ABSTRACT
Strongyloides stercoralis is present worldwide, but its prevalence is still uncertain, mainly due to the lack of sensitivity of diagnostic methods. Molecular techniques are under development, but a standardized protocol is still unavailable. We compared the sensitivity of real-time PCR, using two extraction protocols, with that of the Baermann technique. Samples were collected in the framework of the baseline screening of a randomized clinical trial evaluating moxidectin against S. stercoralis in Lao People's Democratic Republic. Two stool samples from each participant were processed by the Baermann method, and one subsample was processed by PCR. DNA was extracted using the QIAamp DNA stool minikit based on the standard protocol for the QIAamp DNA minikit (QIA) and using a modification of the QIA procedure (POL). Subsequently, all extracted samples were analyzed by real-time PCR. Overall, 95 samples were analyzed by the three diagnostic methods. Sixty-nine (72.6%) samples were positive according to the Baermann method, 25 (26.3%) by the QIA method, and 62 (65.3%) by the POL method. The sensitivities were 86% (95% confidence interval [CI], 76.7 to 92.9), 31.0% (95% CI, 21.3 to 42.6), and 78.0% (95% CI, 66.8 to 86.1) for the Baermann, QIA, and POL methods, respectively. The sensitivities calculated for each day of the Baermann method separately were 60% (48.4 to 70.8%) and 64% (52.2 to 74.2%) for days 1 and 2, respectively. In conclusion, the POL method revealed a good performance and was comparable to the Baermann test performed on two stool samples and superior to the Baermann method performed on one stool sample. Additional studies are needed to standardize a PCR protocol for S. stercoralis diagnosis.
KEYWORDS: PCR, Strongyloides stercoralis, diagnosis
INTRODUCTION
The threadworm Strongyloides stercoralis is known to be present worldwide except for Antarctica (1). However, its real prevalence is still an estimated guess, and epidemiology data vary from 100 million to 300 million infected individuals (2, 3). In the last decade, migration flows and travels to countries where this worm is endemic have changed the geography of infection, contributing to a further increase in spreading of S. stercoralis (2, 4). Initial studies in the 1980s on this parasitic infection were deemed by Genta et al. (5) inspired guesses: data were considered not reliable because knowledge on the presence of S. stercoralis at the country level was poor. This was due mainly to the fact that the diagnostic methods used were unsuitable for accurate detection of S. stercoralis (4, 5). The situation has not changed much since then (1, 6), with S. stercoralis being one of the most underdiagnosed and neglected infections of humans. One of the peculiar aspects of this helminth is that it replicates within the human host, with eggs developing into rhabditiform larvae. Those either can be passed in the stool or can cause autoinfection by developing into infective filariform larvae. Infective larvae can reinfect the host by penetrating either the intestinal mucosa or the perirectal skin.
Weakening of the immune system (due to infection with HIV/AIDS or human T-cell lymphotropic virus [HTLV]) plays an important role in the proliferation of the parasite and maintenance of the infection, which can become disseminated and occasionally fatal (3, 4, 7). Furthermore, the increased iatrogenic immunosuppression secondary to the broader use of corticosteroids and chemotherapy, and organ transplantation also in countries where the worm is endemic, contributes to increase the burden of S. stercoralis infection. This highlights the importance of detecting S. stercoralis infection, which acts synergistically with immunosuppression and considerably increases morbidity and mortality also in countries where this infection is not endemic.
S. stercoralis infections have a different range of clinical presentations, from asymptomatic infection or mild nonspecific symptoms (8) to a life-threatening dissemination of larvae to internal organs (1, 6). In terms of treatment and diagnosis, S. stercoralis infection is considered one of the most neglected diseases among the neglected tropical diseases. Direct methods, such as the Baermann method and Koga agar plate culture, the two WHO-recommended methods, are still the diagnostic methods used most in countries where this infection is endemic, yet they are time-consuming and cumbersome and show only moderate sensitivity (2, 9). In addition, for both methods specific equipment and conditions are crucial for good performance: an incubator, stable electricity, sufficient space for sample incubation with funnels and tubes, and mesh and gauze for incubation (10–12).
Serological methods have recently demonstrated good sensitivity in countries where this infection is not endemic, and they can be easily used in advanced laboratories for diagnosis and screening (13, 14). The main limitation of serology is that it cannot be used to assess drug efficacy. While larva excretion stops a few days after successful treatment (15), serology titers decrease only 6 to 12 months after treatment (16). Therefore, assessment of drug efficacy by serology in areas where the infection is endemic, where a high rate of reinfection is common, is not feasible.
