Abstract
The endoplasmic reticulum (ER) is a key site for lipid biosynthesis and folding of nascent transmembrane and secretory proteins. These processes are maintained by careful homeostatic control of the environment within the ER lumen. Signalling sensors within the ER detect perturbations within the lumen (ER stress) and employ downstream signalling cascades that engage effector mechanisms to restore homeostasis. The most studied signalling mechanism that the ER employs is the unfolded protein response (UPR), which is known to increase a number of effector mechanisms, including autophagy. In this chapter, we will discuss the emerging role of autophagy as a UPR effector pathway. We will focus on the recently discovered selective autophagy pathway for ER, ER-phagy, with particular emphasis on the structure and function of known mammalian ER-phagy receptors, namely FAM134B, SEC62, RTN3 and CCPG1. Finally, we conclude with our view of where the future of this field can lead our understanding of the involvement of ER-phagy in ER homeostasis.
Keywords: Autophagy, ER-phagy, ER stress, ER homeostasis, proteostasis, unfolded protein response
Introduction
The endoplasmic reticulum (ER) is an intracellular organelle that consists of a continuous network of membranous sheets and tubules spanning the cytoplasm. A lipid bilayer segregates the ER lumen from the cytosol. The ER acts as a reservoir for calcium cations (Ca2+). These are maintained at a relatively high concentration within the lumen and can be released during cell signalling responses [1]. The other function of the ER is biosynthesis. ER membranes are divided into two conceptual types, rough and smooth ER, present in different proportions and abundances in different cell lineages, although these are interconnected compartments and gradients of function likely exist. The smooth ER is the site for the biosynthesis of lipids and steroid hormones, and acts as a hub for detoxification enzyme activity [2]. An expansive and specialized smooth ER, the sarcoplasmic reticulum, is present in muscle, wherein it acts as the major calcium store for release during contraction. The rough ER is studded with ribosomes that co-translationally insert nascent polypeptide chains encoding transmembrane or secretory proteins. In the lumen, these proteins fold with the assistance of ER-luminal chaperone proteins, which bind, retain and prevent the aggregation of partially folded substrates [3]. Folding is also aided by post-translational modifications such as N-linked glycosylation, mediated via glycosylases, and intramolecular disulphide bond formation and rearrangement, catalysed by protein disulphide isomerases (PDI) [4,5]. Chaperone binding also prevents secretion from the ER of incompletely folded proteins. The high luminal Ca2+ concentration facilitates chaperone protein function [4]. A distinct redox potential in the ER lumen—a more oxidizing environment than the cytoplasm—optimizes PDI activity [5].
All cells require ER but some specialist types have a particularly heavy demand for certain ER functions. For example, hepatocytes are a major site of lipid synthesis and have expansive smooth ER. Similarly, plasma cells (effector B cells) and exocrine cells, which secrete abundant immunoglobulins and zymogens respectively, have a high abundance of rough ER [6].
ER homeostasis
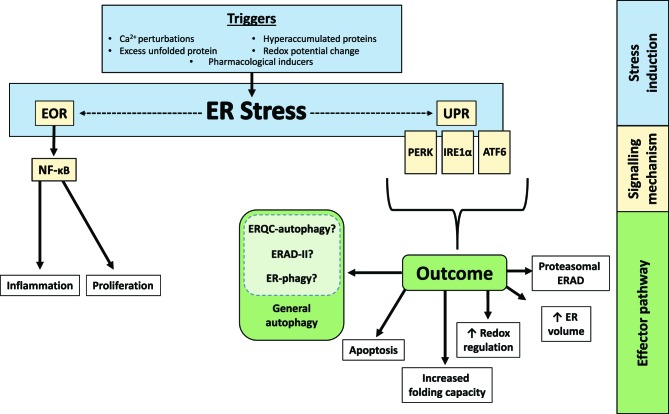
Signalling sensors within the ER detect perturbations within the lumen (ER stress) and employ downstream signalling cascades that engage effector mechanisms to restore homeostasis (Figure 1). These mechanisms include increasing the capacity of the ER, increasing degradation of ER luminal proteins or up-regulating chaperones and luminal protein modification or folding enzymes. This review will focus on ER homeostatic pathways, with a particular emphasis on the emerging role of autophagy as a potential effector mechanism.
Figure 1. Outcomes of ER stress.
Perturbation of ER homeostasis or ‘ER stress’ (blue), is ameliorated by the triggering of signalling cascades (yellow), which in turn engage downstream effector mechanisms (green). Generally speaking, these mechanisms restore homeostasis. This review will discuss these pathways and mechanisms, with particular focus on autophagy as a potential effector and, in further detail, selective autophagic degradation of ER luminal contents or ER (ER-quality control (ERQC)-autophagy, ER-associated degradation (ERAD)-II and ER-phagy).
Physiologically, ER stress occurs upon, for example, changes to luminal Ca2+ concentration, redox status, increased abundance of unfolded proteins and/or hyperaccumulation of proteins. Conditions that produce this include heavy biosynthetic demand, hypoxia, redox stress, deregulated Ca2+ homeostasis and crises such as metabolite insufficiency or low intracellular ATP levels. ER stress can also be produced experimentally with drugs that perturb calcium homeostasis, alter redox status or inhibit glycosylation (Figure 1).
Defects in sensing and signalling pathways downstream of ER stress are associated with numerous pathological conditions including diabetes, non-alcoholic fatty-liver disease, Parkinson’s and Alzheimer’s diseases and cancers such as hepatocellular carcinoma [7,8].
