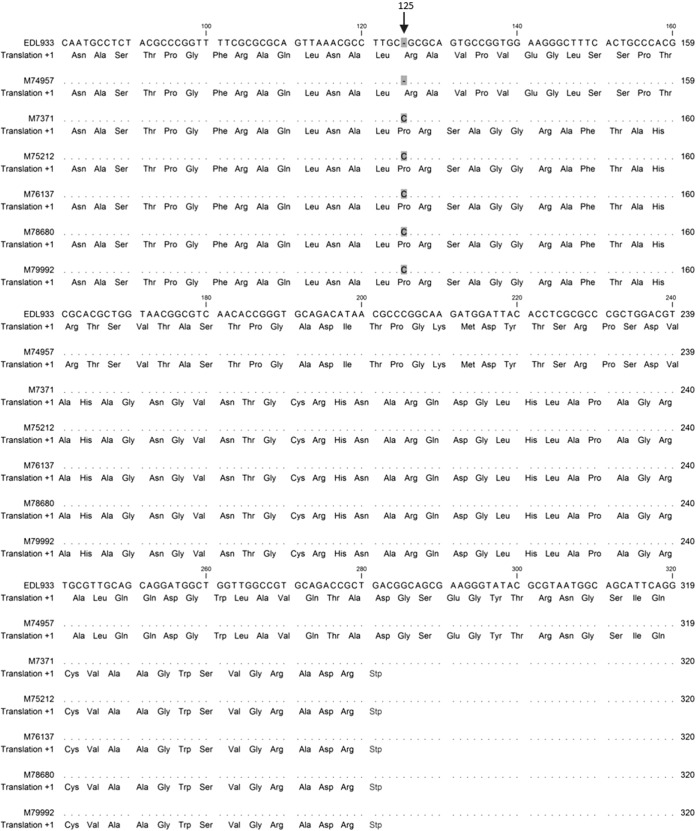
After analyzing ∼100 genes related to both the structure and regulation of the flagellum, the most consistent sequence difference between the reference genomes (EDL933 and Sakai) and the 13 sequenced genomes was identified in the flgF gene, with 10 of the 12 sequenced O157:H-negative genomes containing a single cytosine base insertion at nt 125, inducing a frameshift mutation resulting in an early stop codon at nt 280 to 282 (Fig. 1). Other mutations in these genomes were also noted; however, these were silent and missense mutations, which are less likely to significantly disrupt the expression or function of any flagellum-related genes. The insertion in flgF was absent in 2 of the 10 H-negative genomes, strain M76948, and M77548. In isolate M76948, an isolate from a patient reporting overseas travel, a single base deletion was found in the fliF gene at nt 35 that may be responsible for its nonmotile phenotype, inducing a frameshift mutation resulting in an early stop codon at nt 74 to 76. In isolate M77548, no significant mutations were found (i.e., frameshift or nonsense mutations) that could explain its nonmotile phenotype. As the insertion in the flgF gene was present in the majority of the Australian O157:H-negative genomes examined, further sequence analysis was performed to determine whether this insertion was significantly associated with Australian O157:H-negative isolates.
FIG 1.
Multiple sequence alignments of the flgF gene from H-negative Australian isolates (strains M7371, M75212, M76137, M78680, and M79992), a sequenced H7 isolate (M74957), and the H7 reference genome (EDL933). The insertion of cytosine at nt 125 and the resulting stop codon at nt 280 to 282 distinguish the H-negative isolate from the H7 isolates.
Sequencing analysis results.
To determine the association of the insertion in the flgF gene with the O157:H-negative phenotype in Australian isolates, the flgF gene region was sequenced in 50 additional locally acquired O157 STEC isolates. In total, 63 Australian isolates were analyzed (five H7 and 58 H-negative); of the 58 H-negative isolates, 55 harbored the single cytosine base insertion at nt 125, while none of the 5 H7 motile isolates contained the insertion. Only three H-negative isolates did not harbor the single cytosine base insertion at nt 125 in the flgF gene, of which two were whole-genome sequenced and discussed above (strains M76948 and M77548). As the third isolate had not been whole-genome sequenced, it was not examined further in this study. Chi-square analysis on all 63 isolates demonstrated the flgF insertion to be significantly associated with the H-negative phenotype (P < 0.0001) (Table 1).