Molecular techniques for the diagnosis of S. stercoralis are still under development (17, 18). They offer many advantages compared to the Baermann method: only one sample is needed, the amount of stool required is smaller, multiple infections can be detected (19, 20), fixation of the sample avoids contamination, and, finally, the technique, once standardized, is objective and is quicker to perform. Another advantage of PCR is the fact that DNA from dead or live larvae will be detected, whereas the Baermann method, although analyzing a bigger amount of stool and therefore having a greater chance to detect infection, relies on the fact that the larvae have to be alive in order to migrate into the collection tube.
However, initial studies conducted with PCR on S. stercoralis obtained discordant results (17, 21). In addition, protocols for DNA extraction and PCR are still not well defined, being mainly based on in-house procedures rather than on standardized kits (21). The sensitivity and specificity of PCR methods have been compared with those of traditional direct diagnostic methods (9, 18). The specificity turned out to be very high (2, 9, 17, 22), but the sensitivity was shown to vary. While in some studies PCR, especially real-time PCR (23), showed a better performance than direct methods (23–25), other studies combining direct methods revealed a higher sensitivity (2, 9, 22). In addition, recently PCR has been compared with serology and was shown to be less sensitive (4).
The aim of this study was to compare the sensitivity of real-time PCR with that of the Baermann technique. Moreover, two well-established DNA extraction protocols were evaluated (9, 17). Finally, we evaluated the advantage of a receiver operating characteristic (ROC) analysis for an individual evaluation of cycle threshold (CT) in real-time PCR and its impact on sensitivity and specificity.
MATERIALS AND METHODS
Ethical consideration.
Stool samples were collected in the framework of the baseline screening of an exploratory phase II, randomized, single-blind clinical trial evaluating the safety and efficacy of moxidectin versus ivermectin against S. stercoralis infection (26). The trial was performed between April and June 2016 in the Lao People's Democratic Republic in the district of Pathoumphone, where S. stercoralis infection is endemic.
Ethical clearance was obtained from the Ethics Committee of Northwestern and Central Switzerland (EKNZ; reference no. 15/103) and the Lao Ministry of Health (reference no. 075/2016). The trial is registered with Current Controlled Trials (ISRCTN11983645). Participants 12 to 60 years old were eligible for inclusion in the trial. Written informed consent was collected before enrollment from all participants. At the end of the study, all participants positive for infection were treated according to local guidelines.
Laboratory procedures.
Two stool samples obtained from 95 participants were examined with the Baermann method for the detection of S. stercoralis larvae. The Baermann method was carried out following the WHO standard procedure (27).
A subsample of the first sample of stool (∼200 mg) was preserved in ethanol and shipped to the Swiss Tropical and Public Health Institute (TPH) in Basel, Switzerland, for PCR analyses. Preserved samples were processed with two different protocols for DNA extraction. One DNA extraction (QIA method) was performed using the QIAamp DNA stool minikit (Qiagen; Hilden, Germany) by following the manufacturer's protocol, with minor modifications (9). The second DNA extraction (POL method) was done using the QIAamp DNA minikit with modifications according to Polley et al. (28). In brief, samples were washed once with phosphate-buffered saline (PBS); 400 μl of animal tissue lysis (ATL) buffer with 40 μl of proteinase K was added, followed by 2 h of incubation at 56°C. During this period, the samples were briefly vortexed every 30 min. After incubation, the samples were pelleted and 200 μl of supernatant was processed according to the kit protocol. All samples were analyzed with real-time PCR for detection of S. stercoralis. The 18S rRNA S. stercoralis-specific real-time PCR protocol was conducted using TaqMan GeneExpression MasterMix (Thermo Fisher, Switzerland), sense and antisense primers (5′-to-3′ forward primer, GGA ATT CCA AGT AAA CGT AAG TCA TTA [modified from reference 17], and 5′-to-3′ reverse primer, GTT ACG ACT TTT GCC CGG TTC) and the respective probe (6-carboxyfluorescein [FAM]-TAT ATT AAA TCC TTC CAA TCG CTG TTG-BHQ1) (Eurofin Genomics, Ebersberg, Germany) to amplify a specific 184-bp fragment of S. stercoralis. The thermoprofile on the 7500 ABI real-time machine (Thermo Fisher) was 2 min at 50°C and 10 min at 95°C followed by 45 cycles of 15 s at 95°C and 1 min at 58°C. The specificity of these primers was previously tested on a variety of DNAs from stool samples confirmed by light microscopy at the diagnostic center of the Swiss TPH to be infected with Ascaris lumbricoides, Blastocystis hominis, Cryptosporidium spp., Cyclospora spp., Entamoeba coli, Entamoeba dispar, Entamoeba hartmanni, Entamoeba histolytica, Entamoeba moshkovskii, Endolimax nana, Giardia lamblia, Iodamoeba bütschlii, and Schistosoma mansoni and was found to be 100%. On each real-time PCR plate, we included negative and positive controls with different plasmid concentrations (102, 104, and 106 plasmids/μl) containing an insert with the sequence of the S. stercoralis real-time PCR product. Each DNA sample was further tested for inhibition by addition of 2 μl of a known plasmid concentration (102 plasmids/μl). In case of inhibition, the sample was diluted 1:2 and 1:5 and retested.
