Abstract
Vesicular trafficking events play key roles in the compartmentalization and proper sorting of cellular components. These events have crucial roles in sensing external signals, regulating protein activities and stimulating cell growth or death decisions. Although mutations in vesicle trafficking players are not direct drivers of cellular transformation, their activities are important in facilitating oncogenic pathways. One such pathway is the sensing of external stimuli and signalling through receptor tyrosine kinases (RTKs). The regulation of RTK activity by the endocytic pathway has been extensively studied. Compelling recent studies have begun to highlight the association between autophagy and RTK signalling. The influence of this interplay on cellular status and its relevance in disease settings will be discussed here.
Keywords: autophagy, cancer, endocytosis, receptor tyrosine kinases, signalling
Introduction
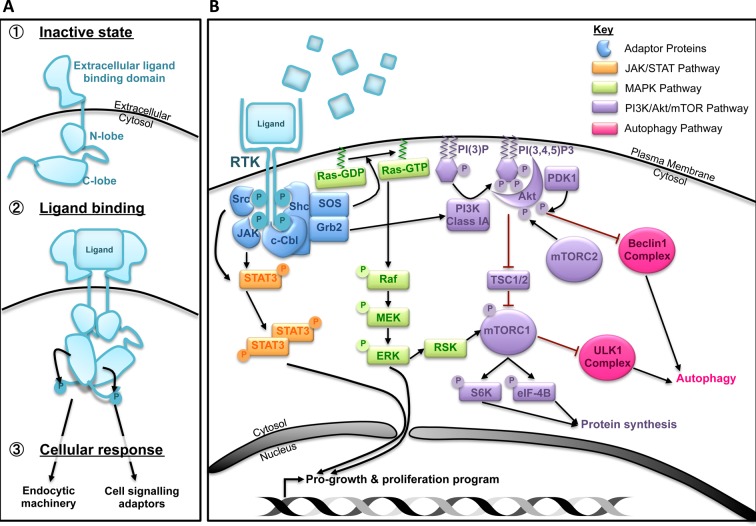
Mammalian cells have acquired a diverse and exquisitely fine-tuned set of plasma membrane receptors that sense changes in the extracellular milieu. One such family of receptors is the receptor tyrosine kinases (RTKs) whose activities influence various cellular processes including proliferation, survival, differentiation and motility [1]. Approximately 58 RTKs have been identified to date and their activities are tightly regulated at multiple stages. Upon ligand binding, dimerized or oligomerized receptors undergo conformational changes resulting in their cross-phosphorylation and activation [1]. Adaptor proteins recognize these phosphorylated residues to co-ordinate downstream intracellular signalling and subsequent endocytic trafficking of RTKs either back to the plasma membrane or to lysosomes for degradation. Active RTKs trigger a plethora of signalling cascades, including the mitogen-activated protein kinase (MAPK), Janus kinase/signal transducer and activator of transcription (JAK/STAT) and phosphoinositol-3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathways (Figure 1) [2]. The role of RTKs in promoting growth and survival is reinforced by the observation that the deregulation of several RTKs (including epidermal growth factor receptor, EGFR, platelet-derived growth factor receptor, PDGFR, insulin-like growth factor-1 receptor, IGF-1R, and hepatocyte growth factor receptor, HGFR or Met) is associated with a number of pathological conditions including cancer.
Figure 1. RTKs signalling overview.
(A) Ligand binding and receptor activation induce downstream signalling and endocytic sorting. (B) A simplified representation of common signalling pathways activated upon RTK stimulation. The outcome of these signals can stimulate cell growth and proliferation through transcriptional and translational activation.
Macroautophagy (herein, referred to as autophagy) is a vesicular trafficking pathway that delivers cellular components to the lysosome system for degradation [3]. Autophagy involves the formation of a lipid bilayer or phagophore, which engulfs cytoplasmic materials and matures to form the autophagosome. Biochemically, autophagy requires the conjugation of ubiquitin-like proteins (the ATG8 family members) to a lipid moiety that anchors them to membranes and facilitates their role in autophagosome maturation and cargo recognition. ATG3, ATG7 and the ATG12–ATG5–ATG16L1 complex can directly mediate this lipid conjugation reaction. Further upstream, the unc-51-like kinase 1 (ULK1) and Vps34 lipid kinase complexes act to relay signals to initiate autophagy. While autophagy can occur constitutively at basal levels, a number of stimuli can up-regulate autophagy including nutrient and energy deprivation, genotoxic response and pathogen infection. Conversely, autophagy is suppressed by nutrient-rich conditions and growth factor availability, which drive cellular proliferation, thereby highlighting the relevance of autophagy as a stress response mechanism.
Cells have developed a variety of methods to integrate RTK activity with autophagic flux, which can provide a switch between catabolism required during times of nutrient scarcity and anabolism when conditions are permissive for cell growth and proliferation. We are coming to understand that the regulation of autophagy and RTK signalling are closely interconnected as both share common signalling and vesicular trafficking pathways. This is an exciting and developing area of research, the results of which show that the interplay of RTKs and autophagy has far-reaching consequences for fundamental cell biology and disease development.
Commonalities of autophagosomal biogenesis and endosomal trafficking
Organelle biogenesis can occur through either the de novo synthesis of new structures or the excision and maturation of precursor structures. While endocytic vesicles bud off precursor structures and mature through the recruitment of small Rab GTPases, autophagosomes require the de novo synthesis of lipid double bilayers originating from various sources of membranes. Multiple cellular origins can contribute to autophagosome membranes including the ER, Golgi apparatus, mitochondria and plasma membrane [3].