TABLE 1.
Chi-square test results for the association of the insertion in the flgF gene between H-negative and H7 isolates
| Type of cell | Insertion present |
Insertion absent |
χ2 | P value | ||
|---|---|---|---|---|---|---|
| Total no. of cells observed (expected) | χ2 for each cell | Total no. of cells observed (expected) | χ2 for each cell | |||
| Motile | 0 (4.37) | 4.37 | 5 (0.63) | 30.01 | 37.34 | P < 0.0001 |
| Nonmotile | 55 (50.63) | 0.38 | 3 (7.37) | 2.59 | ||
Phylogenomic analysis.
The genetic relationship between publicly available international O157 genomes, the Australian H-negative isolates lacking the flgF insertion, and the Australian H-negative isolates containing the flgF insertion was investigated by SNP typing (Fig. 2). A total of 36 genomes were used in the analysis. This comprised the 13 whole-genome-sequenced Australian isolates, including one H7 isolate (M74957), 10 H-negative isolates with the insertion in flgF (strains M7371, M76137, M75212, M78680, M79992, M76796, M75061, M74378, M71373, and M78862), and two H-negative isolates that did not contain the insertion in flgF (M76948 and M77548). A total of 22 genomes for H7 isolates were obtained from NCBI GenBank, all of which did not contain the flgF mutation. An O55:H7 serotype, strain CB9615, was included as an outgroup to root the phylogenomic tree, as the O157:H7 serotype likely emerged from an O55:H7 ancestor (21).
FIG 2.

Phylogenomic tree of serogroup O157 E. coli genomes based on SNPs identified by mapping to the Sakai genome as a reference. The tree was rooted using serotype O55:H7 E. coli strain CB9615. Isolates highlighted in green are H-negative with the insertion in flgF; isolates highlighted in yellow are H-negative without the insertion in flgF.
The phylogenomic tree shows that the H-negative isolates with the insertion in flgF formed a distinct clade separate from both the H7 isolates and the two H-negative isolates without the insertion. These two H-negative isolates, M76948 and M77548, appear genetically distant from the H-negative isolates with the flgF insertion. Overseas travel history was noted for sources of isolate M76948, whereas those of isolate M77548 had no travel history recorded. The H7 isolate, M74957, clustered with other H7 genomes.
DISCUSSION
The nonmotile/H-negative phenotype is an important distinction for Australian isolates of O157 STEC, which are misreported through in silico serotyping as H7/motile, as these isolates typically contain a functional H7 fliC gene. To identify a potential genetic cause for the nonmotile phenotype, genomic comparisons between Australian and international O157 isolates were performed to examine the flagellum-related genetic regions of these strains. The isolates examined in this study were sourced from the Queensland State Reference Laboratory so the majority of cases are spatially associated with Queensland, Australia in particular; however, some patients were noted as originating from other Australian states or reported domestic travel during the incubation period. The O157:H-negative phenotype has previously been reported in other Australian states (6, 7, 22), and therefore the O157:H-negative clone characterized in this study appears to be nationally distributed.
The bacterial flagellum is a highly heterogeneous protein structure that can be divided into three parts—the basal body, the hook, and the filament—which are all connected by a rod structure (23). It is formed through a complex regulatory system that allows for sequential and orderly formation of these three parts. Mutations in any one of the genes involved in the structure or regulation can have a potentially deleterious effect on the formation and/or function of the flagellum. A motile phenotype is therefore dependent on multiple genes.