Statistical analysis.
Data for the amplification curves were entered in an Excel file, and statistical analyses were conducted using Stata 12.0 (Lake Drive College Station, Texas).
The cycle threshold (CT) cutoff value was defined as the number of PCR cycles required for the detection of fluorescence signal of the amplified products to exceed the set threshold value. As a consequence, higher quantities of DNA resulted in lower CT values and vice versa (29).
Because of possible unspecific amplification and to exclude any cross-contamination from highly positive samples, PCR results were considered negative if CT values were more than 40 or if no amplification was detected (18). Because no quantification in the Baermann method was conducted and thus no quantitative correlation with real-time copy numbers was feasible, mean and median copy numbers by DNA extraction method are reported in the supplemental material.
In the absence of a true “gold standard,” we calculated the sensitivity of the methods on the basis of positive results obtained by the Baermann method, performed on 2 days. Assuming that PCR has a specificity of >90% (9, 18, 20, 30), we also calculated sensitivity on the basis of any positive finding with any of the three diagnostic methods. We compared the positivity rates of the tests with the McNemar test. The test was considered significant if its P value was <0.05. Both PCRs were compared separately with the Baermann method, both as the mean of 2 samples collected on 2 different days and for only 1 out of the 2 days of collection. A Wilcoxon rank sum test was used to compare the CT values between microscopy-positive and -negative tests, with a CT of >0.
After establishing that the POL DNA extraction showed a better performance than the QIA extraction when using a cutoff value of 40 cycles, we determined the optimal cutoff for the new test based on its ROC curve with the results obtained by the Baermann method over 2 days by maximizing the index of Youden (i.e., sensitivity + specificity − 1).
RESULTS
Complete data from the three diagnostic methods/protocols were available for 95 participants.
Fifteen (15.8%) participants were classified as negative by all methods and 80 participants were positive by at least one method. Sixty-nine (72.6%) were positive according to the Baermann method based on 2 days of collection, 25 (26.3%) by the QIA method, and 62 (65.3%) by the POL method (Table 1).
TABLE 1.
Results for both extraction methods (QIA and POL) compared to the Baermann method results
| Test and result | No. of samples with indicated Baermann method result |
|||||
|---|---|---|---|---|---|---|
| Days 1 and 2 |
Day 1 |
Day 2 |
||||
| Negative | Positive | Negative | Positive | Negative | Positive | |
| QIA | ||||||
| Negative | 20 | 50 | 37 | 33 | 33 | 37 |
| Positive | 6 | 19 | 10 | 15 | 11 | 14 |
| POL | ||||||
| Negative | 17 | 16 | 23 | 10 | 21 | 12 |
| Positive | 9 | 53 | 24 | 38 | 23 | 39 |
The sensitivities estimated on the basis of a positive Baermann result (2 days; 69/95) were 70% (95% CI, 57 to 80%) and 74% (95% CI, 62 to 84%) for a single Baermann test, 27.5% (95% CI, 16 to 40%) for the QIA method, and 77.0% (95% CI, 65 to 86%) for the POL method (Table 2). The sensitivities estimated on the basis of any positive result obtained by any method (80/95) were 86.3% (95% CI, 76.7 to 92.9%) for 2 days of Baermann method testing, 60% (95% CI, 48.4 to 70.8%) for the Baermann first-day sample, 63.8% (95% CI, 52.2 to 74.2%) for the second-day sample, 31.3% (95% CI, 21.3 to 42.6%) for the QIA method, and 77.5% (95% CI, 66.8 to 86.1%) for the POL method. The McNemar test confirmed that the Baermann method performed on a single day had significantly lower positivity rates than analyses of 2 stool samples by the Baermann and the POL methods (P = 0.02 and 0.03, respectively) (Table 2). The Baermann (2 days) and POL methods were exactly the same in terms of sensitivity, as the POL method could confirm 77% of the Baermann method (2 days)-positive samples and vice versa (Table 2). The two extraction methods were significantly different, and the POL method was significantly more sensitive than the QIA method (P < 0.005). The combination of PCR and direct method showed the best sensitivity (Table 3), reaching 97.5% when the POL method and the Baermann method (2 days) are considered. The POL method combined with a single test by the Baermann method has a sensitivity up to 92.5% (Table 3), which is not significantly different from that for testing 2 samples by the Baermann method plus the POL method (P > 0.005).
TABLE 2.