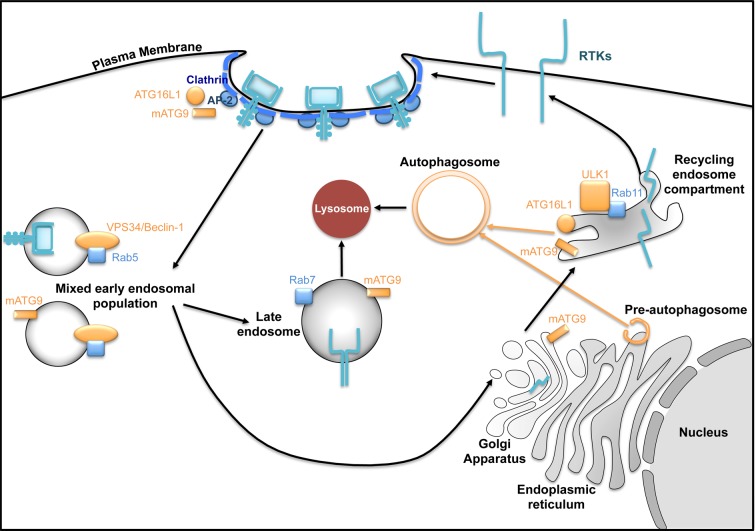
A series of recent studies have uncovered a role for the endocytic pathway in facilitating the nucleation and maturation of preautophagosome structures [4]. For example, the autophagy players ATG9 and ATG16L1 can both interact with the AP-2 adaptor protein and undergo distinct clathrin-mediated endocytosis from the plasma membrane to Rab11-positive recycling endosomes [5]. During autophagy, the ULK1 kinase, which can also localize to Rab11 endosomes, phosphorylates ATG9 and potentiates its redistribution to preautophagosome structures [6,7]. Subsequently, ATG9- and ATG16L1-containing recycling endosomes fuse requiring the activity of VAMP3 [8]. The importance of the endocytic pathway in autophagy is reinforced by the observation that the inhibition of the RTK IGF-1R restricted autophagosome formation potentially through attenuating clathrin-mediated endocytosis and the interaction of ATG16L1 with clathrin heavy chain [9]. Furthermore, the ATG8 family of ubiquitin-like proteins have been reported to interact with a number of the Tre2, Bub2 and Cdc16 (TBC)-1 domain-containing family members which act as GTPase-activating proteins (GAPs) to small Rab GTPases involved in endocytosis [10]. The functional relevance of many of these interactions is starting to unfold. A recent study shows that the interaction between LC3 and TBC1D5, a component of the retromer which plays an important role in the recycling of endocytosed proteins to the Golgi network or plasma membrane, is required for the recycling of the glucose transporter GLUT1/Slc2a1 to the cell surface [11]. TBC1D5 also interacts with ATG9 and, together with AP-2, is required for the proper sorting of ATG9 into autophagosome precursors during autophagy [12]. In addition to contributing to autophagosome biogenesis, the endocytic pathway is also important for autophagosome-lysosome fusion (reviewed elsewhere [4]).
Collectively, these studies indicate associations between autophagy components and the endocytic pathway that influence their mutual activities (Figure 2). Given that the endocytic pathway tightly regulates RTK signalling and the ability of endosomal compartments to contribute to the membrane origin of autophagosomes, it is plausible that RTKs and autophagy components can cross-talk. Current studies are beginning to uncover such commonalities and how they influence these processes.
Figure 2. The endolysosomal system and its connections to RTKs and autophagy.
Following ligand binding, RTKs can undergo clathrin-mediated endocytosis. These vesicles mature into early endosomal populations and can be sorted into either late endosome-lysosome compartment for degradation or recycling endosomes and delivered back to plasma membrane. Autophagy-related proteins overlap with many of these endosomal compartments which are required for autophagosome biogenesis and lysosomal fusion.
RTK signalling regulates autophagy
A wealth of studies has demonstrated the influence of RTK signalling on autophagy regulation with the underlying molecular mechanisms still requiring further investigation. While stimulation of some RTKs (including EGFR, Her2 and FGFR1) have been shown to inhibit autophagy, ligand activation of others (such as Axl, ErbB3/ErbB4, TrkA, Ephrin and VEGFR) can promote autophagy [13–19]. Here, we will delve into the regulation of individual players of the autophagy machinery by RTK signalling of which the best studied are the ULK1 and Beclin-1 complexes.
The ULK1 complex
The ULK1 complex plays a central role in sensing and relaying signals to regulate autophagy. The ULK1 kinase activity is regulated by various post-translational modifications as well as by binding to adapter proteins, including ATG13, FIP200 and ATG101 [20]. The most studied post-translational modification occurs via mTOR complex 1 (mTORC1), which senses changes in nutrients and oxygen availability, growth factors and genotoxic stress. RTKs activate signalling through the PI3K/Akt/mTOR axis and can suppress autophagy through direct inhibitory phosphorylation of ULK1 and ATG13 by mTORC1 [20]. mTORC1 can also indirectly destabilize ULK1 via the phosphorylation of AMBRA1, which inhibits the interaction between AMBRA1 and the E3 ligase TRAF6 thereby preventing the addition of stabilizing Lys63 ubiquitin chains to ULK1 [21]. The activation of mTORC1 by numerous RTKs, e.g. during IGF-1 and FGF stimulation, suppresses autophagy presumably through ULK1 [22,23]. Given that autophagy can proceed in the absence of ULK1 activity, it is possible that RTKs also regulate autophagy through the modulation of downstream autophagy players.
The Beclin-1 complex
The identities of ULK1 kinase substrates required for autophagy induction are only recently starting to unfold. One such substrate is Beclin-1 which exists in two distinct assemblies: the autophagy-specific ATG14L1/AMBRA1/Beclin-1/Vps34 complex and the endocytic modulator UVRAG/Beclin-1/Vps34 [24]. The autophagy-specific complex is required for the lipid kinase activity of Vps34 to produce PtdIns3P (PI3P) and the subsequent recruitment of PI3P sensors, including DFCP1 and WIPI proteins, to thereby initiate autophagy. Phosphorylation of Beclin-1 on Ser14 by ULK1 is required for the activity of Vps34 during autophagy [25]. In addition to regulating Beclin-1 through ULK1, mTOR can also destabilize Beclin-1 through an unknown mechanism involving the mTORC1 and 2 components Raptor and Rictor [26]. Enhanced stability of Beclin-1 has also been observed upon genetic ablation of certain RTKs, such as FGFR [27]. However, whether this requires the deactivation of mTORC1 has not been shown. More directly, Akt has been shown to phosphorylate Beclin-1 on Ser234 and Ser295 resulting in its inhibitory interaction with the cytoskeletal components vimentin and 14-3-3 [28]. Overall, these studies show that Beclin-1 activities can be suppressed via PI3K/Akt/mTOR signalling.
