Abstract
The lysosome plays a pivotal role between catabolic and anabolic processes as the nexus for signalling pathways responsive to a variety of factors, such as growth, nutrient availability, energetic status and cellular stressors. Lysosomes are also the terminal degradative organelles for autophagy through which macromolecules and damaged cellular components and organelles are degraded. Autophagy acts as a cellular homeostatic pathway that is essential for organismal physiology. Decline in autophagy during ageing or in many diseases, including late-onset forms of neurodegeneration is considered a major contributing factor to the pathology. Multiple lines of evidence indicate that impairment in autophagy is also a central mechanism underlying several lysosomal storage disorders (LSDs). LSDs are a class of rare, inherited disorders whose histopathological hallmark is the accumulation of undegraded materials in the lysosomes due to abnormal lysosomal function. Inefficient degradative capability of the lysosomes has negative impact on the flux through the autophagic pathway, and therefore dysregulated autophagy in LSDs is emerging as a relevant disease mechanism. Pathology in the LSDs is generally early-onset, severe and life-limiting but current therapies are limited or absent; recognizing common autophagy defects in the LSDs raises new possibilities for therapy. In this review, we describe the mechanisms by which LSDs occur, focusing on perturbations in the autophagy pathway and present the latest data supporting the development of novel therapeutic approaches related to the modulation of autophagy.
Keywords: Autophagy, Glycogenoses, Lysosomes, Lysosomal storage disorders, Neuronal ceroid lipofuscinoses, Sphingolipidoses
Introduction
Our perspective of the lysosome has shifted remarkably from its standing as a simple, terminal organelle for the degradation of cellular components to become a critical mediator of fundamental metabolic processes. Lysosomes coordinate signals from growth factors and cellular stressors and are sensitive to various metabolites, such as amino acids, glucose, lipids and cholesterol, to pivot cells between anabolic and catabolic processes, including autophagy [1–3]. The importance of the lysosome for cellular function is apparent from the large number of disorders associated with lysosomal failure: collectively known as the lysosomal storage disorders (LSDs), more than 50 inherited conditions affect lysosomal function and many are early-onset and fatal [4–7].
A key cellular homeostatic pathway implicated in several LSDs and myriad human diseases is autophagy. Autophagy is an intracellular degradation pathway essential for cellular survival and organismal health [8–10]. This process is vital for the maintenance of energy and tissue homeostasis by degrading damaged or excess intracellular components such as aggregation-prone proteins, lipids and organelles, and recycling the breakdown products [11]. There are three types of autophagy: macroautophagy (herein referred to as autophagy), microautophagy and chaperone-mediated autophagy (CMA); each type requires functional lysosomes in the final stage for degradation of the cargo [12]. Consequently, disruption of the hydrolytic functions of lysosomes impairs autophagic flux and, conversely, lysosomal function probably requires normal flux through autophagy [13]. Deregulation of autophagy is a common disease mechanism in many LSDs [14,15]. This review will describe the connections between autophagy and the LSDs, highlight common stages of autophagy disrupted in different disorders and discuss autophagy as a potential therapeutic intervention for treating certain LSDs.
