ABSTRACT
The global spread of multidrug-resistant Gram-negative bacteria has led to the return of colistin for treating severe infections. Recently, different plasmid-mediated genes conferring resistance to this drug were described and reported worldwide. International committees (EUCAST/CLSI) reevaluated inconsistencies surrounding colistin antimicrobial susceptibility testing (AST), concluding that broth microdilution (BMD) should serve as the reference method for AST. The development of an accurate, reproducible commercial test based on BMD is therefore highly desirable. SensiTest Colistin (STC), a BMD-based compact 4-test panel containing the lyophilized antibiotic in 7 2-fold dilutions (0.25 to 16 μg/ml) was here compared with the EUCAST-CLSI standard reference method (BMD) and, for some isolates, with the automated Phoenix 100 system (PHX). A total of 353 bacterial strains were evaluated by two different laboratories; 137 isolates were resistant to colistin (19 were intrinsically resistant, 83 harbored the mcr-1 gene). Essential agreement (EA) between STC and BMD was obtained for 339 out of the 353 strains tested (96.0%). Overall categorical agreement was obtained for 349 out of the 353 strains analyzed (98.9%). Two major errors (MEs; 0.93%) and two very major errors (VMEs; 1.46%) were documented. STC appeared to be a simple but highly reliable test with good reproducibility even with panels stored at room temperature or at 35°C. Moreover, STC showed a good performance with strains carrying the mcr-1 gene, with a 98.8% EA. As the secondary endpoint of our study, VMEs for PHX were documented for 6 isolates (10%).
KEYWORDS: multidrug resistance, colistin, MIC, SensiTest Colistin, antimicrobial susceptibility testing, Phoenix 100 system, Liofilchem, Becton Dickinson
INTRODUCTION
The antibiotic properties of polymyxins, originally derived from strains of Paenibacillus (Bacillus) polymyxa, were first described in the 1940s. Formerly studied in the 1950s (1) and used for some years in the treatment of Gram-negative bacterial infections, colistin (polymyxin E) is the drug most widely used in clinical practice. However, with growing concern over significant side effects, colistin lost importance in comparison to emerging more efficacious drugs with less overt toxicity (2) and was removed from use some decades later.
The global spread of multidrug-resistant Gram-negative bacteria (MDRGNB) has led to a distinct limitation in the therapeutic options available. This has seen the return of colistin to the clinical arena (3), albeit it has been reassessed to better define its dosage and daily administration (4). Colistin has often become the last option to treat severe infections caused by MDRGNB, such as Pseudomonas aeruginosa, Acinetobacter baumannii, and carbapenem-resistant Enterobacteriaceae (CRE), being frequently used in a dual-therapy regimen.
Until recently, colistin resistance was always thought to be chromosomally encoded and mutationally acquired, allowing vertical transmission only, and, thus, by its very nature, rare and self-limiting. This kind of colistin resistance is determined by mutations in a wide variety of species-specific mechanisms: mgrB and ccrB in Klebsiella pneumoniae and parS and cprS in P. aeruginosa, in addition to the more widely dispersed two-component systems PhoP-PhoQ and PmrA-PmrB (2, 5–8). Universally, however, these mechanisms involve the reduction of lipopolysaccharide net charges, compromising the binding of cationic polymyxins.
Recently, mobile colistin resistance in the form of the plasmid-mediated mcr-1 gene was described; this gene encodes a phosphoethanolamine transferase (9). Following its initial discovery, mcr-1 has been reported worldwide (10). Currently, at least 12 variants of this gene which encode phosphoethanolamine transferase enzymes that differ from mcr-1 at a single amino acid have been described. Immediately after the discovery of this first gene, other mobile elements significantly different from mcr-1 have been described: mcr-2 was found in Belgium, and mcr-3 was identified in Malaysia, Thailand, and the United States (11, 12). Most recently, mcr-4 was characterized in an Italian strain of Salmonella enterica serovar Typhimurium that was originally isolated in swine in 2013. This gene was also demonstrated to have circulated in the veterinary environment in Belgium and Spain in strains collected in 2015 and 2016 (13). mcr-5 was instead described in Germany in an isolate of Salmonella enterica serovar Paratyphi B, allowing postulation that the transfer of resistance genes from bacterial chromosomes to mobile genetic elements has occurred in multiple independent events (14).