Multiple-activated RTKs have been shown to inhibit autophagy through regulating Beclin-1 [13,27,29]. Of these, only EGFR has been shown to directly phosphorylate Beclin-1 [13]. Upon EGF stimulation, EGFR binds to and phosphorylates Beclin-1 on multiple sites thereby recruiting its inhibitor Rubicon and displacing Vps34. Introducing acidic mutations in Beclin-1 that mimic its phosphorylation by EGFR results in autophagy suppression and enhanced tumorigenesis. The enhanced binding between Beclin-1 and Rubicon during EGF stimulation may conversely regulate EGFR by relieving Rab7 from its inhibitory interaction with Rubicon thus promoting the lysosomal degradation of EGFR [30]. Inactive EGFR can also modulate autophagy. In the absence of ligand stimulation, EGFR forms a complex with the oncoprotein LAMPTM4B and the exocyst component Sec5 which sequesters Rubicon from its inhibitory effects on Beclin-1 and thereby initiates autophagy [31]. Treatment with the EGFR inhibitors erlotinib or gefitinib enhances the association of EGFR with Sec5 and promotes autophagy. Altogether, these studies suggest that active and inactive EGFR can exert opposing effects on the autophagy pathway through regulating Beclin-1 activity.
Further regulation of the Beclin-1 complex during RTK signalling can occur through regulation of Vps34 lipid kinase activity by Rab5. This regulation requires class IA PI3K which produces triphosphate PtdIns(3,4,5)P3 and facilitates the activation of Akt in response to RTK activation [32]. In the absence of growth factors, the catalytic subunit of class IA PI3K, p110β, associates with Rab5 and stabilizes its GTP-bound form which in turn stimulates Vps34-driven autophagy [32]. This suggests that RTK signalling can suppress multiple members of the Beclin-1 complex ensuring only basal levels of autophagy.
Transcriptional regulation of the autophagy machinery
As well as rapidly regulating autophagy via post-translational protein modifications, RTKs exert a sustained control over the pathway by modulating the transcription of several autophagy-related genes. RTK-mediated signalling cascades lead to the modulation of transcription factors, such as transcription factor EB (TFEB), the forkhead transcription factors (FoxO) and STAT3 that have been shown to regulate lysosomal and autophagy gene expression. Phosphorylation of STAT3 on Tyr705, required for its transcriptional activity, is catalysed directly by some RTKs such as VEGF, c-Met and EGFR [33]. Once in the nucleus, STAT3 exerts an autophagy-inhibitory function by down-regulating Vps34 and Beclin-1 expression. Although inhibition of autophagy has also been observed by cytoplasmic and mitochondrial STAT3, autophagy-stimulatory effects of nuclear STAT3 have also been documented. STAT3 can enhance the expression of HIF1A and BNIP3 which promote autophagy through up-regulation of autophagy-related gene expression during hypoxia and relief of Bcl2-mediated inhibition of Beclin-1 respectively (reviewed elsewhere) [33]. On the other hand, the activities of TFEB and FoxO are suppressed by RTK signalling pathways including mTORC1, Akt and ERK [34–37]. Growth factor withdrawal or pharmacological inhibition of RTK signalling leads to the nuclear translocation of TFEB and FoxO and thereby increased expression of lysosomal and autophagy genes. This may have a therapeutic benefit, for example by stimulating the clearance of cellular aggregates in lysosomal storage disorders [38]. Of note, many RTKs can also translocate to the nucleus where they directly influence the activities of transcriptional factors (for example activation of STAT3 by directly binding to EGFR) [39]. However, the affect of nuclear RTKs on autophagy-related gene expression has not yet been determined. Overall, the transcriptional regulation of autophagy by RTK signalling reinforces the long-term suppression of autophagy by these receptors.
Co-operation between autophagy and RTKs
The multitude of connections between the autophagy and RTK pathways is garnering interest regarding the influence of autophagy on RTK signalling. The connections of several RTKs are beginning to be explored with receptors such as VEGFR, c-Met, c-Ret, EGFR and DTK relying on autophagy for their optimal activities [40,41]. Whilst many of these connections are beginning to unfold, the mechanistic focus has rested on c-Met and EGFR signalling, which will be discussed here.
c-Met and adhesion signalling
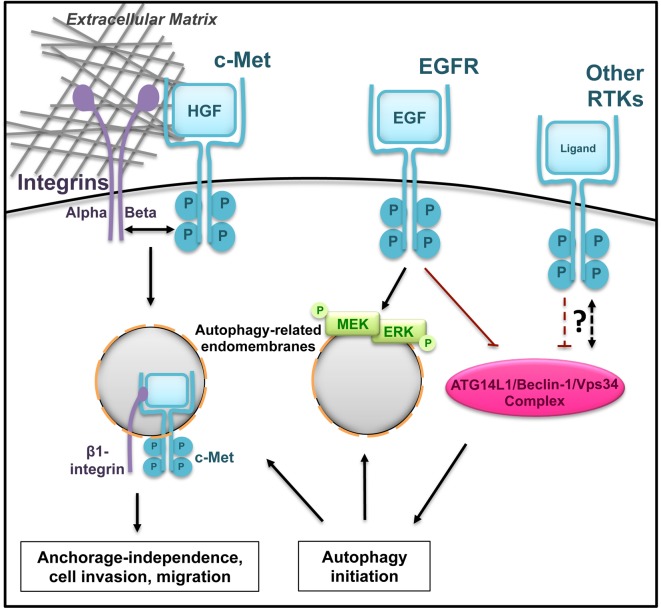
To prevent inappropriate growth when not anchored at their functional site, normal cells engage in a type of cell death called anoikis. To avoid anoikis, cancer cells modulate various cellular pathways including inhibition of cell death regulators, elevation of RTK and integrin signalling and induction of autophagy. Enhanced RTK/integrin signalling and autophagy can stimulate metabolism and maintain cellular energy status to promote growth and survival [42]. During detachment or ligand activation, internalized c-Met, in co-operation with β1 integrin, promotes survival and oncogenic growth in an autophagy-dependent manner [43]. Activated c-Met localizes on LC3-positive vesicles potentially recruiting ERK1/2 (Figure 3). Interestingly, this signalling platform requires the expression of the autophagy players ATG5 and Beclin-1 but not ATG13, suggesting that these structures may differ from canonical autophagosomes and may override RTK-mediated suppression of autophagy via the ULK1 complex. Autophagy also promotes the phosphorylation of c-Met as well as other RTKs in colorectal cancer cell lines implying that autophagy regulates RTK activity further upstream [40].