Autophagy machinery and signalling
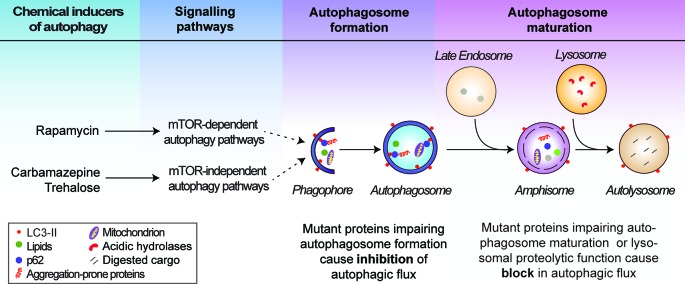
Autophagy encompasses several vesicle fusion events leading to the eventual degradation of its cargo; a dynamic process termed autophagic flux (Figure 1). Multiple autophagy (Atg) genes encoding components of the autophagic machinery are required for the initiation of autophagy, which is marked by the de novo formation of double-membrane structures called phagophores. Two ubiquitin-like conjugation systems involving the Atg5–Atg12–Atg16 complex and phosphatidylethanolamine-conjugated microtubule‐associated protein 1 light chain 3 (LC3-II) are required for the initiation step [16,17]. Cytosolic components such as macromolecules (including specific substrates like p62) and organelles (including mitochondria) are sequestered in the expanding phagophores to form double-membrane vesicles called autophagosomes. LC3-II associates with autophagosome membranes throughout their lifespan and is hence used as a marker for autophagy [18]. Autophagosomes then undergo maturation into autolysosomes by one of two routes: the predominant route is a multi-step process, in which autophagosomes fuse first with late endosomes to form amphisomes, then subsequently with lysosomes; alternatively, autophagosomes can fuse directly with lysosomes [19,20]. This enables delivery of the autophagic cargo to the autolysosomes where these materials are degraded by acidic lysosomal hydrolases (Figure 1). The breakdown products are then exported via lysosomal transporters for recycling [21].
Figure 1. Schematic representation of the autophagy pathway.
Autophagy initiates by the de novo synthesis and elongation of phagophores, which engulf cytosolic materials (autophagic cargo) to form autophagosomes. Autophagosomes predominantly fuse with the late endosomes to form amphisomes and subsequently with the lysosomes to form autolysosomes where the autophagic cargo is degraded by the lysosomal hydrolases. Autophagy can be stimulated by chemical inducers acting via the mTOR-dependent and mTOR-independent pathways regulating autophagy. Defects in autophagic flux at the autophagosome formation and maturation stages are indicated.
There are several molecular mediators of autophagosome maturation, including Rab7, Beclin1–Vps34–Vps15 complex and SNAREs (N-ethylmaleimide-sensitive factor-attachment protein receptors). Beclin1-interacting partners like Atg14L, Ambra1 and UVRAG promote autophagosome maturation whereas Rubicon inhibits this step [20,22,23]. Recently, gene knockout studies in mammalian cells have shown that the Atg8 family proteins (LC3 and GABARAP subfamilies) are crucial for autophagosome–lysosome fusion [24]. GABARAPs were found to preferentially recruit PLEKHM1 (Pleckstrin homology domain containing protein family member 1) [24], which associates with the homotypic fusion and protein sorting (HOPS) complex to mediate autophagosome maturation [25]. The SNARE proteins are critical for membrane tethering and fusion; for example, autophagosomal Syntaxin-17, Atg14 and SNAP-29 interact with late endosomal/lysosomal VAMP8 to mediate autophagosome maturation [26–28]. Upon autolysosome formation, the ATG conjugation system has been suggested to play a role in the degradation of the inner autophagosomal membrane [29]. The late stage of autophagy involving autophagosome maturation is primarily affected in many LSDs; the perturbations in the autophagy pathway in these conditions are described in subsequent sections (Table 1).
Table 1. Overview of defective autophagy in lysosomal storage disorders.