Sensitivities of the three diagnostic methodsa
| Method | % of positive samples (no./total) | % sensitivity in comparison to Baermann (day 1 and 2) positive result (95% CI) (n = 69) | % sensitivity in comparison to any positive test (95% CI) (n = 80) |
|---|---|---|---|
| Baermann, days 1 and 2 | 72.6 (69/95) | 86.3 (76.7–92.9)*° | |
| Baermann, day 1 | 50.5 (48/95) | 69.6 (57.3–80.0) | 60.0 (48.4–70.8)* |
| Baermann, day 2 | 53.7 (51/95) | 74.0 (62.0–84.0) | 63.8 (52.2–74.2)* |
| QIA | 26.3 (25/95) | 27.5 (17.5–39.6) | 31.3 (21.3–42.6)* |
| POL | 65.3 (62/95) | 77.0 (65.1–86.1) | 77.5 (77.0–86.0)*° |
Sensitivity was estimated on the basis of Baermann (collected on 2 days) method positivity and positivity detected by both the Baermann method (days 1 and 2) and PCR. *, statistically significant difference between the Baermann method (days 1 and 2 together and days 1 and 2 separately and QIA and between POL and the Baermann method (days 1 and 2) and the QIA method (P < 0.05); °, no statistically significant difference between the Baermann method (days 1 and 2) and the POL method (P > 0.05).
TABLE 3.
Sensitivity of the combination of Baermann collected on one and two samples and QIA and POL methodsa
| Method | % of positive samples (no./total) | % sensitivity in comparison to any positive test (95% CI) |
|---|---|---|
| Baermann, days 1 and 2, + QIA | 77.9 (74/95) | 93.7 (86.0–97.9) |
| Baermann, days 1 and 2, + POL | 82.0 (78/95) | 97.5 (91.3–99.7) |
| Baermann, day 1, + QIA | 61.0 (58/95) | 72.5 (61.4–81.9) |
| Baermann, day 2, + QIA | 65.3 (62/95) | 77.5 (66.8–86.1) |
| Baermann, day 1, + POL | 75.8 (72/95) | 90.0 (81.2–95.6) |
| Baermann, day 2, + POL | 65.2 (62/95) | 92.5 (84.4–97.2) |
Significant difference between one group of combinations (1-2-5) and the other group of combinations (3-4-6) (P < 0.05).
The median CT values were 36.2 (range, 31.1 to 39.8) and 28.6 (range, 17.3 to 39.5) for the QIA and POL methods, respectively. CT cutoff calculated based on the ROC curve estimated that the optimal threshold was at 30.5 for the POL method. With this definition, the POL method has a sensitivity of 58.0% in confirming Baermann method positivity and a specificity of 88.5% in confirming Baermann method negativity. Similar results were found by analyzing the copy numbers obtained by the extraction method in comparison to the Baermann method (supplemental material).
Figure 1 shows that even if the difference was not statistically significant, PCR-positive but microscopy-negative samples had lower DNA loads (i.e., higher CT values) than PCR-positive samples that were also microscopy positive (P = 0.08, Wilcoxon rank sum test) for both the QIA and POL methods.
FIG 1.
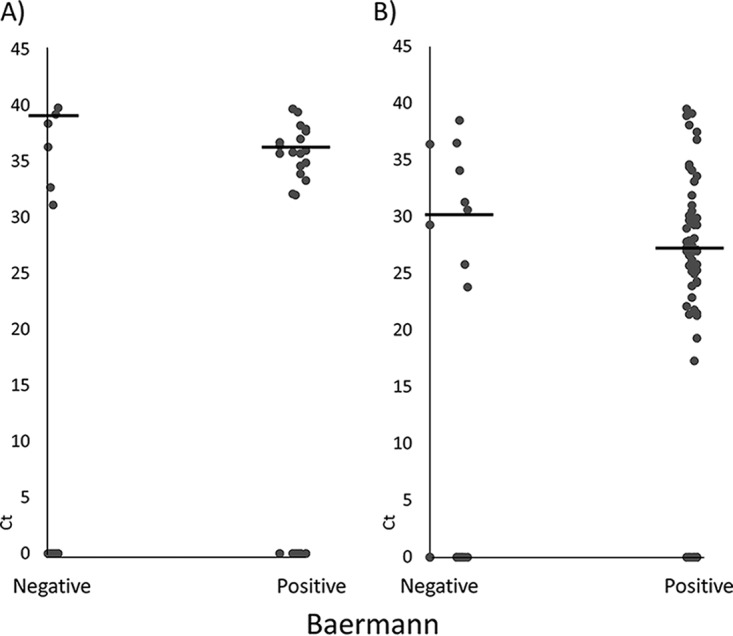
Distribution of CT values among Baermann method positive and negative tests. Microscopy tests were based on two stool samples from consecutive days, and two extraction methods, the QIA (A) and POL (B) methods, were applied to the stool sample for the first day. Horizontal lines represent the means of CT.