Figure 3. Cross-talk between RTK signalling and autophagy.
RTKs and autophagy can mutually regulate their activities. The RTK c-Met co-operates with integrin complexes and localizes to autophagy-related endomembranes to stimulate signalling and cell growth. MEK and ERK also utilize autophagic membranes to promote signalling upon EGF stimulation. Inhibition of autophagy by RTKs has also been reported (shown is the inhibitory phosphorylation of Beclin-1 by EGFR) which provides a negative feedback mechanism. The association of between other RTKs and autophagy have recently started to uncover.
EGFR signalling
Despite the lack of evidence for EGFR recruitment to autophagic membranes [40], functional co-operation has been reported (Figure 3). Singh et al. documented that autophagy is required for optimal MAPK signalling during EGF stimulation [44]. Interestingly, MEK and ERK localized to the cytosolic face of autophagosomes and immunoprecipitated with LC3. Whether EGFR also localizes on these autophagic membranes was not addressed in this study. Furthermore, autophagy can positively regulate EGFR by promoting its stability through the non-RTK, Ack1 [45]. In the absence of ligand binding, Ack1 interacts with autophagy receptors, including p62 and NBR1, within ubiquitin-rich domains. Upon EGF stimulation, Ack1 is displaced from these ubiquitin-rich structures and localizes to early endosomes where it can divert active EGFR from lysosomal degradation. These endosomal compartments are also partially positive for ATG16L1 suggesting that autophagy players can promote oncogenesis by regulating the localization and activation of EGFR [45,46]. The facilitation of EGFR signalling by autophagy may be contradicted by the EGF-mediated suppression of autophagy (described above) providing a negative feedback regulation of this RTK.
In contrast, the Beclin-1 complex has been shown to down-regulate EGFR signalling by promoting PI3P production and the maturation of APPL-positive early endosomes in breast cancer cell lines [47]. By prolonging the residence of EGFR at early endosomes, the absence of Beclin-1 results in sustained Akt/ERK signalling during EGF stimulation. Similarly, genetic manipulations of autophagy-related protein levels (including ATG7 and ATG9A) show that higher levels of autophagy proteins correlate with reduced stability of EGFR or its family member Erb2/Her2 in the presence of their respective inhibitors [48,49]. Whether autophagic flux directly contributes to the degradation of these RTKs has not been fully explored. The opposing findings regarding the regulation of EGFR signalling by autophagy suggest a complex cross-talk between the two pathways that is likely to be context dependent.
The affect of autophagy-RTK cross-talk on health and disease
The interaction between autophagy and RTK signalling is implicated in various physiological and pathological conditions (Table 1). In recent years, much interest has focused on the therapeutic potential of manipulating this connection by targeting one or both the pathways to alleviate the disease. Here we explore the role of RTK-autophagy co-operation in a variety of disease settings and their potential therapeutic targeting.
Table 1. The co-operation between autophagy and RTKs in disease models.
| RTK | Interplay with autophagy | Physiological affect of autophagy-RTK interplay | References |
|---|---|---|---|
| c-Met | Autophagic membranes facilitate c-Met signalling | Enhances tumour growth and metastasis in mouse and zebrafish xenograft models | Barrow-McGee et al. (2016) [43] |
| Autophagy positively regulates c-Met phosphorylation | Promotes cell migration in cultured colorectal cancer cell lines | Lampada et al. (2017) [40] | |
| Epidermal growth factor receptor (EGFR) | EGFR phosphorylates and inhibits Beclin-1 | Beclin-1 phosphorylation increases tumour growth and resistance to EGFR inhibitors in NSCLC xenograft model | Wei et al. (2013) [13] |
| Fms-like tyrosine kinase 3 harbouring internal tandem duplication (FLT3-ITD) | Autophagy promotes FLT3-ITD degradation in the presence of proteasome inhibitors | Autophagy enhances cell death during bortezomib treatment in acute myeloid leukaemia (AML) cells | Larrue et al. (2016 ) [68] |
| Daf-2 (homologue of IGFR) | Mutation in daf-2 induces autophagy in aged worms | Daf-2 mutation extends lifespan in warms in an autophagy-dependent manner | Chang et al. (2017) [83] |
| Axl | Axl signalling induces autophagy and inhibited inflammasome maturation | Axl signalling suppresses LPS- and CCI4-induced acute liver injury in mice but contribution of autophagy not tested | Han et al. (2016) [18] |
| EPHA4 and PDGFR family members | Not studied | Maintenance of neuronal health but relevance of autophagy not studied | Van Hoecke et al. (2012) [73] |
| Hebron et al. (2015) [75] |
A summary of how the co-operation between autophagy and RTK signalling impacts disease and treatment outcome.
Cancer
Oncogenic mutations resulting in RTK overexpression and/or constitutive activation are associated with many cancers [1]. RTKs are therefore considered ideal target candidates for cancer therapy. Yet, despite promising initial antitumour response to small molecule RTK inhibitors (TKIs) and anti-RTK monoclonal antibodies, subsequent tumour relapse has been a recurring problem over the last two decades. Resistance to TKIs can be due to a combination of acquired mutations, kinase-independent activities of these RTKs or activation of survival response pathways such as autophagy [50–54]. The combined use of autophagy antagonists and RTK inhibitors show improved response to treatment in cell culture and xenograft models of solid tumours including breast cancer, ovarian cancer, non-small-cell lung cancer and neuroblastoma [55–59]. Tissue culture and mouse modelling studies also show that genetic inhibition of autophagy suppresses MAPK/Akt/mTOR signalling which correlates with reduced tumour growth thereby suggesting that attenuating autophagy may mirror RTK inhibition [60,61].