| Disease | Gene | Protein | Function | Storage material | Autophagy phenotype | Autophagic flux | Mechanism |
|---|---|---|---|---|---|---|---|
| NEURONAL CEROID LIPOFUSCINOSES | |||||||
| CLN2 | CLN2/TPP1 | Tripeptidyl peptidase 1 | Serine protease | ATPase subunit c, lipofuscin | Inhibition of autophagosome formation; Reduction in autophagosomes and autophagic degradation [60] | Inhibition | Up-regulation of mTOR signalling [60] |
| CLN3 | CLN3 | CLN3 | Unknown function; Lysosomal membrane protein | ATPase subunit c, lipofuscin | Defect in autophagosome maturation; Accumulation of autophagosomes and autophagic cargo [58–60] | Block | Not known; Possibly due to alteration in Ca2+ homeostasis [61] and deregulation of ARF1–Cdc42 pathway [62] |
| CLN5 | CLN5 | CLN5 | Unknown function; Lysosomal protein | ATPase subunit c, lipofuscin | Accumulation of autophagosomes and autophagic cargo [56] | Block | Not known |
| CLN6 | CLN6 | CLN6 | Unknown function; ER membrane protein | ATPase subunit c, lipofuscin | Accumulation of autophagosomes and autophagic cargo [55,57] | Block | Not known |
| CLN7 | CLN7 | CLN7 | Putative lysosomal transporter | ATPase subunit c, lipofuscin | Accumulation of autophagosomes and autophagic cargo [56] | Block | Not known; Possibly due to impairment in lysosomal function [54] |
| CLN10 | CLN10/CTSD | Cathepsin D | Aspartyl protease | ATPase subunit c, saposins A/D, lipofuscin | Accumulation of autophagosomes and autophagic cargo [64,66] | Block | Not known; Possibly due to loss of cathepsin D function [64] |
| SPHINGOLIPIDOSES | |||||||
| Niemann–Pick type C1 | NPC1 | NPC1 | Cholesterol transporter | Unesterified cholesterol, sphingolipids | Defect in autophagosome maturation; Accumulation of autophagosomes and autophagic cargo [73–80] | Block | Disruption in SNARE machinery [73]; Reduction in sphingosine kinase activity and VEGF [80] |
| Niemann–Pick type C2 | NPC2 | NPC2 | Putative role in cholesterol metabolism and transport | Unesterified cholesterol, sphingolipids | Accumulation of autophagosomes and autophagic cargo [84] | Block | Not known; Possibly due to impairment in lysosomal function [84] |
| Gaucher disease | GBA1 | Glucocerebrosidase | Sphingolipid degradation | Glucosylceramide | Defect in autophagosome maturation; Accumulation of autophagosomes and autophagic cargo [91,93–97] | Block | Not known; Possibly due to down-regulation of TFEB and reduction in lysosomes [91] |
| PSAP | Prosaposin, saposin C | Sphingolipid hydrolase cofactor | Glucosylceramide | Defect in autophagosome maturation; Accumulation of autophagosomes and autophagic cargo [92,93] | Block | Not known; Possibly due to reduction in cathepsin B/D activity [92] | |
| Mucolipidosis type IV | MCOLN1 | TRPML1 | Late endo-lysosomal Ca2+ transporter | Gangliosides, phospholipids, mucopolysaccharides | Accumulation of autophagosomes and autophagic cargo [104,105,109] | Block | Not known; Possibly due to impairment in lysosomal function [107] |
| GLYCOGENOSES | |||||||
| Pompe disease | GAA | Acid α-glucosidase | Glycogen degradation | Glycogen | Accumulation of autophagosomes and autophagic cargo [116–118] | Block | Not known; Possibly due to defects in lysosomal acidification [116] |
| Danon disease | LAMP2 | Isoform LAMP2b | Putative role in autophagosome–lysosome fusion | Glycogen | Accumulation of autophagosomes and autophagic cargo [123–126] | Block | Not known; Possibly due to defects in lysosomal function [124] |
| X-linked myopathy with excessive autophagy | VMA21 | VMA21 | Regulates v-ATPase | Glycogen | Accumulation of autophagosomes [128,129] | Block | Not known; Possibly due to defects in lysosomal acidification and function [128] |
The list in Table 1 highlights selected LSDs where defective autophagy has been demonstrated.
Transcriptional regulation of autophagy occurs via the transcription factor EB (TFEB), which drives the expression of genes related to autophagy and lysosomal biogenesis [30,31]. In turn, Ca2+ stored in the lysosomal lumen has been shown to modulate autophagy by promoting the nuclear localization of TFEB through the activation of calcineurin [32]. Various signalling pathways influence autophagy by acting upstream of the autophagic machinery. These include the mechanistic target of rapamycin complex 1 (mTORC1) signalling pathway (through which growth factors, nutrients and energy status influence autophagy) and mTOR-independent pathways (including inositol and IP3, cAMP, Ca2+, calpain) that negatively regulate autophagy [12,33–38]. Both mTOR-dependent and mTOR-independent signalling are amenable to chemical perturbations for modulating autophagy [34,37,39–41] (Figure 1). For instance, the mTORC1 inhibitors like rapamycin and torin1, and mTOR-independent compounds including trehalose and carbamazepine can stimulate autophagy [35,42–44]. Chemical inducers of autophagy have been tested in some LSDs as a possible therapeutic intervention (Table 2) and are described below.