The flgF gene codes for one of four structural proteins that comprise the connecting rod structure, along with flgB, flgC, and flgG. The flgF gene is 756 nt in length and is a part of the large flgBCDEFGHIJ operon, where the early stop codon at nt 280 to 282 resulting from the frameshift mutation in the H-negative isolates may affect the translation of downstream genes (24). These downstream genes are involved in the assembly of the rod (flgG and flgJ) and its supporting ring structures (flgH and flgI), whereas lack of expression of these genes can disrupt the assembly of the rod and furthermore, can prevent the initiation of hook assembly (25). It has been shown that mutants deficient in flagellar rod and ring assembly exhibit reduced levels of FlgE, the primary protein component of the hook (26). The frameshift mutation in the flgF gene identified in the O157:H-negative isolates may be the causative mutation responsible for the H-negative/nonmotile phenotype through disruption of rod assembly and inhibition of hook assembly; however, further study is required to confirm this.
Whether the insertion in the flgF gene is the causative mutation for the nonmotile phenotype is yet to be determined, but this insertion is significantly associated with locally acquired Australian O157:H-negative isolates. The phylogenomic analysis demonstrated that the Australian H-negative isolates with the insertion in the flgF gene represent a clade within the O157 serogroup that is separate from the O157:H7 serotypes.
With the current transition to whole-genome sequencing for in silico serotyping of E. coli isolates, it is important that this change of methods does not compromise STEC epidemiological data. We have shown that 93% of STEC O157 isolates collected between 2007 and 2016 in Queensland, Australia are H-negative and would be misreported as O157:H7 by in silico serotyping, masking the unique epidemiology of Australian STEC cases. Furthermore, with cases of O157:H-negative infections being reported outside Australia (27–29), the transition to in silico serotyping may fail to identify such cases or to detect emerging O157:H-negative epidemiology in other geographical regions. This highlights the importance of validating in silico serotyping bioinformatics approaches against spatially diverse strains to ensure that accuracy and sensitivity is achieved and that genetic targets are appropriate. At times, customized genomics solutions rather than broad genomics tools may be necessary to accommodate the unique geographical epidemiology of strains.
We describe an insertion in the flgF gene that can serve as a useful marker to predict the Australia-associated O157:H-negative phenotype, improving the concordance of in silico serotyping with phenotypic motility and thereby ensuring that epidemiological surveillance is not compromised.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Microbiological Diagnostic Unit, University of Melbourne for isolate serotyping. We thank all past and present members of the Molecular Epidemiology and Reference Microbiology teams, Public Health Microbiology, Forensic and Scientific services for contributing to STEC testing, isolation, and genetic characterization.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01906-17.
REFERENCES
- 1.Majowicz SE, Scallan E, Jones-Bitton A, Sargeant JM, Stapleton J, Angulo FJ, Yeung DH, Kirk MD. 2014. Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog Dis 11:447–455. doi: 10.1089/fpd.2013.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paton JC, Paton AW. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev 11:450–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fakhouri F, Zuber J, Frémeaux-Bacchi V, Loirat C. 2017. Haemolytic uraemic syndrome. Lancet 390:681–696. doi: 10.1016/S0140-6736(17)30062-4. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Nietfeldt J, Ju J, Wise J, Fegan N, Desmarchelier P, Benson AK. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, β-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J Bacteriol 183:6885–6897. doi: 10.1128/JB.183.23.6885-6897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McPherson M, Lalor K, Combs B, Raupach J, Stafford R, Kirk MD. 2009. Serogroup-specific risk factors for Shiga toxin-producing Escherichia coli infection in Australia. Clin Infect Dis 49:249–256. doi: 10.1086/599370. [DOI] [PubMed] [Google Scholar]