Two distinct pathways for response to ER stress have been proposed, comprising the less well-understood ER-overload response (EOR) and the extensively characterized unfolded protein response (UPR). EOR is triggered by hyperaccumulation of ER-resident proteins, not necessarily unfolded protein. Here, the ER releases luminal Ca2+, which stimulates reactive oxygen species production that in turn activates NF-κB signalling [9]. Ultimately, NF-κB up-regulates a variety of transcripts that promote proliferation and inflammation. However, little is known about the mechanisms of EOR, particularly if there is any link with autophagy function. It can inhibit viral protein replication, suggesting that it may act as a rapid cellular antiviral response [10,11]. Moreover, EOR activated by Hepatitis C viral proteins leads to an increase in cancer-related gene expression, suggesting a role in hepatocellular carcinoma progression [11,12].
The UPR acts to restore defective proteostasis upon detection of unfolded protein in the ER lumen. It involves up-regulated transcription of redox enzymes, chaperones (such as binding immunoglobulin protein/78-kDa glucose-regulated protein, abbreviated BiP/Grp78), foldases (chaperones catalysing folding via additional enzymatic activity, including PDIs), glycosylases and, additionally, via the transcription factors C/EBP and SREBP1/2, lipid-synthesizing enzymes, which facilitate ER membrane expansion [13,14]. The UPR has three signalling arms, each emanating from an ER integral membrane sensing protein. These sensors are inositol-requiring enzyme 1α (IRE1α), protein kinase RNA-like ER kinase (PERK) and activating transcription factor (ATF) 6. Upon accumulation of misfolded proteins in the ER lumen, BiP/Grp78 is titrated away from the luminal domains of these sensor proteins, releasing them from their monomeric, inactive states and, in the case of ATF6, allowing transit to the Golgi.
IRE1α possesses endoribonuclease activity that splices X-box binding protein 1 (XBP1) transcripts, permitting generation of the transcription factor XBP1-S, which up-regulates genes such as ER luminal chaperones and ER-associated degradation (ERAD) machinery component [15]. Up-regulation of chaperones increases the capacity of the ER to deal with unfolded protein. Until recently, ERAD—the removal from the ER of proteins by retrotranslocation into the cytosol and destruction at the proteasome—was thought to be the major mechanism to degrade ER proteins. This process promotes ER homeostasis independently from, but also as an effector of, the UPR, given the transcriptional up-regulation of components of its machinery by the latter [15,16]. ERAD (or ERAD-I) should not be confused with ERAD-II, one of several terms coined recently to describe phenomena where lysosomal, rather than proteasomal, activity appears to result in degradation of ER luminal content (see ‘Autophagy and its role in ER homeostasis’ section). Finally, IRE1α endonuclease activity can also act to degrade secretory protein mRNAs present at the ER in a process termed regulated IRE1α-dependent decay of mRNAs (RIDD), rebalancing protein production and folding capacity [17].
ATF6 differs from other UPR sensors, as it is released from BiP/Grp78 binding to undergo anterograde transport from the ER to the Golgi after ER stress, where it is cleaved by proteases (S1P and S2P, Site 1 and Site 2 proteases) to produce a transcriptionally active polypeptide that can now translocate to the nucleus [18]. Activated ATF6 up-regulates the transcription of ER chaperones, ERAD components and XBP1 mRNA [19–21]. The latter illustrates that the three arms of the UPR may feedback upon one another.
PERK activation occurs with slower kinetics than ATF6 and IRE1α activation upon acute ER stress [22,23]. PERK is a serine-threonine protein kinase that phosphorylates the translation initiation protein, eukaryotic initiation factor-2α (eIF2α), at Ser51, in order to arrest global translation. This results in fewer new protein molecules entering the ER. However, paradoxically, some mRNAs are translationally up-regulated under these conditions, such as ATF4. ATF4 can up-regulate the transcription of C/EBP homologous protein (CHOP) which can then up-regulate growth arrest and DNA damage inducible 34 (GADD34) protein levels. CHOP is a transcription factor that drives pro-apoptotic gene transcription, whereas GADD34 protein dephosphorylates eIF2α, providing negative feedback within the stress response [24,25].
Often, if the UPR fails to resolve ER stress, then activation of cell death by CHOP or other mechanisms such as IRE1α-driven, TRAF2-dependent activation of JNK, and consequent apoptosis, occurs [26]. However, some cell types, both primary and cancerous, are able to withstand constitutive, low-level ER stress and maintain viability, with concomitant chronic up-regulation of UPR transcripts. Some specialized cell types may exhibit UPR-like responses in the differentiated state. Examples include plasma [27,28], osteoblast [29], pancreatic acinar [6], Paneth (intestinal lysozyme granule secreting cells) [30], hepatocyte [31,32] and salivary gland [6] cells, which have an expanded ER relative to their precursor cell state. For example, XBP1 up-regulation, which in turn generates XBP1-S and leads to the production of molecules required to populate the expanded ER, such as chaperones, is required for plasma cell differentiation [33–35].