DISCUSSION
S. stercoralis infection is considered the most neglected of the neglected tropical diseases (1). Its worldwide prevalence is underestimated (2, 3), mainly due to an asymptomatic course of infection and low sensitivity of diagnostic methods (31, 32). Currently the examination of multiple samples with combined techniques is used to improve diagnostic power (32, 33).
Hence, there is a need for alternative, standardized methods. We evaluated one of the standard coprological methods (the Baermann method) with a molecular approach, using two different DNA extraction methods. Lately, several studies have been performed with PCR in order to find a suitable alternative for S. stercoralis detection, but to our knowledge there have been only a few studies comparing different DNA isolation techniques (21). Among these, Repetto et al. describe a modification to the standard QIAamp stool minikit method that performs better than the original protocol (25). The same group reported an in-house extraction method which was more sensitive than the commercial Qiagen kit (21).
If only 1 day of Baermann method testing is taken into consideration, the POL method is more sensitive (78% versus 60%), while the QIA method (31% versus 60 to 64%) shows lower sensitivity than the Baermann method. Moreover, we have seen that the combination of the POL method and 2 days of Baermann method testing reaches a high sensitivity, but also the POL method with only 1 day of Baermann method testing has a sensitivity of 90%. Hence, it would be feasible to increase sensitivity and shorten the sample collection with a combination of PCR (POL extraction method) and the Baermann method on a single stool sample. The stool amount analyzed by the Baermann method being around 50 to 100 times larger than for molecular methods further explains the observed discrepancies between the methods, as the probability of the presence of a larva is higher when more stool is analyzed.
Our results are in accordance with reports from elsewhere that PCR performs well using fecal samples and that no consecutive samples are needed to improve the power of the diagnostic technique (9, 25, 34). We found that the POL method revealed a significantly higher sensitivity (78% versus 31%) than the widely and traditionally used QIA method. The POL method is similar to the technique of helminth DNA extraction described by Verweij (35) except the missing bead mill procedure, which was not available in our laboratories at the time of the study. The POL method was the only alternative method (with good results for protozoans) known, which could be used without a cell lysing instrument (9, 18, 22, 36).
In the literature, results on the performance of the QIA extraction method are controversial, with some authors reporting a sensitivity of PCR similar to that of microscopy and others reporting lower or higher sensitivity for the PCR than for microscopy (9, 18, 37, 38). Surprisingly, we observed a low performance of the QIA method. As reported previously (18), one possible explanation for lower sensitivity of the QIA method might be the short lysis period (5 min at 95°C) not being sufficient to lyse helminth eggs and worms. In our samples, an external inhibition control detected S. stercoralis plasmid DNA well; therefore, we suppose that there are no stool-related substances that inhibit the reaction (18).
The observed difference in sensitivities of the combined methods is interesting. Both extraction methods in combination with day 1 and day 2 Baermann testing show a range of sensitivities, suggesting a variety in day-to-day larva output. This is a limitation of the microscopic technique, which loses power of detection if only one sample is taken into consideration, whereas molecular methods have relatively good sensitivity even for single-day detection. In this regard, it would have been relevant to compare 2 days of Baermann testing with 2 days of PCR analyses to confirm that PCR is not affected by larva output. Although this would be interesting, the performance of 2 PCRs is too expensive to be considered for field application. Another advantage of molecular diagnosis over the Baermann method is that a differentiation between hookworm and S. stercoralis larvae is sometime difficult. Since our study was embedded in a larger clinical trial, data obtained by the Kato-Katz technique were available and coinfections were thoroughly checked. Moreover, samples were analyzed shortly after collection in order to minimize the risk of hatching of hookworm eggs.
One limitation of our study is that larva counting and larva staging were not performed; therefore, a correlation between intensity of infection and CT values was not feasible. In previous studies, intensity of infection was, as expected, inversely proportional to CT values (12).
A long-debated aspect of molecular diagnostics is the CT cutoff: in the literature it is frequently set at 40 (17, 18, 37), but some authors set it at 35 (19) or do not mention it (9, 30, 34). A high CT cutoff often results in unspecific amplification, which is more likely at later real-time PCR cycles, and possible cross-contamination of highly positive samples. We calculated sensitivity using 40 as the CT cutoff, but considering the ROC curve between the Baermann and POL methods, we observed that a different threshold (30.5) might be more suitable for our study. On the other hand, if we adjusted the CT cutoff to 30.5, the sensitivity of both PCR methods would further decrease. This consideration demonstrates that more research is needed in order to reach standardization.