Table 2. Beneficial effects of the chemical inducers of autophagy in models of lysosomal storage disorders.
| Autophagy inducer | Mechanism of autophagy induction | LSD | Beneficial effects in LSD models |
|---|---|---|---|
| mTOR-DEPENDENT AUTOPHAGY INDUCER | |||
| Rapamycin [42] | Inhibition of mTORC1 [42] | NPC1 | Rescue of autophagic flux and improvement in cell viability in mutant Npc1 MEFs [73], mouse neurons with Npc1 knockdown [73] and NPC1 patient iPSC-derived neuronal and hepatic cells [74] |
| NPB | Reduction in mitochondrial ROS and lipid droplets, and induction of lysosomal exocytosis in NPB patient B lymphocytes [148] | ||
| PD | Reduction in muscle glycogen in Gaa-deficient mice when treated together with recombinant human GAA [167]; Improved autophagic flux and GAA maturation in Pompe disease patient myotubes [117] | ||
| GD | Improvement in lifespan and locomotor activity in GD Drosophila model [97] | ||
| mTOR-INDEPENDENT AUTOPHAGY INDUCER | |||
| Trehalose [44] | Inhibition of SLC2A glucose transporters [145]; Activation of TFEB by Akt inhibition [144] | NPC1 | Rescue of autophagic flux and improvement in cell viability in NPC1 patient iPSC-derived neurons [74] |
| CLN3 | Clearance of ceroid lipopigment deposits in CLN3 patient fibroblasts, and attenuation of neuropathology and extension of lifespan in Cln3-deficient mice [144] | ||
| Carbamazepine [35] | Reduction in inositol and IP3 levels [35] | NPC1 | Rescue of autophagic flux and improvement in cell viability in NPC1 patient iPSC-derived neurons and hepatic cells [74] |
| Lithium [35] | Inhibition of IMPase and reduction in inositol and IP3 levels [35] | NPC1 | Rescue of autophagic flux in mutant Npc1 MEFs [73] |
| CLN3 | Rescue of autophagic flux and improvement in cell viability in mutant Cln3 cerebellar cells [146] | ||
| L-690,330 [35] | Inhibition of IMPase and reduction in inositol and IP3 levels [35] | CLN3 | Rescue of autophagic flux and improvement in cell viability in mutant Cln3 cerebellar cells [146] |
| Verapamil [36] | Inhibition of L-type Ca2+ channel and reduction in cytosolic Ca2+ [36] | NPC1 | Rescue of autophagic flux and improvement in cell viability in NPC1 patient iPSC-derived neurons [74] |
| BRD2716, BRD5631, BRD34009 [142] | Unknown | NPC1 | Rescue of autophagic flux and improvement in cell viability in NPC1 patient iPSC-derived neurons [142] |
Abbreviations: CLN, ceroid lipofuscinosis neuronal disease; GD, Gaucher disease; IMPase, inositol monophosphatase; IP3, inositol 1,4,5-trisphosphate; iPSC, induced pluripotent stem cells; LSD, lysosomal storage disorder; mTORC1, mechanistic target of rapamycin complex I; NPB, Niemann–Pick type B disease; NPC1, Niemann–Pick type C1 disease; PD, Pompe disease; SLC2A, Solute carrier 2A; TFEB, Transcription factor EB.