Autophagy and its role in ER homeostasis
One of the more recent effector mechanisms proposed for the UPR is macroautophagy (hereafter autophagy). Autophagy seals off portions of the cytosol within double-membraned vesicles, called autophagosomes. These fuse with lysosomes, wherein their cargo is degraded [36]. The core, conserved elements of this process are covered in depth elsewhere in this book. Here, we will briefly recap two of the core autophagy protein complexes required for understanding of the discussion further on in this chapter. Focal adhesion kinase family kinase-interacting protein 200-kDa (FIP200) is a key autophagy-related (ATG) protein in most forms of autophagy, which assists in nucleation of autophagosomes via scaffolding unc-51-like kinase 1/2 (ULK1/2) kinase activity, which phosphorylates other proteins required for autophagy [37]. It has been known for some time that the ER can platform formation of autophagosomes, potentially at sites of mitochondrial contact [38–40], generating a cradle from which the isolation membrane, the precursor membrane to the autophagosome, extrudes. The isolation membrane is where ‘early’ ATG protein complexes, including those containing FIP200, are concentrated. The subsequent extension of the tubular isolation membrane and self-enclosure results in the distinctive double lipid bilayer structure of the mature autophagosome. Downstream of ULK1 activity, a key protein grouping is ATG8-family members, of which there are six paralogues in mammals. These are microtubule-associated protein 1A/1B-light chain 3 (LC3) A, B and C and γ-aminobutyric acid receptor associated protein (GABARAP), GABARAPL1 and GABARAPL2 [41]. These ubiquitin-like proteins are lipidated and thus covalently attached to nascent autophagosomal membranes.
Autophagy can be a non-selective mechanism that degrades general cytosol. However, autophagy is frequently selective in nature, targeting damaged organelles or aberrant intracellular structures, referred to as ‘cargo’, such as peroxisomes [42], mitochondria [43,44] and lysosomes [45], ubiquitinated protein aggregates and foreign pathogens [38,39,46–48]. A key molecular component of a given selective autophagy pathway is the cargo receptor(s). The canonical form of this receptor class in mammals bridges cargo to ATG8 family protein(s) by binding both simultaneously. ATG8 binding is mediated via linear peptide regions called LC3-interacting region (LIR) motifs [48,49]. A prime example of this is the well-known receptor protein, p62/SQSTM1, which links cargo to ATG8, for example during cytosolic protein aggregate autophagy.
It is becoming evident that ER stress signals can lead to an increase in general autophagy action (Figure 2). Tunicamycin and thapsigargin, two pharmacological inducers of the UPR, drive autophagosome formation in the human neuroblastoma cell line SK-N-SH and increased LC3 lipidation in mouse embryonic fibroblast (MEF) cells, dependent upon the IRE1 sensor [50]. In human glioblastoma and several adenocarcinoma cell lines, it was shown that ER stress downstream of hypoxia results in transcriptional up-regulation of MAP1LC3B (LC3B) and ATG5, via ATF4 and CHOP [51]. Here, the induced autophagy had a prosurvival role. When autophagy was induced by leucine starvation in MEFs, a range of core autophagy genes and cytosolic cargo receptors were identified as PERK-dependent ATF4 transcriptional targets, including Map1lc3b, Atg5, Atg3, Atg7, Atg10, Atg12, Atg16l1, Becn1, Gabarap, Gabarapl2, p62 and Nbr1 [52]. Other pharmacologic ER stressors inducing general autophagic flux include A23187, thapsigargin, tunicamycin, brefeldin A (HCT116 human colon and DU145 human prostate carcinoma cells, and MEFs), cocaine (A172 human astrocytoma cells) and 14-deoxy-11,12-didehydroandrographolide (T47D human breast carcinoma cells) [53–55]. The selectivity of autophagy was addressed in HCT116 and DU145 cells via ultrastructural characterization of autophagosomes, which were shown to contain a variety of cargo, suggesting a general up-regulation of relatively non-selective autophagy [53]. Interestingly, in yeast, the specific activity of Atg1, the orthologue of ULK1, is increased upon ER stress, without evident transcriptional induction [56]. This has not yet been observed in mammals.
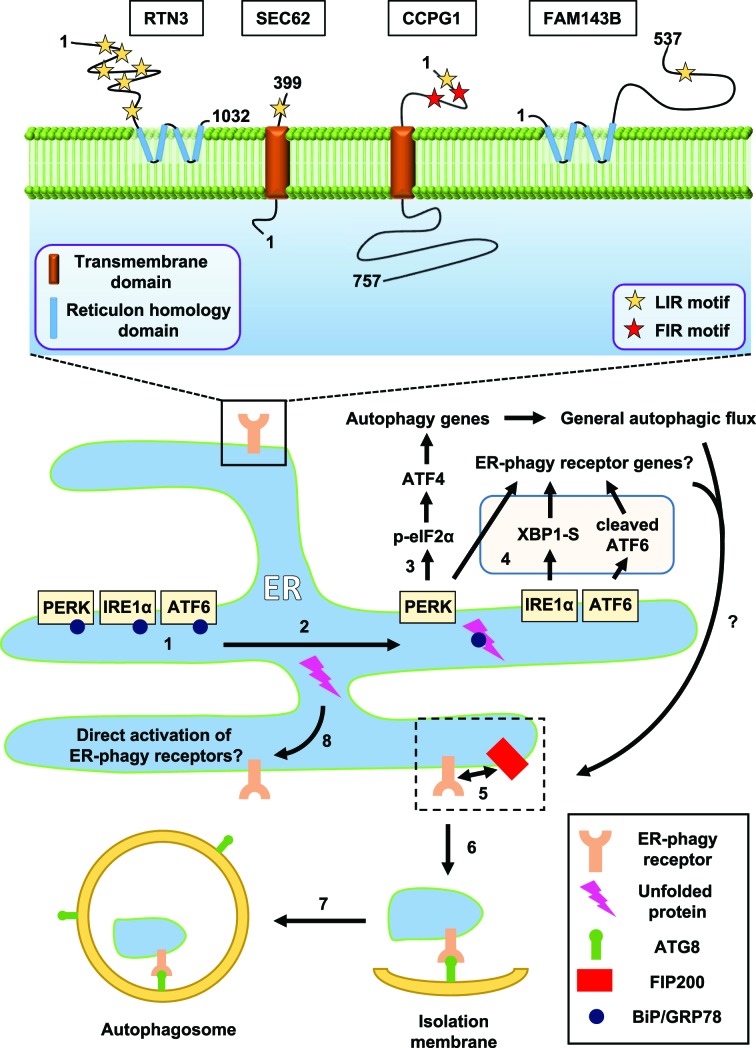
Figure 2. Engagement of autophagy by ER stress and molecular model for ER-phagy events.