Autophagy activation during TKI treatment may occur following the relief of RTK-mediated inhibition of autophagy, as discussed above or as a consequence of cell death induction or altered metabolism [54,62]. The mechanism of how autophagy prevents drug-induced cell death and contributes to drug resistance remains largely unknown. In the context of RTK signalling, it has been shown that treatment of patients with one RTK inhibitor can result in the amplification of another receptor [63]. Here, the ability of autophagy to facilitate RTK signalling may provide one mechanism through which it contributes to drug resistance. For instance, a subset of colorectal cancer patients who developed resistance to anti-EGFR monoclonal antibody therapy harbour amplification of Met [64]. The role of autophagy in supporting c-Met signalling suggests that its targeting may be beneficial during combinational therapy. Alternatively, the influence of autophagy activation on cell survival may occur indirectly through maintenance of cellular homoeostasis (e.g. clearance of damaged organelles such as mitochondria as a result of chemotherapy treatment), the absence of which can be catastrophic [65].
A growth suppressive function of autophagy during RTK inhibition has also been reported [66,67]. Phosphorylation of Beclin-1 by EGFR inhibits autophagy and promotes tumour growth and resistance to erlotinib treatment [13]. Autophagy-mediated degradation of the Fms-like tyrosine kinase 3 harbouring internal tandem duplication (FLT3-ITD) can suppress tumorigenesis during treatment with the proteasome inhibitor bortezomib in an acute myeloid leukaemia (AML) model [68]. Autophagy may also engage in supporting cell death induction where it can potentially mediate caspase-independent forms of cell death or regulate cell death machinery [69]. In cases of RTKs with potential tumour suppression functions (including EphB2 and TrkA), it has been shown that receptor activation is associated with enhanced autophagy and non-apoptotic cell death [15,17]. The molecular mechanism underlying autophagy induction by these RTKs or what distinguishes tumour suppressive from tumour promoting autophagy requires further investigation.
Conclusions derived from combined autophagy and RTK inhibition studies are complicated by the fact that many cell culture and xenograft models are limited by the use of homogeneous cell clones whereas most solid tumours are heterogeneous. This suggests that the combined inhibition of RTK and autophagy may produce an intratumoural heterogeneous response. Additionally, clinically approved compounds that inhibit or activate autophagy are not specific therefore it is possible that autophagy-independent mechanisms contribute to the response of cells to TKIs. Furthermore, inhibiting autophagy at different stages (initiation compared with lysosomal degradation) may exert opposing effects on tumour cells. With such discrepancies considered, it is likely that different cancers will vary in their response to combined inhibition of RTK and autophagy. As early phase clinical trials are currently undergoing (including combinations of hydroxychloroquine along TKIs such as sunitinib and erlotinib in solid tumours), it is yet to be determined whether our current understanding of the biological relevance of autophagy and RTK inhibition in cancer treatment will benefit clinical outcome [70].
Other models
In contrast with the potential benefit of inhibiting autophagy in some cancers, enhanced autophagy may constitute a health benefit in alternative diseases. One example is the autophagy homoeostatic function required for neuronal health largely mediated through the clearance of protein aggregates [71]. While the treatment with non-RTK inhibitors and the associated clearance of aggregates by autophagy has yielded promising results in neurodegenerative disease models, the intriguing possibility of repurposing RTK inhibitors to induce autophagy in these settings has not been fully explored [72–75]. Similarly, RTK inhibition has been used to promote selective degradation of microbes by autophagy (known as xenophagy) [76]. Autophagy stimulation during Mycobacterium tuberculosis infection limits the expansion of the bacterium in macrophages [77]. Treatment with the EGFR inhibitor gefitinib activated autophagy and partially limited M. tuberculosis infection in mice [78]. Several additional RTK inhibitors have been shown to restrict pathogen infection but whether their action is mediated by autophagy remains to be investigated to determine the benefits of repurposing TKIs to activate autophagy in specific disease settings [79–82].
Autophagy is considered to be beneficial in delaying the onset of aging. In a Caenorhabditis elegans model for extended lifespan induced by mutating daf-2, a homologue of mammalian IGF-1R, higher levels of autophagy were detected in aged worms [83]. Whole body RNAi of ATG18 (C. elegans homologue of WIPI) suppressed lifespan extension of daf-2 mutant worms. The implication of these studies in a mammalian setting remains to be confirmed. Reduced IGF-1 levels in mice correlated with an increase in autophagy and loss in heart weight during starvation implying that enhanced autophagy may have deleterious effects [22]. Furthermore, conflicting results show that prolonged IGF-1R inhibition in mammalian cells may instead attenuate autophagy by suppressing the contribution of plasma membrane to autophagosome biogenesis [9]. Together these studies suggest that the implication of autophagy in such aging models requires further investigation.
Conditions where RTK signalling induces autophagy have also been manipulated to ameliorate inflammation. Ligand activation of the RTK Axl correlated with transcriptional enhancement of autophagy and suppression of NLRP3 inflammasome activity, while the related RTKs DTK and MERTK did not influence autophagy [18]. Such signalling from Axl restricted acute hepatic injury in mice during carbon tetrachloride or lipopolysaccharide administration suggesting an overall advantage of autophagy activation in restricting the immune response.
Altogether, these studies suggest that autophagy can respond to changes in RTK signalling in vivo. However, whether autophagy mediates the physiological implications of RTK modulation and its benefits in additional disease models remain to be determined.