Lysosomal storage diseases associated with defective autophagy
LSDs are caused by abnormal lysosomal function leading to accumulation of undegraded metabolites [4–7]. The composition of accumulated materials in the lysosomes varies substantially between the LSDs and, while all LSDs are inherited conditions and in many cases mutations are found in genes encoding lysosomal proteins, the types of proteins affected are also varied. As a consequence, the underlying cell biology changes occurring in the LSDs may vary, but each results (directly or indirectly) in reduced clearance of aggregates and diminished cellular homeostasis and survival. Autophagic dysregulation is commonly found in LSDs but again, a spectrum of defects is seen at various stages of the autophagic pathway in different LSDs (Table 1) [14,15]. Here, we will concentrate on some of the better-studied LSDs and detail the different autophagic defects identified.
Neuronal ceroid lipofuscinosis
While defective autophagy has been implicated in the pathology of many different disorders, including cancer and various cardiovascular, metabolic, pulmonary and infectious diseases [10,45], the nervous system appears to be particularly susceptible [46,47]. This is likely due to a combination of the long-lived nature of post-mitotic neurons placing particular stress on protein-clearing processes, the extreme polarization of many neurons and the high metabolic requirements of neurons leading to higher levels of oxidative damage in lysosomes via the Fenton reaction. Consistent with this, dysregulated autophagy has been identified in almost every late-onset neurodegenerative disorder [34,47,48], and similar defects are now being recognized in the neuronal ceroid lipofuscinoses (NCLs), a sub-group of the LSDs that are collectively the most common causes of childhood-onset neurodegeneration [5,49,50]. To date, 13 disease-causing genes have been identified to cause NCL pathology. Several of these genes encode lysosomal proteins, including soluble enzymes/proteins (CLN1/PPT1, CLN2/TPP1, CLN5, CLN10/CTSD and CLN13/CTSF) and membrane proteins (CLN3, CLN7/MFSD8 and CLN12/ATP13A2), but others include endoplasmic reticulum (ER) membrane proteins (CLN6 and CLN8), cytosolic proteins (CLN4/DNAJC5 and CLN14/KCTD7) and one expressed in the secretory pathway (CLN11/GRN) [51–53]. The diverse nature of these proteins, their substrates or cargos and locations within the cell are reflective of the LSDs as a whole: multiple different cell biological processes are probably affected but with each terminating in common pathology. However, as autophagic perturbations start to be identified in NCL disease models (Figure 2), the possibility that each disease hinges on defective autophagy becomes more plausible.
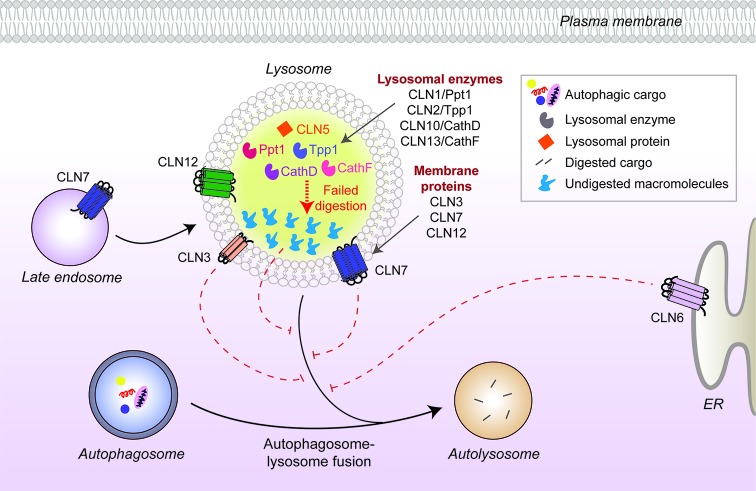
Figure 2. CLN protein distribution and their link to autophagy defects in neuronal ceroid lipofuscinoses.
Many CLN proteins reside in the lysosomal matrix (CLN1, 2, 5, 10, 13) or at the lysosomal membrane (CLN3, 7), while others localize to different cellular compartments such as the ER membrane (CLN6). Disease-causing mutations in some of the CLN proteins inhibit autophagosome maturation (dashed red lines) and block autophagic flux, but the underlying mechanisms are unknown. Mutated lysosomal hydrolases (CLN1, 2, 10, 13) are unable to degrade autophagic cargo, which subsequently accumulate and impair lysosomal function.