Almost contemporarily with the emergence and dissemination of mcr-1, international committees (a EUCAST/CLSI joint working group) sought to address the inconsistencies surrounding the antimicrobial susceptibility testing (AST) of colistin. They concluded that broth microdilution (BMD) should serve as the reference method for testing susceptibility to colistin/polymyxin compounds. Owing to the large size and cationic nature of polymyxins, disk diffusion and gradient diffusion have been demonstrated to be unreliable (15). Additionally, agar dilution is not logistically feasible in clinical settings. Therefore, it follows that the development of a commercial test for colistin AST based on BMD is highly desirable.
Here we present a clinical evaluation of SensiTest Colistin (STC; Liofilchem, Italy), a compact 4-test panel containing the lyophilized antibiotic in 7 2-fold dilutions (0.25 to 16 μg/ml) with one additional well as a growth control. The system is proposed to evaluate colistin susceptibility using a BMD method that complies with the recommendations of international standards (i.e., CLSI, EUCAST) in a simpler and less time-consuming way.
The results of STC were compared with those of the classical BMD technique, performed according to the recommendations of CLSI and EUCAST and used as a “gold standard,” and, for some isolates, also with the results of an automated system, the Phoenix 100 system (PHX; Becton Dickinson, USA).
MATERIALS AND METHODS
Strain collections.
Three different sets of isolates were analyzed by two different laboratories (the Clinical Microbiology Laboratory, Reggio Emilia, Italy [center A] and the Department of Medical Microbiology and Infectious Diseases, Cardiff, UK [center B]). A total of 353 bacterial strains were evaluated.
Out of the 353 strains, 216 were collected in Italy and analyzed in center A. Of these 216 strains, 159 isolates were prospectively collected from blood culture samples and analyzed as fresh isolates. Fifty-seven of the 216 strains belonged to laboratory collections of peculiar clinical isolates that had been collected in the previous 2 years and stored at −80°C in microbeads (Pro-Lab Diagnostics, USA) and were selected on the basis of the following characteristics: 45 were isolates of the Enterobacteriaceae that had reduced susceptibility to colistin (7 of them were Escherichia coli isolates carrying the mcr-1 gene, as demonstrated using an in-house PCR) and 12 were carbapenem-resistant K. pneumoniae strains.
Fifty-nine out of the 353 strains were collected over the previous 3 years as part of a clinical study in Tanta Teaching Hospital, Egypt, and were stored in the laboratory in Cardiff at −80°C in microbeads (Pro-Lab Diagnostics). Of these, 20 Enterobacteriaceae strains were colistin resistant (according to EUCAST breakpoints) with an undefined mechanism and 39 strains were colistin susceptible. These strains were evaluated in center B.
Seventy-five out of the 353 strains had been collected in Thailand within the last 12 months and were also stored in the laboratory in Cardiff, again, at −80°C using microbeads. All these strains harbored the mcr-1 gene, which was confirmed by PCR. Also these isolates were analyzed in center B.
Finally, three reference strains (E. coli ATCC 25922, P. aeruginosa ATCC 27853, E. coli NCTC 13846) were included as controls and tested to validate the different experimental sessions in both laboratories. Details about the isolates analyzed in the present study are shown in Table 1.
TABLE 1.