(1) Under normal ER homeostasis, the ER luminal chaperone protein BiP/GRP78 binds to the UPR sensor proteins PERK, IRE1α and ATF6. (2) Upon the presence of ER stress caused by unfolded protein (pink lightning), BiP/GRP78 dissociates from UPR sensors and binds to the unfolded protein, thus activating ER stress sensors. (3) PERK is able to up-regulate the transcription of numerous autophagy genes and cargo receptors through its effector transcription factors ATF4 and CHOP, resulting in an increase in general autophagic flux. (4) Any of the UPR sensors could hypothetically increase the transcription in ER-phagy receptor genes. In the case of CCPG1, this indeed occurs. (5) In the case of CCPG1 protein, an interaction with FIP200 is required for recruitment of ER into autophagosomes. It is unclear whether this happens at the ER surface (depicted) or at a latter stage of the pathway. FIP200 is rarely found on the inner surface of autophagosomes, arguing for the former. (6) The ER becomes scissioned. Concomitant with this, ER-phagy receptors bind to ATG8 proteins via their LIR motifs, linking fragmented ER to the isolation membrane. (7) The isolation membrane grows and encloses to form an autophagosome, which will eventually fuse with the lysosome and degrade the ER fragment. (8) It is hypothetically possible that ER stress directly engages and activates ER-phagy receptors independent of transcriptional induction and UPR sensors. To date, there are four described mammalian ER-phagy receptors (top panel). All receptors share the common characteristic of at least one cytosolic LIR motif (yellow star). CCPG1 possesses additional cytosolic FIP200-interacting region (FIR) motifs (red star).
None of the above studies addressed whether autophagy directly regulates ER homeostasis or showed that autophagy could participate in selective degradation of the ER. However, loss of all autophagy function by knockout of conditional Atg7 flox alleles in T lymphocytes (Lck-Cre) or Atg5 flox alleles in B lymphocytes (CD19-Cre) resulted in expanded ER and elevated ER stress signalling, suggesting a role for autophagy in ER homeostasis [57,58]. Additionally, when wild-type yeast were treated with the UPR inducer DTT, ultrastructural analysis showed, for one of the first times, that ER in any organism could be selectively sequestered in autophagic-like vacuoles [59]. These early observations generated the idea that autophagy might both regulate ER function and even do so by direct action on the ER. In the latter instance, three pathways for putative direct regulation of ER homeostasis by selective autophagy are described, with the terms ‘ER-quality control autophagy’ (‘ERQC autophagy’), ERAD-II and ER-phagy proposed. The degree of overlap between these processes is currently unclear. ER-phagy is presently the mechanistically best-described pathway.
ERQC autophagy was used as a term to describe a mammalian process in which disease-associated, conformer mutants of proteins are removed from the lumen by autophagy without large portions of the ER itself being degraded [60,61]. An example of this is the degradation of the mutant form of the G-protein-coupled receptor E90K-GnRHR [61]. There is little mechanistic information on this phenomenon. A potentially similar phenomenon was reported in mammals for removal of insoluble molecules of mutant protein from the ER via a lysosomal route, for which the term ERAD-II was proposed, in analogy to proteasomal ERAD (or ERAD-I) [62].
ER-phagy involves the sequestration of portions of ER cisternae into autophagosomes and occurs in both yeast and mammals (Figure 2). Potential examples include the ER stress associated inhibition of mTOR and incorporation of ER into autophagosomes in Listeria-infected phagocytes, which may ameliorate ER stress and death [63]. Indeed, it is presumed that general up-regulation of autophagy capacity, as described above, would translate into a commensurate greater rate of ER-phagy, but this remains unproven. However, recent findings that have given the phenomenon of ER-phagy mechanistic credence and have allowed direct testing of its role in cellular function and pathology, are the discovery of ER-phagy-specific cargoes and cargo receptors, both in yeast [64,65] and mammals [66–69]. We will examine predominantly mammalian ER-phagy receptors below to illustrate their mechanisms of action and explore their potential integration into ER stress responses.
Four main mammalian ER-phagy receptors have been identified in vitro, namely FAM134B (U2OS and MEF cells), SEC62 (HeLa and MEF cells), RTN3 (U2OS and MEF cells) and CCPG1 (HeLa and A549 cells) [66–69]. These share some common and divergent principles of action (Table 1). Additionally, two ER-phagy receptors exist in yeast, Atg39 and Atg40 [65], which work along broadly similar principles (Table 1). Other well-known autophagy receptor proteins, such as p62/SQSTM1, may possibly have a role in ER homeostasis, although it is not clear that this is due to any direct function in ER-phagy [70]. In the majority of the above cases, ultrastructural analyses have shown that ER-phagy receptors drive the sequestration of isolated fragments of ER into an autophagosomal lumen defined by discrete, delimiting membrane(s) [65–68].