Several studies in NCL mouse models have indicated deregulation of autophagy in vivo. A recent study in Cln7-deficient mice reported accumulation of autophagosomes and autophagic substrates coupled with lysosomal dysfunction in the brain, suggesting that a block in autophagic flux may be occurring [54]. A similar build-up of autophagic compartments and substrates was also seen in mouse models of CLN5 and CLN6 diseases, as well as in CLN6 patient-derived fibroblasts [55–57]. The most common form of NCL is Batten disease, caused by mutations in the membrane protein, CLN3 [51]. Defective autophagosome maturation associated with increased autophagic and lysosomal compartments was observed in the Cln3-deficient mouse that models Batten disease, and also in patient fibroblasts as well as patient-specific induced pluripotent stem cell (iPSC)-derived neuronal cells [58–60]. The mechanism underpinning the changes to autophagy in CLN3-deficient cells is not fully understood, and the functions of the CLN3 protein remain unknown, but one potential mechanism may be due to altered Ca2+ homeostasis and deregulation of the ARF1–Cdc42 pathway identified in CLN3 mutant cells that can impact on vesicular trafficking [61,62].
Several forms of NCL are caused by mutations in lysosomal proteases, including Ppt1 (CLN1), Ppt2 (CLN2) and Cathepsin D (CLN10) [53,63]. Loss of cathepsin function commonly leads to dysregulated autophagy, presumably by reducing autophagic flux due to defective cargo clearance. Autophagosomes and mitochondrial proteins accumulate in cultured neurons of mice lacking cathepsin D or B and L [64–66], while in vivo, cathepsin D-deficient mice exhibit widespread accumulation of storage material in the lysosomes (indicating lysosomal dysfunction) and accumulation of autophagosomes. These changes likely lead to the neurodegeneration present in these mice that mimics the human disease [66]. However, a reduction in autophagic flux has been reported in CLN2 patient fibroblasts due to inhibition of autophagosome formation, which is linked to increased ROS and Akt–mTOR signalling pathway [60].
Sphingolipidoses
Sphingolipids are a major class of lipids enriched in the nervous system and are critical for neural development and function [67,68]. Sphingolipid turnover is therefore tightly regulated with their degradation mediated by a multi-step process requiring numerous lysosomal hydrolases [69]. The sphingolipidoses are a class of LSDs caused by deficiencies in functional hydrolases, culminating in the accumulation of wholly or partially undegraded sphingolipids [68]. Members of the sphingolipidoses include Niemann–Pick, Gaucher and Fabry diseases, mucolipidosis, and GM1/2 gangliosidoses such as Tay-Sachs and Sandhoff diseases [68]. Alterations in autophagy have been reported in some of the sphingolipidoses; the most extensively studied being Niemann–Pick type C1 (NPC1) disease.
Niemann–Pick disease
Niemann–Pick disease is a recessively inherited neurodegenerative condition characterized by an accumulation of unesterified cholesterol deposits in various tissues, particularly the brain. The disease is subdivided into types A, B, C1 and C2. Types A and B are caused by mutations in SMPD1, leading to loss of acid sphingomyelinase activity, whereas types C1 and C2 are due to defective cholesterol transport [70]. Ninety-five per cent of cases are attributed to mutations in the lysosomal cholesterol transporter NPC1, with the lysosomal glycoprotein NPC2 that possibly aids in cholesterol trafficking affected in the remainder [71,72].