In conclusion, we found that the POL method outperforms the QIA method for S. stercoralis DNA detection in stool samples. We demonstrated that the combination of two diagnostic methods, with one sample tested by both the Baermann and POL methods, reaches a sensitivity of 90%, which is significantly higher than that of PCR or the Baermann method only. Our study confirmed that PCR has advantages over the Baermann method in terms of time to perform the method, possibility to standardize the protocol and to conduct multiple analyses with the same sample, and the small quantity of fecal material needed to perform the method.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the participants from 2 villages in the district of Pathoumphone, Champasack Province, Lao People's Democratic Republic, for providing the samples. We acknowledge the support of technical staff from the Centre for Malariology, Parasitology and Entomology, Department of Parasitology, University of Health Sciences, Ministry of Health, Vientiane, and the Champasack Provincial Malaria Station, who helped with the stool sample examinations; we also acknowledge the Champasack Provincial Health Department and Pathoumphone District Health Office for their collaboration and support. We are grateful to Ingrid Felger for providing the laboratories and equipment for PCR analysis.
J.K., B.B., and R.W. designed the study; B.B., R.W., S.S., K.P., S.X., and K.K. performed field work and sample analyses; C.S. performed statistical analysis; and B.B., J.K., and R.W. wrote the manuscript.
This study was supported by the European Research Council (ERC-2013-CoG 614739-A_HERO).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01941-17.
REFERENCES
- 1.Olsen A, van Lieshout L, Marti H, Polderman T, Polman K, Steinmann P, Stothard R, Thybo S, Verweij JJ, Magnussen P. 2009. Strongyloidiasis—the most neglected of the neglected tropical diseases? Trans R Soc Trop Med Hyg 103:967–972. doi: 10.1016/j.trstmh.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Schär F, Trostdorf U, Giardina F, Khieu V, Muth S, Marti H, Vounatsou P, Odermatt P. 2013. Strongyloides stercoralis: global distribution and risk factors. PLoS Negl Trop Dis 7:e2288. doi: 10.1371/journal.pntd.0002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisoffi Z, Buonfrate D, Montresor A, Requena-Méndez A, Muñoz J, Krolewiecki AJ, Gotuzzo E, Mena MA, Chiodini PL, Anselmi M, Moreira J, Albonico M. 2013. Strongyloides stercoralis: a plea for action. PLoS Negl Trop Dis 7:e2214. doi: 10.1371/journal.pntd.0002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buonfrate D, Mena MA, Angheben A, Requena-Mendez A, Muñoz J, Gobbi F, Albonico M, Gotuzzo E, Bisoffi Z, COHEMI Project Study Group . 2015. Prevalence of strongyloidiasis in Latin America: a systematic review of the literature. Epidemiol Infect 143:452–460. doi: 10.1017/S0950268814001563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genta RM. 1989. Global prevalence of strongyloidiasis: critical review with epidemiologic insights into the prevention of disseminated disease. Rev Infect Dis 11:755–767. doi: 10.1093/clinids/11.5.755. [DOI] [PubMed] [Google Scholar]
- 6.Mejia R, Nutman TB. 2012. Screening, prevention, and treatment for hyperinfection syndrome and disseminated infections caused by Strongyloides stercoralis. Curr Opin Infect Dis 25:458–463. doi: 10.1097/QCO.0b013e3283551dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montes M, Sawhney C, Barros N. 2010. Strongyloides stercoralis: there but not seen. Curr Opin Infect Dis 23:500–504. doi: 10.1097/QCO.0b013e32833df718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tachamo N, Nazir S, Lohani S, Karmacharya P. 2016. Strongyloidiasis in the immunocompetent: an overlooked infection. J Community Hosp Intern Med Perspect 6:32038. doi: 10.3402/jchimp.v6.32038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker SL, Piraisoody N, Kramme S, Marti H, Silué KD, Panning M, Nickel B, Kern WV, Herrmann M, Hatz CF, N′Goran EK, Utzinger J, von Müller L. 2015. Real-time PCR for detection of Strongyloides stercoralis in human stool samples from Côte d'Ivoire: diagnostic accuracy, inter-laboratory comparison and patterns of hookworm co-infection. Acta Trop 150:210–217. doi: 10.1016/j.actatropica.