Table 1. Known ER-phagy receptors in yeast and mammals, and their known functions and characteristics.
| Receptor | General description | AIM/LIR | Other ATG-interaction motifs | Physiological role | Reticulon homology domain (RHD) | Transmembrane protein | Region of ER |
|---|---|---|---|---|---|---|---|
| Yeast (Saccharomyces cerevisiae) | |||||||
| Atg39 | Localized to the peripheral ER and nuclear envelope. Also participates in nucleophagy as it encapsulates nuclear contents as well as ER membranes | WNLV | Atg11BR | Regulates perinuclear ER and nuclear morphology | No | Yes | Perinuclear |
| Atg40 | Localized predominantly at cytoplasmic and cortical ER. Facilitates the loading of ER sheets and tubules into autophagosomes | YDFM | Unknown (proposed Atg11 interaction) | Regulates ER morphology | Yes | Yes | Perinuclear, cortical and cytoplasmic |
| Mammals (Homo sapiens) | |||||||
| FAM134B | Promotes the remodelling and scission of ER sheets through its reticulon domain | FELL | Unknown | Health of sensory neurons | Yes | No | Sheets |
| SEC62 | Delivers portions of ER into autophagosomes following ER stress in a process termed ‘recovER-phagy’ | FEMI | Unknown | Unknown | No | Yes | Unknown |
| RTN3 | Promotes the remodelling and scission of ER tubules through its reticulon domain | FTLL | Unknown | Unknown | Yes | No | Tubules |
| YSKV | |||||||
| FEVI | |||||||
| WDLV | |||||||
| FEEL | |||||||
| YDIL | |||||||
| CCPG1 | Delivers portions of ER to autophagosomes in response to ER-stress induction | WTVI | Two FIR motifs | Luminal proteostasis of exocrine pancreas (acinar cells) | No | Yes | Unknown |
The bold-underlined residues are key residues required for ATG8/LC3 binding. Abbreviations: AIM, Atg8-interacting motif (yeast equivalent of an LIR); ATG11BR, Atg11-binding region; FIR, FIP200-interacting region.
ER-phagy receptors are basally ER resident proteins, either transmembrane with associated luminal and cytosolic domains or anchored within ER membranes, and in both instances exposing at least one LIR motif to the cytosol (Figure 2). The LIR motif in all instances is required for ER sequestration into autophagosomes [66–69]. RTN3 possesses a total of six LIR motifs, all of which contribute to ATG8 binding. CCPG1 also contains two novel FIP200-interacting region (FIR) motifs [69]. As FIR motifs are required for ER-phagy, FIP200 may also thus have a distinct role in cargo selection in addition to its known role in regulating ULK1 activity (Figure 2).
FAM134B, CCPG1 and RTN3 all act to trim ER content, thus maintaining ER morphology [66–69]. Notably, both FAM134B and RTN3 (and yeast Atg40) share the presence of a reticulon homology domain (RHD), which consists of two hairpin helices that anchor the protein to ER membranes. The presence of the RHD facilitates membrane curvature and potentially mediates ER scission to facilitate ER-phagy [65–67]. RTN3 resides in peripheral ER tubules whereas FAM134B is found on perinuclear ER sheets and these subdomain resident receptors act to drive local ER-phagy [67]. For instance, RTN3 specifically drives tubular ER-phagy. Both FAM134B and RTN3 can drive ER-phagy in vitro that is stimulated by the general autophagic stimulus of amino acid starvation. In vitro, CCPG1 may also participate in ER-stress driven ER-phagy, after DTT treatment. Finally, Sec62 participates specifically in ER-phagy during recovery from ER stressors, clearing fragments of ER enriched in now redundant ER chaperones [68]. This process is distinguished within the larger set of ER-phagy responses by the term ‘recovER-phagy’. These different mechanistic routes for ER-phagy, targeting distinct regions of the ER and at different stages of the ER stress response, suggest functional specialization of ER-phagy pathways, which may have further relevance in vivo where the ER exhibits large differences in form and function between different tissue types (see below).
There is emerging evidence for direct links between homeostatic ER responses and ER-phagy from in vivo models. Investigations of the physiological role of autophagy have generally involved ablation of all autophagy function by conditional knockout of core Atg genes in various tissues in mice, using tissue-specific, promoter-driven recombinases, typically Cre or tamoxifen-inducible Cre-ERT2. Summarizing, autophagy loss per se generally disrupts ER morphology and size, and produces stress responses. In Paneth cells, Atg7 or Atg16l were seen to be required to restrain IRE1α activation after ER stress induced by experimental XBP1 down-regulation. Loss of this action leads to intestinal inflammation [71]. Furthermore, knockout of Atg5 or Atg7 in pancreatic acinar cells produces ER dilation, stress, inhibition of secretory protein transcription, cell death and inflammation [72,73], although not all reports agree with these findings [74]. However, it is important to note that the contribution of ER-phagy is not precisely interrogated when core autophagy genes are deleted; ER pathology may be an indirect consequence of damaged mitochondria or protein aggregate accumulation, and consequent effects on bioenergetics and signalling. The construction of ER-phagy receptor mutant mice has begun to allow exploration of this issue. Fam134b−/− mice exhibit dilated ER and Golgi within peripheral sensory neurons and cell death [66]. Intriguingly, FAM134B is mutated in human families with heritable sensory neuropathy and these mutations ablate ER-phagy function [66]. In Ccpg1 hypomorphic animals, the acinar cells within the exocrine pancreas exhibit distended ER and insoluble ER luminal protein accumulation, as well as elevated UPR [69]. Few other tissues are reported to be affected by loss of ER-phagy receptor function, indicating that physiologically important ER-phagy receptors remain undiscovered or untested. This observation befits the functional specialization of ER within different cell lineages.
Future directions
In the authors’ view, a major source of outstanding questions in the field of mammalian ER homeostasis centre upon the mechanism and function of ER-phagy.