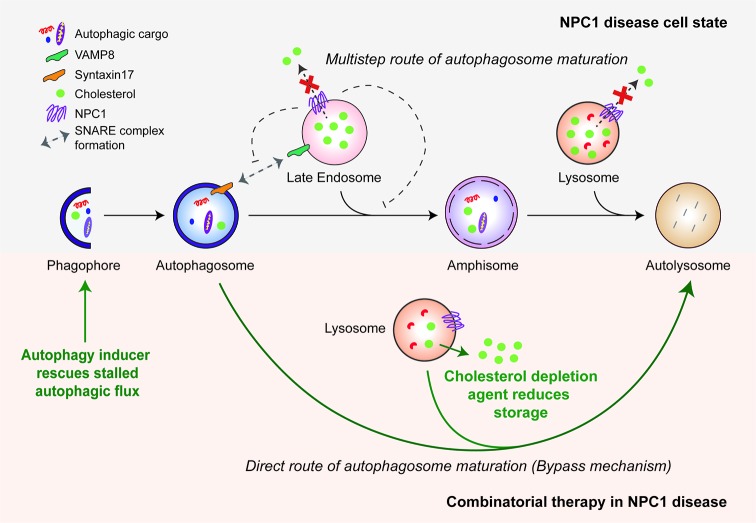
A number of studies have demonstrated a major role for autophagy in the pathology of NPC1 disease and potentially in treatment options. Various experimental platforms, including Npc1 mutant mice and disease-relevant cells (such as neurons) differentiated from human embryonic stem cells (hESCs) with NPC1 knockdown or patient-derived iPSCs, have revealed an accumulation of autophagosomes and lysosomes both in vivo and in vitro [73–80]. We previously reported that this autophagy phenotype is related to a block in autophagic flux (Figure 3). Consistent with this, we characterized a failure in the Syntaxin17/VAMP8 SNARE machinery that retards amphisome formation and significantly stalls the multi-step route of autophagosome maturation [73]. While it is possible that loss-of-function of the NPC1 protein, which normally resides on the late endosomal/lysosomal compartments, directly perturbs amphisome formation, accumulation of lysosomal cholesterol has also been reported to cause aberrant sequestration of SNAREs and prevent autophagosome maturation [81]. Another study, although consistent with dysfunctional autophagosome maturation, reported a distinct mechanism in which diminished sphingosine kinase activity and reduced levels of vascular endothelial growth factor (VEGF) lead to the accumulation of sphingosine that, in turn, impairs autophagic flux [80]. In addition to these mechanisms, depletion of lysosomal Ca2+ stores due to sphingosine storage was seen in NPC1 disease models, and this could possibly influence autophagy via calcineurin or calpain [32,36,82,83]. Similar to the autophagy phenotype in NPC1 disease, a block in autophagic flux has been reported in NPC2 disease as evident by the accumulation of autophagosomes and autophagic substrates as well as impaired lysosomal activity in Npc2-knockdown mouse adipocytes [84].
Figure 3. Autophagy defects in NPC1 disease and the bypass mechanism of autophagosome maturation for restoring autophagic flux.
Mutant NPC1 protein prevents cholesterol efflux from the endo-lysosomal compartments and impairs autophagosome maturation in the multi-step route due to failure in the SNARE machinery. Induction of autophagy by chemical inducers bypasses this block and restores autophagic flux via direct autophagosome–lysosome fusion. A combinatorial treatment strategy is shown with cholesterol depletion agents. The green arrows indicate therapeutic effects of autophagy induction and cholesterol depletion.
The reduced cell viability seen in NPC1 mouse and patient-specific iPSC models has been attributed to dysregulated autophagy [73,74,78,80]. Importantly, since autophagy regulates the clearance of lipids including cholesterol (via lipophagy), autophagy-deficient Atg5–/– cells also exhibited accumulation of these materials similar to the cellular phenotypes observed in NPC1 disease [73,85]. Therefore, the underlying autophagy defects in NPC1 disease can further aggravate the accumulation of lipids and cholesterol, and thus, defective autophagy should be considered a major disease mechanism in Niemann–Pick disease.