Study resultsa
| Strain | Total no. of isolates (no. colistin resistantb) | No. of isolates for which MIC was: |
% EA | No. (%) of strains for which the following were obtained: |
|||
|---|---|---|---|---|---|---|---|
| Same | ±1 dilution | CA | ME | VME | |||
| Acinetobacter baumannii | 6 (0) | 1 | 5 | 100.0 | 6 (100) | 0 | 0 |
| Acinetobacter speciesc | 4 (1) | 3 | 1 | 100.0 | 4 (100) | 0 | 0 |
| Citrobacter koseri | 1 (0) | 0 | 1 | 100.0 | 1 (100) | 0 | 0 |
| Enterobacter aerogenes | 4 (0) | 1 | 1 | 50.0 | 3 (75.0) | 1 | 0 |
| Enterobacter cloacae complex | 4 (2) | 1 | 3 | 100.0 | 4 (100) | 0 | 0 |
| Escherichia coli | 205 (89) | 80 | 119 | 97.1 | 204 (99.5) | 0 | 1 |
| Hafnia alvei | 6 (6) | 3 | 2 | 83.3 | 6 (100) | 0 | 0 |
| Klebsiella oxytoca | 6 (0) | 2 | 4 | 100.0 | 6 (100) | 0 | 0 |
| Klebsiella pneumoniae | 76 (23) | 34 | 38 | 94.7 | 75 (98.7) | 1 | 0 |
| Leclercia adecarboxylata | 1 (0) | 0 | 1 | 100.0 | 1 (100) | 0 | 0 |
| Morganella morganii | 2 (2) | 2 | 0 | 100.0 | 2 (100) | 0 | 0 |
| Proteus mirabilis | 5 (5) | 5 | 0 | 100.0 | 5 (100) | 0 | 0 |
| Providencia species | 2 (2) | 2 | 0 | 100.0 | 2 (100) | 0 | 0 |
| Pseudomonas aeruginosa | 19 (0) | 9 | 10 | 100.0 | 19 (100) | 0 | 0 |
| Salmonella species | 7 (2) | 3 | 3 | 85.7 | 6 (85.7) | 0 | 1 |
| Serratia marcescens | 4 (4) | 4 | 0 | 100.0 | 4 (100) | 0 | 0 |
| Shigella species | 1 (1) | 1 | 0 | 100.0 | 1 (100 | 0 | 0 |
| Total | 353 | 151 | 188 | 96.0 | 349 (98.9) | 2 | 2 |
| Colistin susceptible | 216 | 81 | 124 | 94.9 | 214 (99.1) | 2 | NA |
| Colistin resistant (not due to mcr-1) | 54 | 43 | 9 | 96.3 | 53 (98.1) | NA | 1 |
| Colistin resistant (due to mcr-1) | 83 | 27 | 55 | 98.8 | 82 (98.8) | NA | 1 |
| Total | 353 | 151 | 188 | 96 | 349 (98.9) | 2 | 2 |
EA, essential agreement; CA, categorical agreement; ME, major errors; VME, very major errors; NA, not applicable.
Colistin resistance was defined according to EUCAST breakpoints.
These included one strain each of Acinetobacter ursingii, A. lwoffii, A. junii, and A. nosocomialis.
STC. (i) Strain analysis.
Briefly, a 0.5 McFarland suspension of the microorganism to be tested was prepared in a solution of 0.90% (wt/vol) NaCl (saline) and then diluted 1:20, always in saline, obtaining solution A. Solution A (0.4 ml) was then added to the 3.6-ml tube of Mueller-Hinton broth II provided in the STC kit, obtaining solution B. One hundred microliters of solution B was then dispensed into each well in a row. The STC panels were then incubated at 36 ± 2°C for 16 to 20 h in ambient air. The results were read visually with the naked eye by two different operators, by using bright, indirect lighting against a dark background. The presence of growth in the growth control well was considered first, allowing the test to be considered valid. For MIC determination, bacterial growth was considered the presence of turbidity, a button at the bottom of the well, or pinpoint colonies in the broth. Different lots of the panels were tested during the study period.
(ii) Evaluation of reproducibility and stability.
Tests for reproducibility and stability were performed in center A. Six different strains were chosen: the three quality control (QC) reference strains (which included the E. coli NCTC 13846 mcr-1-positive strain), two E. coli strains (one, named CSR-55, with an MIC of 8 μg/ml with an undefined resistance mechanism and the other, named CSR-57, with an MIC of 2 μg/ml), and one carbapenem-resistant K. pneumoniae strain with an MIC of ≥16 μg/ml due to mechanisms different from mobile determinants. All the isolates were tested as replicates 10 times.
The stability of the product was assessed in two ways. First, three different boxes containing 32 panels were stored at 4°C (as suggested by the manufacturer), at room temperature, and at 35°C. The six different strains described above were tested on the day after the arrival of the panels (time zero [T0]) and 1 week after (T7), 2 weeks after (T14), 1 month after (T30), and 3 months after (T90) the delivery of the products.