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Knopp S, Mgeni AF, Khamis IS, Steinmann P, Stothard JR, Rollinson D, Marti H, Utzinger J. 2008. Diagnosis of soil-transmitted helminths in the era of preventive chemotherapy: effect of multiple stool sampling and use of different diagnostic techniques. PLoS Negl Trop Dis 2:e331. doi: 10.1371/journal.pntd.0000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anamnart W, Pattanawongsa A, Intapan PM, Maleewong W. 2010. Factors affecting recovery of Strongyloides stercoralis larvae: an approach to a newly modified formalin-ether concentration technique for diagnosis of strongyloidiasis. J Clin Microbiol 48:97–100. doi: 10.1128/JCM.01613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ketzis JK. 2017. Limitations to the adoption of a standardized Strongyloides stercoralis diagnostic method: case study in the Caribbean. Acta Trop 170:178–183. doi: 10.1016/j.actatropica.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Bisoffi Z, Buonfrate D, Sequi M, Mejia R, Cimino RO, Krolewiecki AJ, Albonico M, Gobbo M, Bonafini S, Angheben A, Requena-Mendez A, Muñoz J, Nutman TB. 2014. Diagnostic accuracy of five serologic tests for Strongyloides stercoralis infection. PLoS Negl Trop Dis 8:e2640. doi: 10.1371/journal.pntd.0002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Formenti F, Buonfrate D, Prandi R, Marquez M, Caicedo C, Rizzi E, Guevara AG, Vicuña Y, Huerlo FR, Perandin F, Bisoffi Z, Anselmi M. 2016. Comparison of S. stercoralis serology performed on dried blood spots and on conventional serum samples. Front Microbiol 7:1778. doi: 10.3389/fmicb.2016.01778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schär F, Hattendorf J, Khieu V, Muth S, Char MC, Marti HP, Odermatt P. 2014. Strongyloides stercoralis larvae excretion patterns before and after treatment. Parasitology 141:892–897. doi: 10.1017/S0031182013002345. [DOI] [PubMed] [Google Scholar]
- 16.Page WA, Dempsey K, McCarthy JS. 2006. Utility of serological follow-up of chronic strongyloidiasis after anthelminthic chemotherapy. Trans R Soc Trop Med Hyg 100:1056–1062. doi: 10.1016/j.trstmh.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Verweij JJ, Canales M, Polman K, Ziem J, Brienen EAT, Polderman AM, van Lieshout L. 2009. Molecular diagnosis of Strongyloides stercoralis in faecal samples using real-time PCR. Trans R Soc Trop Med Hyg 103:342–346. doi: 10.1016/j.trstmh.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Knopp S, Salim N, Schindler T, Karagiannis Voules DA, Rothen J, Lweno O, Mohammed AS, Singo R, Benninghoff M, Nsojo AA, Genton B, Daubenberger C. 2014. Diagnostic accuracy of Kato-Katz, FLOTAC, Baermann, and PCR methods for the detection of light-intensity hookworm and Strongyloides stercoralis infections in Tanzania. Am J Trop Med Hyg 90:535–545. doi: 10.4269/ajtmh.13-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llewellyn S, Inpankaew T, Nery SV, Gray DJ, Verweij JJ, Clements ACA, Gomes SJ, Traub R, McCarthy JS. 2016. Application of a multiplex quantitative PCR to assess prevalence and intensity of intestinal parasite infections in a controlled clinical trial. PLoS Negl Trop Dis 10:e0004380. doi: 10.1371/journal.pntd.0004380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Incani RN, Ferrer E, Hoek D, Ramak R, Roelfsema J, Mughini-Gras L, Kortbeek T, Pinelli E. 2017. Diagnosis of intestinal parasites in a rural community of Venezuela: advantages and disadvantages of using microscopy or RT-PCR. Acta Trop 167:64–70. doi: 10.1016/j.actatropica.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Repetto SA, Alba Soto CD, Cazorla SI, Tayeldin ML, Cuello S, Lasala MB, Tekiel VS, González Cappa SM. 2013. An improved DNA isolation technique for PCR detection of Strongyloides stercoralis in stool samples. Acta Trop 126:110–114. doi: 10.1016/j.actatropica.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Sultana Y, Jeoffreys N, Watts MR, Gilbert GL, Lee R. 2013. Real-time polymerase chain reaction for detection of Strongyloides stercoralis in stool. Am J Trop Med Hyg 88:1048–1051. doi: 10.4269/ajtmh.12-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharifdini M, Mirhendi H, Ashrafi K, Hosseini M, Mohebali M, Khodadadi H, Kia EB. 2015. Comparison of nested polymerase chain reaction and real-time polymerase chain reaction with parasitological methods for detection of Strongyloides stercoralis in human fecal samples. Am J Trop Med Hyg 93:1285–1291. doi: 10.4269/ajtmh.15-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saugar JM, Merino FJ, Martín-Rabadán P, Fernández-Soto P, Ortega S, Gárate T, Rodríguez E. 2015. Application of real-time PCR for the detection of Strongyloides spp. in clinical samples in a reference center in Spain. Acta Trop 142:20–25. doi: 10.1016/j.actatropica.