Concluding remarks
The interplay of autophagy and RTKs appears to be complex and context dependent. Given that there is no direct method to monitor autophagic flux in vivo or in patient samples, it is possible that the association between RTKs and autophagy may be influenced by additional factors not modelled using existing experimental approaches. This may be particularly relevant in light of our current understanding of the roles of RTK activation and autophagy in supporting cancer progression. For these reasons, targeting autophagy to influence RTKs and vice versa requires careful experimental investigation and points towards the need for personalized medicine. Future studies utilizing genetic means to specifically target autophagy and the development of autophagy-specific modulators will be required to address these issues before advancing from the bench to the clinic.
Summary
RTKs can directly and indirectly regulate autophagy.
Transcription of autophagy players can be suppressed by RTK signalling.
Autophagy facilitates signalling from certain RTKs.
The interaction of RTKs and autophagy can be manipulated to impact cell fitness and disease development.
Acknowledgments
We apologize to those whose original research we were unable to acknowledge directly.
Abbreviations
- Ack1
activated cdc42 kinase 1
- AMBRA1
activating molecule in beclin-1-regulated autophagy 1
- AP-2
adaptor protein 2
- APPL
amyloid-beta-like protein precursor
- ATG
autophagy-related protein
- Bcl-2
B-cell lymphoma 2
- BNIP3
BCL2 interacting protein 3
- DFCP1
double-FYVE containing protein 1
- EGFR
epidermal growth factor receptor
- EphB2
ephrin-type B receptor 2
- ER
endoplasmic reticulum
- ERK1/2
extracellular signal-related kinases 1/2
- FGFR1
fibroblast growth fractor receptor 1
- FIP200
FAK family kinase-interacting protein of 200kDa
- FoxO
forkhead transcription factor
- GLUT1
glucose transporter 1
- HER2
human epidermal growth factor receptor 2
- HIF1A
hypoxia-inducible factor 1 alpha
- IGF-1R
insulin-like growth factor-1 receptor
- LAPTM4B
lysosomal-associated transmembrane protein 4B
- LC3
microtubule-associated proteins 1A/1B light chain 3B
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MERTK
tyrosine-protein kinase Mer precursor
- NBR1
next to BRCA1 gene 1
- mTOR
mammalian target of rapamycin
- mTORC
mTOR complex
- TrkA
tropomyosin receptor kinase A
- PI3K
phosphoinositol-3-kinase
- RTK
receptor tyrosine kinase
- STAT
signal transducer and activator of transcription
- TBC1D5
Tre2, Bub2, Cdc16 1 domain family member 5
- TFEB
transcription factor EB
- TKI
tyrosine kinase inhibitor
- TRAF6
TNF receptor-associated factor 6
- ULK1
unc-51-like kinase 1
- Unc-51
uncoordinated protein 51
- UVRAG
UV radiation resistance-associated gene
- VAMP3
vesicle-associated membrane protein 3
- VEGFR
vascular endothelial growth factor receptor
- Vps34
vacuolar sorting protein 34
- WIPI
WD repeat domain phosphoinositide-interacting protein
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Cancer Research UK Fellowship [grant number C52370/A21586 (to N.G.)].
References
- 1.Lemmon M.A. and Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendoza M.C., Er E.E. and Blenis J. (2011) The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 36, 320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb C.A., Yoshimori T. and Tooze S.A. (2013) The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774 [DOI] [PubMed] [Google Scholar]
- 4.Amaya C., Fader C.M. and Colombo M.I. (2015) Autophagy and proteins involved in vesicular trafficking. FEBS Lett. 589, 3343–3353 [DOI] [PubMed] [Google Scholar]
- 5.Ravikumar B., Moreau K., Jahreiss L., Puri C. and Rubinsztein D.C. (2010) Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12, 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longatti A., Lamb C.A., Razi M., Yoshimura S., Barr F.A. and Tooze S.A. (2012) TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J. Cell Biol. 197, 659–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou C., Ma K., Gao R., Mu C., Chen L., Liu Q. et al. (2017) Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 27, 184–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puri C., Renna M., Bento C.F., Moreau K. and Rubinsztein D.C. (2013) Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154, 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renna M., Bento C.F., Fleming A., Menzies F.M., Siddiqi F.H., Ravikumar B. et al. (2013) IGF-1 receptor antagonism inhibits autophagy. Hum. Mol. Genet. 22, 4528–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popovic D., Akutsu M., Novak I., Harper J.W., Behrends C. and Dikic I. (2012) Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32, 1733–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy S., Leidal A.M., Ye J., Ronen S.M. and Debnath J. (2017) Autophagy-dependent shuttling of TBC1D5 controls plasma membrane translocation of GLUT1 and glucose uptake. Mol. Cell 67, 84–95.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popovic D. and Dikic I (2014) TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep. 15, 392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y., Zou Z., Becker N., Sumpter R., Xiao G., Kinch L. et al. (2013) EGFR-mediated beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 154, 1269–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chukkapalli S., Amessou M., Dilly A.K., Dekhil H., Zhao J., Liu Q. et al. (2014) Role of the EphB2 receptor in autophagy, apoptosis and invasion in human breast cancer cells. Exp. Cell Res. 320, 233–246 [DOI] [PubMed] [Google Scholar]
- 15.Kandouz M., Haidara K., Zhao J., Brisson M.-L. and Batist G. (2010) The EphB2 tumor suppressor induces autophagic cell death via concomitant activation of the ERK1/2 and PI3K pathways. Cell Cycle 9, 398–407 [DOI] [PubMed] [Google Scholar]
- 16.Schmukler E., Shai B., Ehrlich M. and Pinkas-Kramarski R. (2012) Neuregulin promotes incomplete autophagy of prostate cancer cells that is independent of mTOR pathway inhibition. PLoS ONE 7, e36828, 10.1371/journal.pone.0036828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen K., Wagner B., Hamel W., Schweizer M., Haag F., Westphal M. et al. (2007) Autophagic cell death induced by TrkA receptor activation in human glioblastoma cells. J. Neurochem. 103, 259–275 [DOI] [PubMed] [Google Scholar]