Each single panel allows testing of up to four strains; if less than 4 tests are performed, the manufacturer provides a film with which to seal the inoculated rows (in order to prevent any leakage of contaminated fluids) and to return the panel into its own desiccant envelop, which is placed in a refrigerator. The stability of the panels used at the different times was established by analyzing the six different strains described above with six different panels at day 0 (T0) (inoculum in the first row) and then after 5 (T5), 13 (T13), and 19 (T19) days, in the meantime storing the panels in a refrigerator and then inoculating the second (at T5), third (at T13), and fourth (at T19) rows.
The expected values of the MICs were those established by the EUCAST Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion (version 7.0, valid from 1 January 2017) for the control strains or a ±1 dilution of the MIC obtained in five previously performed BMD experimental sessions for the other strains.
Broth microdilution.
BMD was performed according to the ISO standard method (20776-1), which has been demonstrated to work well for Enterobacteriaceae, P. aeruginosa, and Acinetobacter species. Colistin sulfate salt was bought from Sigma-Aldrich (Merck KGaA, Germany) as a lyophilized powder (100 mg, 15,000 U/mg) that was resuspended in distilled water. Vials with a final concentration of 1,024 μg/ml were stored at −80°C until the different test sessions were performed. The cation-adjusted Mueller-Hinton broth (CAMHB) used was ready-for-use Mueller-Hinton II medium (Liofilchem, Italy). The trays bought from Nuova Aptaca, Italy, were made of plain polystyrene and not treated in any way before use. No additives (Tween 80 or other surfactants) were added in any part of the testing process.
For any working session, accounting for 20 isolates, 11 working concentrations of colistin (range, 0.064 μg/ml to 64 μg/ml in 2-fold dilutions) were prepared in separate tubes containing CAMHB, according to the dilution scheme proposed by CLSI (16). Fifty microliters of each intermediate concentration was dispensed into the wells of the microwell plates. For each strain tested, a positive growth control was included in the first well of the plate.
Quality control strains were tested in the first session; each further experimental session was validated by using one of these strains added on rotation.
Isolated bacterial colonies were selected from an 18- to 24-h blood agar culture and transferred to a CAMHB tube. The broth was incubated overnight at 35 to 37°C, the turbidity was adjusted to a 0.5 McFarland standard, and the suspension was then diluted in broth to obtain a final bacterial concentration of 5 × 105 CFU/ml.
Finally, 50 μl of the bacterial suspension was added to each well of the 96-well microplates, which were incubated in an ambient air incubator at 35°C ± 2°C for 16 to 20 h. The MICs were determined as the lowest concentration that completely inhibited the bacterial growth in the wells.
Phoenix 100 system.
The tests with the Phoenix 100 system were performed in center A, according to the manufacturer's recommendations as a part of the standard of care for the patients from whom the 216 clinical isolates were obtained. PHX was also performed on the 3 QC strains, for a total of 219 isolates tested. The NMIC-417 card was used for the present study.
Discrepant results.
Samples with discrepant results that showed a disagreement between BMD and STC with more than a 2-fold dilution were retested, with both methods being performed in at least two other different experimental sessions. The results of each test that were confirmed in more than two experimental sessions were considered confirmed, and the MIC value was used for the final analysis.
Agreement between methods.
Essential agreement (EA) was defined as the agreement within plus or minus 1 2-fold dilution of the MIC determined by STC with the MIC determined by the reference method (BMD). EA determination was evaluated exactly for all the values below 16 μg/ml (values of ≥16 μg/ml were considered in agreement). EA between PHX and BMD was not analyzed due to the narrow MIC range tested by the PHX card (only 3 dilutions from 1 to 4 mg/liter).
For discrepant results, we evaluated the categorical agreement (CA), i.e., when the results did not change the categorization of the isolates (considered susceptible or resistant) or the occurrence of major errors (ME; the BMD result was susceptible and the STC or the PHX result was resistant) or very major errors (VME; the BMD result was resistant and the STC or the PHX result was susceptible) (17).
RESULTS
The study results are summarized in Table 1. All the tests performed were considered valid (that is, growth was present in the growth control well). Only in a few cases was the evaluation of growth challenging, due to the presence of pinpoint colonies scattered into the broth. That occurred mainly for Hafnia alvei strains. Examples of the STC panels are shown in Fig. 1.
FIG 1.