One question: is whether there is a relationship between ER-phagy and mechanistically opaque processes such as ERQC autophagy and ERAD-II? What degree of overlap in players and mechanisms is there here? Additionally, what molecular determinants mark an ER-phagy receptor, other than LIR motifs? For example, CCPG1 additionally possesses FIR motifs for recognition of the ER by autophagy. Linked to this, it is also crucial to consider whether ER-phagy receptors merely mark ER membrane for degradation or play an active role in scissioning ER membrane to permit sequestration or in sensing ER stress. As a potential example of the former, FAM134B and RTN3 have reticulon domains that might assist ER membrane breakage. In the latter category, some of these receptors have luminal polypeptide regions and it might be conjectured that they could directly sense changes in redox, luminal [Ca2+] or unfolded protein in parallel with canonical sensors. In the case of CCPG1, the ER luminal domain contains a coiled-coil forming region, and it is possible that regulated multimerization/clustering could modify function, as suggested for RTN3 [67,69]. Cytosolic domains might also play a role within the downstream relays of ER stress signalling pathways. For example, some cargo receptors acting in other selective autophagy paradigms bear phosphoregulable LIR motifs [75,76].
Does ER stress specifically trigger ER-phagy, non-selective autophagy or a combination of both, and in which tissue types and to what ultimate purpose? The finding that the UPR can transcriptionally up-regulate CCPG1 provides a mechanistic link between ER stress and selective autophagy of the ER. The Ccpg1 promoter also binds the transcription factor MIST1 [77], which is predominantly expressed in professional secretory cells such as pancreatic acinar cells. MIST1 expression is required for correct differentiation here and is itself dependent upon the IRE1α-XBP1 arm of the UPR [78,79], providing a potential tissue-specific link between CCPG1 up-regulation and its role in ER homeostasis. It is therefore of key importance to dissect whether ER stress response pathways can regulate the activity of other ER-phagy receptors, either to activate them acutely or as is likely for CCPG1 in vivo, also in part to mediate their tissue-specific expression.
It is difficult to say with exactitude how ER-phagy mediates ER homeostasis at a detailed mechanistic level. It is possible that ER-phagy acts to remodel and rebalance the different regions of the ER to meet fluctuating biosynthetic demand. Alternatively, perhaps proteostatic defects result in localized accumulation of unfolded or aggregated protein, by transport mechanisms within the ER lumen, such that specific portions of ER are ‘sacrificed’ in a piecemeal manner in order to maintain unfolded protein at a manageable level within the lumen. There is also little information on rough ER compared with smooth ER as a cargo of autophagy. Different pathways and different receptors may participate, again pointing to likely tissue-specific divergence of these mechanisms in vivo.
Finally, another potentially ER-phagy related mechanism that specialist cells might use to respond to ER stress is secretory autophagy. Paneth cells secrete ER luminal lysozyme via the ER to defend against pathogens. Here, ER stress, triggered by bacterial invasion, results in an increase in secretory autophagy of lysozyme, i.e. the use of the autophagy machinery to build secretory vesicles containing the lysozyme, rather than utilization of the default secretory pathway via the Golgi [80]. Investigations are required to determine if ER-phagy receptors play a role here.
Conclusion
The UPR and other ER stress response signalling pathways engage multiple effector pathways. A recently emerging effector is autophagy and, of particular interest, ER-phagy. Taken together, the evidence points to a pivotal role for ER-phagy in normal ER homeostasis and overall cell health, particularly in specialized tissue types in vivo. Crucially, ER-phagy helps ameliorate the effects of ER stress through the degradation of ER membranes, removal of ER luminal protein aggregates and/or removal of ER-chaperone proteins. Furthermore, ER-phagy also acts to trim ER content, helping maintain the dynamism that is characteristic of this key organelle. Future work will surely expand the molecular components and role of this pathway in currently known and new tissue types.
Summary
ER is the key site for the folding of nascent transmembrane and secretory proteins.
The lumen of ER has a specialized environment to ensure the fidelity of this process.
Signalling sensors within the ER lumen detect stress and employ downstream cascades to engage effector mechanisms and restore homeostasis.
The major signalling cascade employed by the ER is the UPR, which translationally and transcriptionally engages a variety of effector mechanisms.
Selective degradation of the ER by autophagy occurs via a process termed as ER-phagy.
There are currently four known mammalian ER-phagy receptors; FAM134B, SEC62, RTN3 and CCPG1.
ER-phagy receptors possess LIR motifs, allowing interactions with ATG8 family proteins.
ER-phagy has important roles in the physiology of secretory cells in vivo.
General autophagic flux and direct ER-phagy might both be transcriptionally up-regulated by the UPR.