- 18.Han J., Bae J., Choi C.Y., Choi S.P., Kang H.S., Jo E.K. et al. (2016) Autophagy induced by AXL receptor tyrosine kinase alleviates acute liver injury via inhibition of NLRP3 inflammasome activation in mice. Autophagy 12, 2326–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domigan C.K., Warren C.M., Antanesian V., Happel K., Ziyad S., Lee S. et al. (2015) Autocrine VEGF maintains endothelial survival through regulation of metabolism and autophagy. J. Cell Sci. 128, 2236–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong P.-M., Puente C., Ganley I.G. and Jiang X. (2013) The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy 9, 124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nazio F., Strappazzon F., Antonioli M., Bielli P., Cianfanelli V., Bordi M. et al. (2013) mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15, 406–416 [DOI] [PubMed] [Google Scholar]
- 22.Troncoso R., Vicencio J.M., Namchenko A., Kawashima Y., Del Campo A., Toro B. et al. (2012) Energy-preserving effects of IGF-1 antagonize starvation-induced cardiac autophagy. Cardiovasc. Res. 93, 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin X., Zhang Y., Liu L., McKeehan W.L., Shen Y., Song S. et al. (2011) FRS2α is essential for the fibroblast growth factor to regulate the mTOR pathway and autophagy in mouse embryonic fibroblasts. Int. J. Biol. Sci. 7, 1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itakura E., Kishi C., Inoue K. and Mizushima N. (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell R.C., Tian Y., Yuan H., Park H.W., Chang Y.Y., Kim J. et al. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Whiteman M.W., Lian H., Wang G., Singh A., Huang D. et al. (2009) A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J. Biol. Chem. 284, 21412–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan H., Li Z.M., Shao J., Ji W.X., Xia W. and Lu S. (2017) FGF2/FGFR1 regulates autophagy in FGFR1-amplified non-small cell lung cancer cells. J. Exp. Clin. Cancer Res. 36, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R.C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G. et al. (2012) Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J., Hou W., Lu C., Goldstein L.A., Stolz D.B., Watkins S.C. et al. (2013) Interaction between Her2 and Beclin-1 proteins underlies a new mechanism of reciprocal regulation. J. Biol. Chem. 288, 20315–20325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Q., Westphal W., Wong K.N., Tan I. and Zhong Q. (2010) Rubicon controls endosome maturation as a Rab7 effector. Proc. Natl. Acad. Sci. U.S.A. 107, 19338–19343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan X., Thapa N., Sun Y. and Anderson R.A. (2015) A kinase-independent role for EGF receptor in autophagy initiation. Cell 160, 145–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dou Z., Pan J.A., Dbouk H.A., Ballou L.M., DeLeon J.L., Fan Y. et al. (2013) Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol. Cell 50, 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You L., Wang Z., Li H., Shou J., Jing Z., Xie J. et al. (2015) The role of STAT3 in autophagy. Autophagy 11, 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S. et al. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S. et al. (2007) FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]
- 36.Mammucari C., Milan G., Romanello V. et al. (2007) FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- 37.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S. et al. (2012) A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmieri M., Pal R., Nelvagal H.R., Lotfi P., Stinnett G.R., Seymour M.L. et al. (2017) mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 8, 14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M.-K. and Hung M.-C. (2015) Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J 282, 3693–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lampada A., O’Prey J., Szabadkai G., Ryan K.M., Hochhauser D. and Salomoni P. (2017) mTORC1-independent autophagy regulates receptor tyrosine kinase phosphorylation in colorectal cancer cells via an mTORC2-mediated mechanism. Cell Death Differ. 24, 1045–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H.-B., Yang S., Weng H.Y., Chen Q., Zhao X.L., Fu W.J. et al. (2017) Autophagy-induced KDR/VEGFR-2 activation promotes the formation of vasculogenic mimicry by glioma stem cells. Autophagy 13, 1528–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fung C., Lock R., Gao S., Salas E. and Debnath J (2008) Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol. Biol. Cell 19, 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrow-McGee R., Kishi N., Joffre C., Menard L., Herveiu A., Bakhouche B.A. et al. (2016) Beta 1-integrin-c-Met cooperation reveals an inside-in survival signalling on autophagy-related endomembranes. Nat. Commun. 7, 11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Lopez N., Athonvarangkul D., Mishall P. et al. , and (2013) Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 4, 2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones S., Cunningham D.L., Rappoport J.Z. and Heath J.K. (2014) The non-receptor tyrosine kinase Ack1 regulates the fate of activated EGFR by inducing trafficking to the p62/NBR1 pre-autophagosome. J. Cell Sci. 127, 994–1006 [DOI] [PubMed] [Google Scholar]
- 46.Yue X., Song W., Zhang W., Chen L., Xi Z., Xin Z. et al. (2008) Mitochondrially localized EGFR is subjected to autophagic regulation and implicated in cell survival. Autophagy 4, 641–649 [DOI] [PubMed] [Google Scholar]
- 47.Rohatgi R.A., Janusis J., Leonard D., Bellve K.D., Fogarty K.E., Baehrecke E.H. et al. (2015) Beclin 1 regulates growth factor receptor signaling in breast cancer. Oncogene 34, 5352–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.So K.S., Kim C.H., Rho J.K., Kim S.Y., Choi Y.J., Song J.S. et al. (2014) Autophagosome-mediated EGFR down-regulation induced by the CK2 inhibitor enhances the efficacy of EGFR-TKI on EGFR-mutant lung cancer cells with resistance by T790M. PloS ONE 9, e114000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nunes J., Zhang H., Angelopoulos N., Chhetri J., Osipo C., Grothey A. et al. (2016) ATG9A loss confers resistance to trastuzumab via c-Cbl mediated Her2 degradation. Oncotarget 7, 27599–27612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han W., Pan H., Chen Y., Sun J., Wang Y., Li J. et al. (2011) EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS ONE 6, e18691, 10.1371/journal.pone.0018691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eimer S., Belaud-Rotureau M.A., Airiau K., Jeanneteau M., Laharanne E., Veron N. et al. (2011) Autophagy inhibition cooperates with erlotinib to induce glioblastoma cell death. Cancer Biol. Ther. 11, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 52.Li X., Lu Y., Pan T. and Fan Z. (2010) Roles of autophagy in cetuximab-mediated cancer therapy against EGFR. Autophagy 6, 