(Left) Results of an STC test. The well on the right of the red line indicates the value of the MIC for the isolate. (Right) Results of another STC test. Arrows, pinpoint colonies of H. alvei.
Agreement between STC and BMD.
Essential agreement (EA) was obtained for 339 out of the 353 strains tested (96.0%). In particular, for 151 strains the same MIC value was obtained by the two methods, whereas a difference of ±1 dilution was documented for 188 isolates.
Discrepancies of 2 or more dilutions were documented for 14 strains: 6 strains from the Italian clinical collection, 7 strains from those collected in Egypt, and 1 mcr-1 isolate collected in Thailand. These included 6 Escherichia coli isolates, 4 K. pneumoniae isolates, 2 Enterobacter aerogenes isolates, 1 strain of H. alvei, and 1 isolate of Salmonella species. The discrepancy did not change the strains' clinical categorization in 12 of these cases; the other 2 cases were 1 isolate of Enterobacter aerogenes (MICs, 0.25 μg/ml by BMD and 4 μg/ml by STC) and one Salmonella species (MICs, 4 μg/ml by BMD and 1 μg/ml by STC). Overall categorical agreement (CA) was obtained for 349 out of the 353 strains analyzed (98.9%). Two MEs were recognized for one strain of E. aerogenes (as described above) and one strain of K. pneumoniae (MICs of 2 μg/ml by BMD and 4 μg/ml by STC), whereas two VMEs were documented for one strain each of Salmonella species (as described above) and E. coli (MICs, 4 μg/ml by BMD and 2 μg/ml by STC). The rate of MEs for STC was 0.92% (2 isolates out of the 216 susceptible strains), whereas the rate of VMEs was 1.46% (2 out of 137 isolates).
Reproducibility of STC.
The six strains gave a total of 60 replicates. Total agreement among the replicates (i.e., the same MIC) was documented in 50 out of the 60 tests performed (83.3%), whereas the EA was 100%.
Stability of STC.
Stability tests on the STC panels were performed across different time periods with the STC panels stored at different temperatures, as shown in Table 2. All the tests fell into agreement within ±1 2-fold dilution. Only one was out of range: the MIC at T7 of the reference strain NCTC 13846 (E. coli harboring the mcr-1 gene) for the panels stored at 4°C, which was 2 dilutions higher than expected.
TABLE 2.
Stability tests using panels stored at different temperatures for 3 months
| Panel storage temp and strain | MIC (μg/ml) |
|||||
|---|---|---|---|---|---|---|
| Expected range | T0 | T7 | T14 | T30 | T90 | |
| 4°C | ||||||
| ATCC 25922 | 0.25–2a | 1 | 0.5 | 0.25 | 0.5 | 0.5 |
| ATCC 27853 | 0.5–4a | 1 | 1 | 1 | 1 | 1 |
| CSR-55 | 4–16 | 16 | 8 | 8 | 8 | 8 |
| CSR-57 | 1–4 | 4 | 4 | 2 | 4 | 4 |
| CSR-68 | 8–32 | ≥16 | ≥16 | ≥16 | ≥16 | ≥16 |
| NCTC 13846 | 2–8a | 4 | 16 | 4 | 4 | 8 |
| Negative control | NGb | NG | NG | NG | NG | |
| Room temp | ||||||
| ATCC 25922 | 0.25–2a | 0.5 | 0.5 | 0.5 | 1 | 1 |
| ATCC 27853 | 0.5–4a | 1 | 1 | 1 | 1 | 1 |
| CSR-55 | 4–16 | 8 | 16 | 8 | 16 | 8 |
| CSR-57 | 1–4 | 4 | 4 | 2 | 4 | 4 |
| CSR-68 | 8–32 | ≥16 | ≥16 | ≥16 | ≥16 | ≥16 |
| NCTC 13846 | 2–8a | 4 | 4 | 4 | 4 | 4 |
| Negative control | NG | NG | NG | NG | NG | |
| 35°C | ||||||
| ATCC 25922 | 0.25–2a | 0.5 | 0.5 | 0.5 | 1 | 1 |
| ATCC 27853 | 0.5–4a | 1 | 1 | 1 | 1 | 2 |
| CSR-55 | 4–16 | 8 | 8 | 8 | 8 | ≥16 |
| CSR-57 | 1–4 | 4 | 4 | 4 | 4 | 4 |
| CSR-68 | 8–32 | ≥16 | ≥16 | ≥16 | ≥16 | ≥16 |
| NCTC 13846 | 2–8a | 4 | 4 | 4 | 4 | 8 |
| Negative control | NG | NG | NG | NG | NG | |
According to EUCAST Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion (version 7.0, valid from 1 January 2017).