Abbreviations
- ATF
activating transcription factor
- ATG
autophagy-related
- Bip/Grp78
binding immunoglobulin protein/78-kDa glucose-regulated protein
- CHOP
C/EBP homologous protein
- eIF2α
eukaryotic initiation factor-2α
- EOR
ER-overload response
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERQC
ER quality control
- FIR
FIP200-interacting region
- GADD34
growth arrest and DNA damage inducible 34
- IRE1α
inositol-requiring enzyme 1α
- LC3
light chain 3
- LIR
LC3-interacting region
- MEF
mouse embryonic fibroblast
- PDI
protein disulphide isomerase
- PERK
protein kinase RNA-like ER kinase
- RHD
reticulon homology domain
- UPR
unfolded protein response
- XBP1
X-box binding protein 1
Funding
This work was supported by the Cancer Research UK Career Development Fellowship [grant number CRUK-A12825 (to S.W.)]; and the BBSRC Project [grant number BB/N000315/1].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Berridge M.J. (1998) Neuronal calcium signaling. Neuron 21, 13–26 [DOI] [PubMed] [Google Scholar]
- 2.Baumann O. and Walz B. (2001) Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205, 149–214 [DOI] [PubMed] [Google Scholar]
- 3.Brodsky J.L. and Skach W.R. (2011) Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr. Opin. Cell Biol. 23, 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braakman I. and Hebert D.N. (2013) Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, a013201, 10.1101/cshperspect.a013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulleid N.J. (2012) Disulfide bond formation in the mammalian endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 4, a013219, 10.1101/cshperspect.a013219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee A.H., Chu G.C., Iwakoshi N.N. and Glimcher L.H. (2005) XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24, 4368–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M. and Kaufman R.J. (2016) Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 [DOI] [PubMed] [Google Scholar]
- 8.Lindholm D., Korhonen L., Eriksson O. and Koks S. (2017) Recent insights into the role of unfolded protein response in ER stress in health and disease. Front. Cell Dev. Biol. 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pahl H.L. and Baeuerle P.A. (1995) A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-kappa B. EMBO J. 14, 2580–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pahl H.L. and Baeuerle P.A. (1995) Expression of influenza virus hemagglutinin activates transcription factor NF-kappa B. J. Virol. 69, 1480–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong L., Li S., Huang M., Xiong Y., Zhang Q., Ye L. et al. (2015) The roles of endoplasmic reticulum overload response induced by HCV and NS4B protein in human hepatocyte viability and virus replication. PLoS ONE 10, e0123190, 10.1371/journal.pone.0123190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong L., Li S., Yu X., Fang X., Xu A., Huang M. et al. (2016) Hepatitis C virus and its protein NS4B activate the cancer-related STAT3 pathway via the endoplasmic reticulum overload response. Arch. Virol. 161, 2149–2159 [DOI] [PubMed] [Google Scholar]
- 13.Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S. and Walter P. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 14.Lee J.S., Mendez R., Heng H.H., Yang Z.Q. and Zhang K. (2012) Pharmacological ER stress promotes hepatic lipogenesis and lipid droplet formation. Am. J. Transl. Res. 4, 102–113 [PMC free article] [PubMed] [Google Scholar]
- 15.Lee A.H., Iwakoshi N.N. and Glimcher L.H. (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vembar S.S. and Brodsky J.L. (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollien J. and Weissman J.S. (2006) Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107 [DOI] [PubMed] [Google Scholar]
- 18.Ye J., Rawson R.B., Komuro R., Chen X., Dave U.P., Prywes R. et al. (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364 [DOI] [PubMed] [Google Scholar]
- 19.Wu J., Rutkowski D.T., Dubois M., Swathirajan J., Saunders T., Wang J. et al. (2007) ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell 13, 351–364 [DOI] [PubMed] [Google Scholar]
- 20.Yoshida H., Matsui T., Yamamoto A., Okada T. and Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H. et al. (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]
- 22.Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B. et al. (2007) IRE1 signaling affects cell fate during the unfolded protein response. Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutkowski D.T., Arnold S.M., Miller C.N., Wu J., Li J., Gunnison K.M. et al. (2006) Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4, e374, 10.1371/journal.pbio.0040374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding H.P., Zhang Y., Scheuner D., Chen J.J., Kaufman R.J. and Ron D. (2009) Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proc. Natl. Acad. Sci. U.S.A. 106, 1832–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R. et al. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K. et al. (2002) ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H. et al. (2004) XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93 [DOI] [PubMed] [Google Scholar]
- 28.Gass J.N., Gifford N.M. and Brewer J.W. (2002) Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277, 49047–4905 [DOI] [PubMed] [Google Scholar]
- 29.Saito A., Ochiai K., Kondo S., Tsumagari K., Murakami T., Cavener D.R. et al. (2011) Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J. Biol. Chem. 286, 4809–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaser A., Lee A.H., Franke A., Glickman J.N., Zeissig S., Tilg H. et al. (2008) XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee A.H., Scapa E.F., Cohen D.E. and Glimcher L.H. (2008) Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutkowski D.T., Wu J., Back S.H., Callaghan M.U., Ferris S.P., Iqbal J. et al. (2008) UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell 15, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Anken E R.E.P., Maggioni C., Mezghrani A., Sitia R., Braakman I. et al. (2003) Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity 18, 243–253 [DOI] [PubMed] [Google Scholar]
- 34.Hu C.C., Dougan S.K., McGehee A.M., Love J.C. and Ploegh H.L. (2009) XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 28, 1624–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd D.J., McHeyzer-Williams L.J., Kowal C., Lee A.H., Volpe B.T., Diamond B. et al. (2009) XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J. Exp. Med. 206, 2151–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ktistakis N.T. and Tooze S.A. (2016) Digesting the expanding mechanisms of autophagy. Trends Cell Biol. 26, 624–635 [DOI] [PubMed] [Google Scholar]
- 37.Lin M.G. and Hurley J.H. (2016) Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 39, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A. et al. (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T. and Yamamoto A. (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 [DOI] [PubMed] [Google Scholar]
- 40.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N. et al. (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 [DOI] [PubMed] [Google Scholar]