1066–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X. and Fan Z. (2010) The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res. 70, 5942–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weihua Z., Tsan R., Huang W.C., Wu Q., Chiu C.H., Fidler I.J. et al. (2008) Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell 13, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dragowska W.H., Weppler S.A., Wang J.C., Wong L.Y., Kapanen A.I., Rawji J.S. et al. (2013) Induction of autophagy is an early response to gefitinib and a potential therapeutic target in breast cancer. PLoS ONE 8, e76503, 10.1371/journal.pone.0076503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Z., He K., Ma Q., Yu Q., Liu C., Ndege I. et al. (2017) Autophagy inhibitor facilitates gefitinib sensitivity in vitro and in vivo by activating mitochondrial apoptosis in triple negative breast cancer. PLoS ONE 12, e0177694, 10.1371/journal.pone.0177694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeVorkin L., Hattersley M., Kim P., Ries J., Spowart J., Angelsio M.S. et al. (2017) Autophagy inhibition enhances sunitinib efficacy in clear cell ovarian carcinoma. Mol. Cancer Res. 15, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aveic S., Pantile M., Saydel A., Esposito M.R., Zanon C., Li G. et al. (2016) Combating autophagy is a strategy to increase cytotoxic effects of novel ALK inhibitor entrectinib in neuroblastoma cells. Oncotarget 7, 5646–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cufí S., Vazquez-Martin A., Oliveras-Ferraros C., Corominas-Faja B., Cuyas E., Lopez-Bonet E. et al. (2013) The anti-malarial chloroquine overcomes primary resistance and restores sensitivity to trastuzumab in HER2-positive breast cancer. Sci. Rep. 3, 2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gammoh N., Fraser J., Puente C., Syred H.M., Kang H., Ozawa T. et al. (2016) Suppression of autophagy impedes glioblastoma development and induces senescence. Autophagy, 12 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karsli-Uzunbas G., Guo J.Y., Price S., Teng X., Laddha S.V., Khor S. et al. (2014) Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 4, 914–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Makinoshima H., Takita M., Matsumoto S., Yagishita A., Owada S., Esumi H. et al. (2014) Epidermal growth factor receptor (EGFR) signaling regulates global metabolic pathways in EGFR-mutated lung adenocarcinoma. J. Biol. Chem. 289, 20813–20823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y. and Fu L. (2011) Mechanisms of acquired resistance to tyrosine kinase inhibitors. Acta Pharm. Sin. B 1, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bardelli A., Corso S., Bertotti A., Hobor S., Valtorta E., Siravegna G. et al. (2013) Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 3, 658–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levy J.M.M., Towers C.G. and Thorburn A. (2017) Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y., Henson E.S., Xiao W., Huang D., McMillan-Ward E.M., Israels S.J. et al. (2016) Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy 12, 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fung C., Chen X., Grandis J.R. and Duvvuri U (2012) EGFR tyrosine kinase inhibition induces autophagy in cancer cells. Cancer Biol. Ther. 13, 1417–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larrue C., Saland E., Boutzen H., Vergez F., David M., Joffre C. et al. (2016) Proteasome inhibitors induce FLT3-ITD degradation through autophagy in AML cells. Blood 127, 882–892 [DOI] [PubMed] [Google Scholar]
- 69.Yonekawa T. and Thorburn A (2013) Autophagy and cell death. Essays Biochem. 55, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chude C.I. and Amaravadi R.K. (2017) Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int. J. Mol. Sci. 18, 1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rubinsztein D.C., Bento C.F. and Deretic V. (2015) Therapeutic targeting of autophagy in neurodegenerative and infectious diseases. J. Exp. Med. 212, 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hebron M.L., Lonskaya I. and Moussa C.E.-H. (2013) Nilotinib reverses loss of dopamine neurons and improves motor behavior via autophagic degradation of -synuclein in Parkinson’s disease models. Hum. Mol. Genet. 22, 3315–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Hoecke A., Schoonaert L., Lemmens R., Timmers M., Staats K.A., Laird A.S. et al. (2012) EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat. Med. 18, 1418–1422 [DOI] [PubMed] [Google Scholar]
- 74.Zheng Y., Wang Q., Xiao B., Lu Q., Wang Y. and Wang X. (2012) Involvement of receptor tyrosine kinase Tyro3 in amyloidogenic APP processing and β-amyloid deposition in Alzheimer’s disease models. PLoS ONE 7, e39035, 10.1371/journal.pone.0039035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hebron M. and Moussa E.-H. (2015) Two sides of the same coin: tyrosine kinase inhibition in cancer and neurodegeneration. Neural Regen. Res. 10, 1767–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deretic V., Saitoh T. and Akira S (2013) Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I. and Deretic V. (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 78.Stanley S.A., Barczak A.K., Silvis M.R., Luo S.S., Sogi K., Vokes M. et al. (2014) Identification of host-targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS Pathog. 10, e1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Napier R.J., Rafi W., Cheruvu M., Powell K.R., Zaunbrecher M.A., Bornmann W. et al. (2011) Imatinib-sensitive tyrosine kinases regulate mycobacterial pathogenesis and represent therapeutic targets against tuberculosis. Cell Host Microbe 10, 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar N., Sharma N.R., Ly H., Parslow T.G. and Liang Y. (2011) Receptor tyrosine kinase inhibitors that block replication of influenza a and other viruses. Antimicrob. Agents Chemother. 55, 5553–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lutz M.A., Gervais F., Bernstein A., Hattel A.L. and Correll P.H. (2002) STK receptor tyrosine kinase regulates susceptibility to infection with Listeria monocytogenes. Infect. Immun. 70, 416–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Burton E.A., Plattner R. and Pendergast A.M. (2003) Abl tyrosine kinases are required for infection by Shigella flexneri. EMBO J. 22, 5471–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang J.T., Kumsta C., Hellman A.B., Adams L.M. and Hansen M (2017) Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. eLife 6, e18459, 10.7554/eLife.18459 [DOI] [PMC free article] [PubMed] [Google Scholar]