NG, no growth.
The STC also appeared to be stable if MICs were evaluated at different times for the different rows of the same panel (Table 3).
TABLE 3.
Stability tests using the same panel inoculated on different days
| Strain | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| Expected range | Single-panel replicates |
||||
| T0 | T5 | T13 | T19 | ||
| ATCC 25922 | 0.25–2a | 0.5 | 0.5 | 1 | 1 |
| ATCC 27853 | 0.5–4a | 1 | 1 | 2 | 2 |
| CSR-55 | 4–16 | 16 | 8 | 8 | 8 |
| CSR-57 | 1–4 | 4 | 4 | 4 | 4 |
| CSR-68 | 8–32 | ≥16 | ≥16 | ≥16 | ≥16 |
| NCTC 13846 | 2–8a | 4 | 4 | 4 | 4 |
According to the EUCAST Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion (version 7.0, valid from 1 January 2017).
Agreement between BMD and PHX.
PHX showed an overall agreement with BMD for 212 out of the 219 isolates analyzed (96.8%). The seven strains with discordant results resulted in one ME for a P. aeruginosa strain and six VMEs (two for E. coli [one strain harboring the mcr-1 gene], two for Salmonella species, one for an H. alvei strain, and one for a K. pneumoniae strain), accounting for 10% of the 60 colistin-resistant isolates tested.
DISCUSSION
Colistin is often considered the last resource for the treatment of severe Gram-negative bacterial infections, in particular, those caused by P. aeruginosa, A. baumannii, and CRE (3). This compound has no activity against Gram-positive bacterial strains or against some Gram-negative bacteria, such as Burkholderia cepacia, Elizabethkingia meningoseptica, H. alvei, Morganella morganii, Proteus species, Providencia species, Serratia marcescens, and Yersinia pseudotuberculosis (2, 18, 19).
Immediately after the recent discovery of a plasmid-mediated mechanism of resistance that more often conveys low-level resistance (MICs, 4 to 8 μg/ml) (9), strains (mostly E. coli) harboring the gene have been described worldwide (10, 20). More recently, other mobile genetic resistance traits have been described (11–14). Thus, the spread of colistin-resistant microorganisms has become a matter of concern worldwide, and the ECDC published in June 2016 a document calling for a rapid risk assessment in order to control the spread of plasmid-mediated colistin resistance in Enterobacteriaceae (https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/enterobacteriaceae-risk-assessment-diseases-caused-by-antimicrobial-resistant-microorganisms-europe-june-2016.pdf). This document highlights different actions for the implementation of surveillance strategies and antimicrobial stewardship about this topic. Moreover, the development of improved laboratory methods for the determination of the correct colistin MIC and molecular mcr-1 detection is considered beneficial and necessary (21).
For colistin, a reliable evaluation of MICs, together with the possibility of therapeutic drug monitoring (TDM), is also necessary for the correct management of patients, given colistin's potential significant adverse effects, in order to avoid toxic effects while maintaining sufficient antibacterial activity (21).
A joint EUCAST and CLSI subcommittee issued recommendations in July 2016 (subsequently confirmed in updates of August and November 2016 and June 2017) confirming that broth microdilution, using untreated polystyrene, is so far the only valid method and that disk diffusion does not work because of the poor diffusion of the large colistin molecule (15, 22). In this document, the adhesive properties related to the cationic nature of colistin were emphasized, suggesting that the BMD test should be performed by using cation-adjusted Mueller-Hinton broth without additives (in particular, no polysorbate 80 or other surfactants) with trays of untreated polystyrene. Colistin may adhere to BMD plates; this issue should be taken into account when developing new devices. In cases of major discrepancies with the result expected, verification of the free concentrations of colistin in the test panels may be warranted. The trays used to produce the STC panels are made of plain, untreated polystyrene, compliant with EUCAST-CLSI guidelines. The results obtained in our various STC experiments were congruent with those of BMD, and so it was decided that evaluation of the free concentrations of colistin in these cases was unnecessary.