- 41.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y. and Yoshimori T. (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 42.Kim P.K., Hailey D.W., Mullen R.T. and Lippincott-Schwartz J. (2008) Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U.S.A. 105, 20567–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heo J.M., Ordureau A., Paulo J.A., Rinehart J. and Harper J.W. (2015) The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 60, 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L. et al. (2015) The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T. et al. (2013) Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurston T.L., Ryzhakov G., Bloor S., von Muhlinen N. and Randow F. (2009) The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 47.Zheng Y.T., Shahnazari S., Brech A., Lamark T., Johansen T. and Brumell J.H. (2009) The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183, 5909–5916 [DOI] [PubMed] [Google Scholar]
- 48.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H. et al. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 49.Birgisdottir A.B., Lamark T. and Johansen T. (2013) The LIR motif - crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 [DOI] [PubMed] [Google Scholar]
- 50.Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S. et al. (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26, 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rouschop K.M., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K. et al. (2010) The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 120, 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.B’Chir W., Maurin A.C., Carraro V., Averous J., Jousse C., Muranishi Y. et al. (2013) The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 41, 7683–7699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding W.X., Ni H.M., Gao W., Hou Y.F., Melan M.A., Chen X. et al. (2007) Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 282, 4702–4710 [DOI] [PubMed] [Google Scholar]
- 54.Periyasamy P., Guo M.L. and Buch S. (2016) Cocaine induces astrocytosis through ER stress-mediated activation of autophagy. Autophagy 12, 1310–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan H.K., Muhammad T.S.T. and Tan M.L. (2016) 14-Deoxy-11,12-didehydroandrographolide induces DDIT3-dependent endoplasmic reticulum stress-mediated autophagy in T-47D breast carcinoma cells. Toxicol. Appl. Pharmacol. 300, 55–69 [DOI] [PubMed] [Google Scholar]
- 56.Yorimitsu T., Nair U., Yang Z. and Klionsky D.J. (2006) Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281, 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia W., Pua H.H., Li Q.J. and He Y.W. (2011) Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J. Immunol. 186, 1564–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pengo N., Scolari M., Oliva L., Milan E., Mainoldi F., Raimondi A. et al. (2013) Plasma cells require autophagy for sustainable immunoglobulin production. Nat. Immunol. 14, 298–305 [DOI] [PubMed] [Google Scholar]
- 59.Bernales S., McDonald K.L. and Walter P. (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4, e423, 10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teckman J.H. and Perlmutter D.H. (2000) Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am. J. Physiol. Gastrointest. Liver Physiol. 279, G961–G974 [DOI] [PubMed] [Google Scholar]
- 61.Houck S.A., Ren H.Y., Madden V.J., Bonner J.N., Conlin M.P., Janovick J.A. et al. (2014) Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol. Cell 54, 166–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujita E., Kouroku Y., Isoai A., Kumagai H., Misutani A., Matsuda C. et al. (2007) Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II). Hum. Mol. Genet. 16, 618–629 [DOI] [PubMed] [Google Scholar]
- 63.Moretti J., Roy S., Bozec D., Martinez J., Chapman J.R., Ueberheide B. et al. (2017) STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell 10.1016/j.cell.2017.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lipatova Z. and Segev N. (2015) A role for macro-ER-phagy in ER quality control. PLoS Genet. 11, e1005390, 10.1371/journal.pgen.1005390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mochida K., Oikawa Y., Kimura Y., Kirisako H., Hirano H., Ohsumi Y. et al. (2015) Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362 [DOI] [PubMed] [Google Scholar]
- 66.Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M. et al. (2015) Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358 [DOI] [PubMed] [Google Scholar]
- 67.Grumati P., Morozzi G., Holper S., Mari M., Harwardt M.I., Yan R. et al. (2017) Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 6, 10.7554/eLife.25555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fumagalli F., Noack J., Bergmann T.J., Cebollero E., Pisoni G.B., Fasana E. et al. (2016) Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 18, 1173–1184 [DOI] [PubMed] [Google Scholar]
- 69.Smith M.D., Harley M.E., Kemp A.J., Wills, J., Lee, M., Arends, M. et al. (2017) CCPG1, a non-canonical autophagy cargo receptor essential for pancreatic ER proteostasis. Dev. Cell., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang H., Ni H.M., Guo F., Ding Y., Shi Y.H., Lahiri P. et al. (2016) Sequestosome 1/p62 protein is associated with autophagic removal of excess hepatic endoplasmic reticulum in mice. J. Biol. Chem. 291, 18663–18674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adolph T.E., Tomczak M.F., Niederreiter L., Ko H.J., Bock J., Martinez-Naves E. et al. (2013) Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Diakopoulos K.N., Lesina M., Wormann S., Song L., Aichler M., Schild L. et al. (2015) Impaired autophagy induces chronic atrophic pancreatitis in mice via sex- and nutrition-dependent processes. Gastroenterology 148, 626–638.e17 [DOI] [PubMed] [Google Scholar]
- 73.Antonucci L., Fagman J.B., Kim J.Y., Todoric J., Gukovsky I., Mackey M. et al. (2015) Basal autophagy maintains pancreatic acinar cell homeostasis and protein synthesis and prevents ER stress. Proc. Natl. Acad. Sci. U.S.A. 112, E6166–E6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hashimoto D., Ohmuraya M., Hirota M., Yamamoto A., Suyama K., Ida S. et al. (2008) Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J. Cell Biol. 181, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khaminets A., Behl C. and Dikic I. (2016) Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6–16 [DOI] [PubMed] [Google Scholar]
- 76.Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R. et al. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tian X., Jin R.U., Bredemeyer A.J., Oates E.J., Blazewska K.M., McKenna C.E. et al. (2010) RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol. Cell. Biol. 30, 1269–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hess D.A., Humphrey S.E., Ishibashi J., Damsz B., Lee A.H., Glimcher L.H. et al. (2011) Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology 141, 1463–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huh W.J., Esen E., Geahlen J.H., Bredemeyer A.J., Lee A.H., Shi G. et al. (2010) XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology 139, 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bel S., Pendse M., Wang Y., Li Y., Ruhn K.A., Hassell B. et al. (2017) Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 10.1126/science.aal4677 [DOI] [PMC free article] [PubMed] [Google Scholar]