- 6.Mellor GE, Sim EM, Barlow RS, D'Astek BA, Galli L, Chinen I, Rivas M, Gobius KS. 2012. Phylogenetically related Argentinean and Australian Escherichia coli O157 isolates are distinguished by virulence clades and alternative Shiga toxin 1 and 2 prophages. Appl Environ Microbiol 78:4724–4731. doi: 10.1128/AEM.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasant BR, Stafford RJ, Jennison AV, Bennett SM, Bell RJ, Doyle CJ, Young JR, Vlack SA, Titmus P, El Saadi D, Jarvinen KAJ, Coward P, Barrett J, Staples M, Graham RMA, Smith HV, Lambert SB. 2017. Mild illness during outbreak of Shiga toxin-producing Escherichia coli O157 infections associated with agricultural show, Australia. Emerg Infect Dis 23:1686–1689. doi: 10.3201/eid2310.161836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkins C. 2015. Whole-genome sequencing data for serotyping Escherichia coli—it's time for a change! J Clin Microbiol 53:2402–2403. doi: 10.1128/JCM.01448-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Rothemund D, Curd H, Reeves PR. 2003. species-wide Variation in the Escherichia coli flagellin (H-antigen) gene. J Bacteriol 185:2936–2943. doi: 10.1128/JB.185.9.2936-2943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald DM, Bonocora RP, Wade JT. 2014. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet 10:e1004649. doi: 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews S. 2014. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 14.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gama-Castro S, Salgado H, Santos-Zavaleta A, Ledezma-Tejeida D, Muñiz-Rascado L, García-Sotelo JS, Alquicira-Hernández K, Martínez-Flores I, Pannier L, Castro-Mondragón JA, Medina-Rivera A, Solano-Lira H, Bonavides-Martínez C, Pérez-Rueda E, Alquicira-Hernández S, Porrón-Sotelo L, López-Fuentes A, Hernández-Koutoucheva A, Moral-Chávez VD, Rinaldi F, Collado-Vides J. 2016. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 44:D133–D143. doi: 10.1093/nar/gkv1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dallman TJ, Ashton PM, Byrne L, Perry NT, Petrovska L, Ellis R, Allison L, Hanson M, Holmes A, Gunn GJ, Chase-Topping ME, Woolhouse MEJ, Grant KA, Gally DL, Wain J, Jenkins C. 2015. Applying phylogenomics to understand the emergence of Shiga-toxin-producing Escherichia coli O157:H7 strains causing severe human disease in the UK. Microb Genom 1:e000029. doi: 10.1099/mgen.0.000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott EJ, Robins-Browne RM, O'Loughlin EV, Bennett-Wood V, Bourke J, Henning P, Hogg GG, Knight J, Powell H, Redmond D. 2001. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch Dis Child 85:125–131. doi: 10.1136/adc.85.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Ochman H. 2007. Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci U S A 104:7116–7121. doi: 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton A. 1966. Effect of nonsense mutations on translation of the lactose operon of Escherichia coli. Cold Spring Harbor Symp Quant Biol 31:181–187. doi: 10.1101/SQB.1966.031.01.026. [DOI] [PubMed] [Google Scholar]
- 25.Cohen EJ, Hughes KT. 2014. Rod-to-hook transition for extracellular flagellum assembly is catalyzed by the l-ring-dependent rod scaffold removal. J Bacteriol 196:2387–2395. doi: 10.1128/JB.01580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonifield HR, Yamaguchi S, Hughes KT. 2000. The flagellar hook protein, FlgE, of Salmonella enterica serovar Typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J Bacteriol 182:4044–4050. doi: 10.1128/JB.182.14.4044-4050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos RFC, Nascimento JDS, Geimba MP, Hessel CT, Tondo EC. 2017. First report of human gastroenteritis caused by Escherichia coli O157:NM in Brazil. Foodborne Pathog Dis 14:665–666. doi: 10.1089/fpd.2017.2296. [DOI] [PubMed] [Google Scholar]
- 28.Mag T, Nogrady N, Herpay M, Toth I, Rozgonyi F. 2010. Characterisation of verotoxin-producing Escherichia coli strains isolated from human patients in Hungary over a 7-year period. Eur J Clin Microbiol Infect Dis 29:249–252. doi: 10.1007/s10096-009-0836-z. [DOI] [PubMed] [Google Scholar]
- 29.Karch H, Bielaszewska M. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J Clin Microbiol 39:2043–2049. doi: 10.1128/JCM.39.6.2043-2049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.