Moreover, issues about the correct MIC estimate with some automated instruments have been recently highlighted. EUCAST suggests to the users of semiautomated devices to apply rigorous QC, checking with the manufacturer whether or not they are confident that their method to evaluate colistin gives correct results (22). Regarding automated systems, the performances of the Vitek-2 (bioMérieux, Marcy l'Etoile, France) and MicroScan (Beckman Coulter, CA, USA) systems against CRE and mcr-1-positive isolates have recently been evaluated, resulting in a high rate of VMEs for Vitek-2 (36% versus 4% for MicroScan), whereas no MEs were documented for Vitek-2 and 15.8% MEs were demonstrated for MicroScan (23). The high rate of VMEs with the Vitek-2 AST-GN colistin (cs01n) system compared to the results of agar dilution (the reference method used for cs01n development) and compared to those of BMD was also demonstrated by an internal investigation performed by bioMérieux (bioMérieux, communication to customers).
These data express the pertinence of the development of a new diagnostic tool for clinical microbiology laboratories. Nevertheless, standard BMD methods are labor-intensive and do not fall in the routine practice of the majority of the clinical microbiology laboratories.
SensiTest Colistin appears to be a simple but highly reliable test to assess colistin susceptibility. The preparation procedure is easy and fast, and evaluation of the results is simple, reflecting that of the BMD methodology. With this device, it is possible to test a single drug, and therefore, it may be used as a second-line assay for laboratories which use automated instruments in their daily practice without redundancies for other antibiotics. Even if the panels are customized for testing four separate isolates at distinct time points, the MIC given by STC appeared to be stable.
In our assessment, the device appeared to be highly reproducible. We tested the reproducibility of the panels stored at different temperatures, reflecting conditions that the STC panels may experience in real-life use under particularly challenging conditions, such as those that may occur in developing countries, with possible troubleshooting due to transport and storage.
As an ancillary result of our study, we demonstrated an overall good agreement between BMD and the Phoenix 100 system, even with the limitation that PHX was performed as the standard of care a few days before the execution of the BMD. Among the 219 isolates also tested with PHX, the VMEs involved 2 strains each of E. coli (1 mcr-1-positive isolate) and Salmonella species, 1 strain of H. alvei, and 1 strain of K. pneumoniae. The rate of VMEs for PHX was 10% in our study (6 isolates out of the 60 colistin-resistant strains analyzed with the automated instrument).
STC was recently evaluated by the EUCAST committee in June 2017, together with other commercially available BMD techniques. In that study, STC was tested on 75 strains: the EA appeared to be 88%, with 7 MEs and 1 VME (22). In our study, the test performance was better, with EA of 96.0% (2 MEs and 2 VMEs). A direct comparison between these data and those from our study appears to be difficult because of a likely disparity between the EUCAST strains tested and our clinically representative, geographically varied test panel.
In our experience, STC showed a very good performance with strains carrying the mcr-1 gene, with 98.8% EA (Table 1). For some strains, such as H. alvei, it is important to carefully evaluate any growth in the wells, as it may present as pinpoint microcolonies, as was the case for this species (Fig. 1). However, it is likely that the recent caveat that this microorganism be reclassified as constitutively resistant to colistin (19) may render in future the evaluation of colistin susceptibility for these strains redundant.
Another possible advantage of using STC is that, being an open system, it is possible to evaluate also the minimal bactericidal concentration (MBC) after the reading is obtained by spotting 1 to 10 μl onto a Mueller-Hinton agar plate (Fig. 2) and considering the growth after 24 h.
FIG 2.