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 25.Repetto SA, Ruybal P, Solana ME, López C, Berini CA, Alba Soto CD, Cappa SMG. 2016. Comparison between PCR and larvae visualization methods for diagnosis of Strongyloides stercoralis out of endemic area: a proposed algorithm. Acta Trop 157:169–177. doi: 10.1016/j.actatropica.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Barda B, Sayasone S, Phongluxa K, Xayavong S, Keoduangsy K, Odermatt P, Puchkov M, Huwyler J, Hattendorf J, Keiser J. 2017. Efficacy of moxidectin versus ivermectin against Strongyloides stercoralis infections: a randomized controlled non-inferiority trial. Clin Infect Dis 65:276–281. doi: 10.1093/cid/cix278. [DOI] [PubMed] [Google Scholar]
- 27.García L, Bruckner D. 2001. Diagnostic medical parasitology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 28.Polley SD, Boadi S, Watson J, Curry A, Chiodini PL. 2011. Detection and species identification of microsporidial infections using SYBR Green real-time PCR. J Med Microbiol 60:459–466. doi: 10.1099/jmm.0.026781-0. [DOI] [PubMed] [Google Scholar]
- 29.Basuni M, Muhi J, Othman N, Verweij JJ, Ahmad M, Miswan N, Rahumatullah A, Aziz FA, Zainudin NS, Noordin R. 2011. A pentaplex real-time polymerase chain reaction assay for detection of four species of soil-transmitted helminths. Am J Trop Med Hyg 84:338–343. doi: 10.4269/ajtmh.2011.10-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dacal E, Saugar JM, Soler T, Azcárate JM, Jiménez MS, Merino FJ, Rodríguez E. 23 January 2017. Parasitological versus molecular diagnosis of strongyloidiasis in serial stool samples: how many? J Helminthol doi: 10.1017/S0022149X17000050. [DOI] [PubMed] [Google Scholar]
- 31.de Kaminsky RG. 1993. Evaluation of three methods for laboratory diagnosis of Strongyloides stercoralis infection. J Parasitol 79:277–280. doi: 10.2307/3283519. [DOI] [PubMed] [Google Scholar]
- 32.Steinmann P, Zhou X-N, Du Z-W, Jiang J-Y, Wang L-B, Wang X-Z, Li L-H, Marti H, Utzinger J. 2007. Occurrence of Strongyloides stercoralis in Yunnan Province, China, and comparison of diagnostic methods. PLoS Negl Trop Dis 1:e75. doi: 10.1371/journal.pntd.0000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khieu V, Srey S, Schär F, Muth S, Marti H, Odermatt P. 2013. Strongyloides stercoralis is a cause of abdominal pain, diarrhea and urticaria in rural Cambodia. BMC Res Notes 6:200. doi: 10.1186/1756-0500-6-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amor A, Rodriguez E, Saugar JM, Arroyo A, López-Quintana B, Abera B, Yimer M, Yizengaw E, Zewdie D, Ayehubizu Z, Hailu T, Mulu W, Echazú A, Krolewieki AJ, Aparicio P, Herrador Z, Anegagrie M, Benito A. 2016. High prevalence of Strongyloides stercoralis in school-aged children in a rural highland of north-western Ethiopia: the role of intensive diagnostic work-up. Parasit Vectors 9:617. doi: 10.1186/s13071-016-1912-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verweij JJ. 2014. Application of PCR-based methods for diagnosis of intestinal parasitic infections in the clinical laboratory. Parasitology 141:1863–1872. doi: 10.1017/S0031182014000419. [DOI] [PubMed] [Google Scholar]
- 36.Kaisar MMM, Brienen EAT, Djuardi Y, Sartono E, Yazdanbakhsh M, Verweij JJ, Supali T, Van Lieshout L. 2017. Improved diagnosis of Trichuris trichiura by using a bead-beating procedure on ethanol preserved stool samples prior to DNA isolation and the performance of multiplex real-time PCR for intestinal parasites. Parasitology 144:965–974. doi: 10.1017/S0031182017000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong MD, Karsenti N, Lau R, Ralevski F, Cheema K, Burton L, Klowak M, Boggild AK. 2016. Strongyloidiasis in Ontario: performance of diagnostic tests over a 14-month period. Travel Med Infect Dis 14:625–629. doi: 10.1016/j.tmaid.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Meurs L, Polderman AM, Vinkeles Melchers NVS, Brienen EAT, Verweij JJ, Groosjohan B, Mendes F, Mechendura M, Hepp DH, Langenberg MCC, Edelenbosch R, Polman K, van Lieshout L. 2017. Diagnosing polyparasitism in a high-prevalence setting in Beira, Mozambique: detection of intestinal parasites in fecal samples by microscopy and real-time PCR. PLoS Negl Trop Dis 11:e0005310. doi: 10.1371/journal.pntd.0005310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