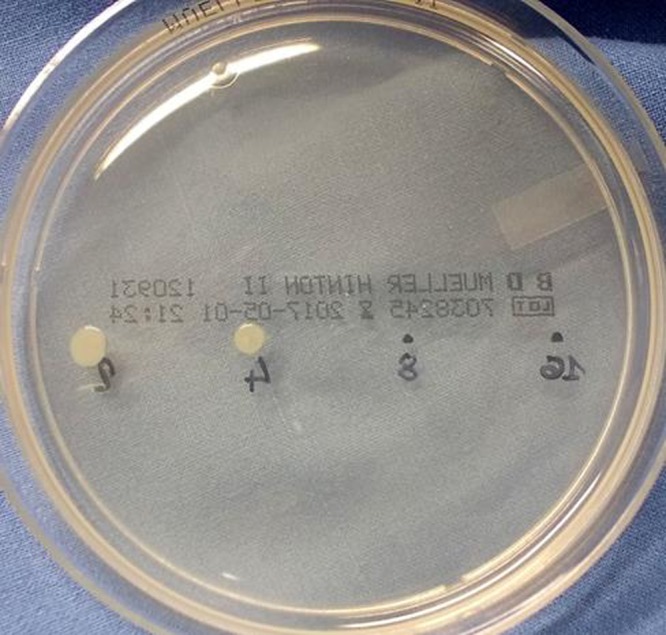
Evaluation of the bactericidal effect by spotting 1 μl of the resuspended wells of a strain having an MIC of 8 mg/ml (the MIC is equal to the MBC).
In the present study, a limited number of A. baumannii and P. aeruginosa strains were tested, and further evaluation with these isolates may be of value. However, even if further assessments, perhaps through in-field/in-clinic studies, may be warranted, our preliminary findings suggest that STC could be proposed both as a first-line test for selected specimens and as a confirmatory test adjacent to automated screening.
ACKNOWLEDGMENTS
This work was partially supported by grants from the Scientific Committee of the IRCCS Arcispedale Santa Maria Nuova, Reggio Emilia, to F.B.
SensiTest Colistin panels were kindly provided by Liofilchem.
We have no conflicts of interest to declare.
REFERENCES
- 1.Koyama Y, Kurosasa A, Tsuchiya A, Takakuta K. 1950. A new antibiotic ‘colistin’ produced by spore-forming soil bacteria. J Antibiot 3:457–458. [Google Scholar]
- 2.Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin Microbiol Rev 21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 4.Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, Silveira FP, Forrest A, Nation RL. 2011. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 55:3284–3294. doi: 10.1128/AAC.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeannot K, Bolard A, Plesiat P. 2017. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 49:526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 6.Kato A, Latifi T, Groisman EA. 2003. Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc Natl Acad Sci U S A 100:4706–4711. doi: 10.1073/pnas.0836837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JY, Park YK, Chung ES, Na IY, Ko KS. 2016. Evolved resistance to colistin and its loss due to genetic reversion in Pseudomonas aeruginosa. Sci Rep 6:25543. doi: 10.1038/srep25543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Hoiby N. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 11.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21(27):pii=30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 12.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22(31):pii=30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 15.EUCAST. 2016. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
- 16.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed CLSI document M100-S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.FDA. 2009. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems, p 1–42. FDA, Rockville, MD. [Google Scholar]
- 18.EUCAST. 2016. EUCAST expert rules, intrinsic resistance and exceptional phenotypes, version 3.1. http://www.eucast.org/expert_rules_and_intrinsic_resistance/.
- 19.Jayol A, Saly M, Nordmann P, Menard A, Poirel L, Dubois V. 2017. Hafnia, an enterobacterial genus naturally resistant to colistin revealed by three susceptibility testing methods. J Antimicrob Chemother 72:2507–2511. doi: 10.1093/jac/dkx154. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Tian GB, Zhang R, Shen Y, Tyrrell JM, Huang X, Zhou H, Lei L, Li HY, Doi Y, Fang Y, Ren H, Zhong LL, Shen Z, Zeng KJ, Wang S, Liu JH, Wu C, Walsh TR, Shen J. 2017. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis 17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 21.Caniaux I, van Belkum A, Zambardi G, Poirel L, Gros MF. 2017. MCR: modern colistin resistance. Eur J Clin Microbiol Infect Dis 36:415–420. doi: 10.1007/s10096-016-2846-y. [DOI] [PubMed] [Google Scholar]
- 22.EUCAST. 2017. Antimicrobial susceptibility testing of colistin—problems detected with several commercially available products. www.eucast.org/ast_of_bacteria/warnings/#c13111.
- 23.Chew KL, La MV, Lin RTP, Teo JWP. 2017. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive Enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol 55:2609–2616. doi: 10.1128/JCM.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]