Gaucher disease
Gaucher disease is the most common form of sphingolipidosis [86], and is caused by a deficiency in glucocerebrosidase (GCase) activity which catalyses the final step in glycosphingolipid degradation [87]. The loss of enzyme function in Gaucher disease directly impairs this process leading to accumulation of glucocerebroside and widespread pathology that particularly affects the liver and, in severe cases, the CNS [88]. Gaucher disease is most frequently caused by mutations in the GBA1 gene which encodes GCase [87]; however, there are rare cases of Gaucher disease caused instead by mutations in the PSAP gene [89,90]. PSAP encodes the lysosomal glycoprotein prosaposin, the precursor saposins A-D, which are essential cofactors for various sphingolipid hydrolases. Saposin C is required for GCase activity, and thus mutations in PSAP that lead to saposin C deficiency also cause Gaucher disease [89,90].
Various lines of evidence suggest defective autophagy in Gaucher disease. Impaired autophagosome maturation and down-regulation of TFEB, including a reduction in lysosomal gene expression, were found in neurons differentiated from patient-specific iPSCs [91]. A similar impairment in autophagosome degradation was also seen in primary fibroblasts deficient in saposin C; in this case associated with reduced cathepsin B/D activity [92]. Mouse models of Gaucher disease have also revealed similar autophagy phenotypes: accumulation of various autophagic cargo such as dysfunctional mitochondria, ubiquitinated protein aggregates, insoluble α-synuclein and p62, together with autophagosomes and lysosomes were found in the brain or neurons and astrocytes cultured from mice deficient for Gba, Psap or glucosylceramidase [93–96]. Recently, a Drosophila model of neuropathic Gaucher disease generated by knocking out the Gba gene revealed severe lysosomal defects, GCase accumulation and a block of autophagic flux in the brain, resulting in reduced lifespan, neurodegeneration and age-dependent locomotor deficits [97]. Together, multiple model systems highlight deregulation of autophagy in Gaucher disease.
Mucolipidosis type IV
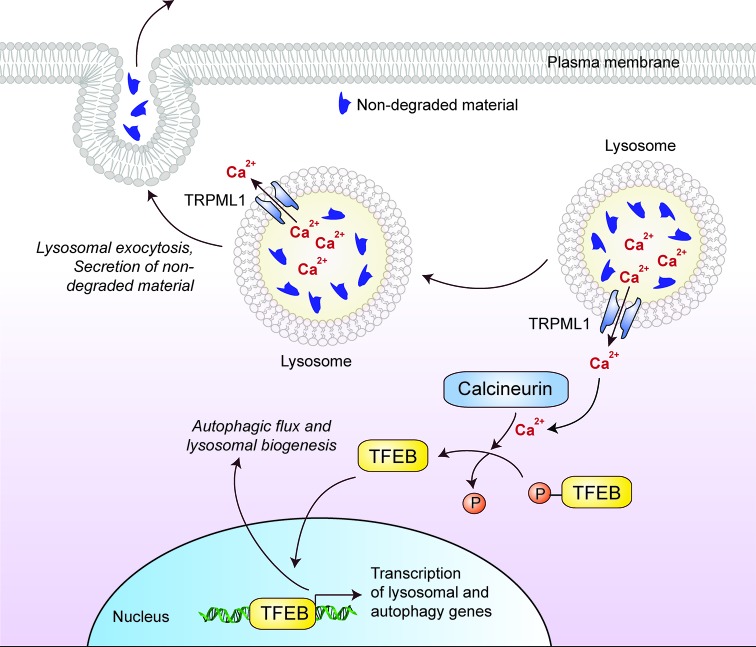
Mucolipidosis type IV (MLIV) is a neurodegenerative condition, caused by mutations in the MCOLN1 gene that result in the loss of TRPML1 function [98,99]. TRPML1 is an inward rectifying, non-selective cation channel of the transient receptor potential family that transports divalent cations, such as Ca2+, Fe2+ and Zn2+, from the lysosomal intraluminal space into the cytosol [100–102]. TRPML1 and TFEB positively regulate each other and together have the potential to impact on autophagy: Ca2+ release by TRPML1 activates the Ca2+-dependent phosphatase, Calcineurin, which mediates dephosphorylation-dependent nuclear translocation of TFEB leading, in turn, to increased expression of several lysosomal and autophagy-related proteins including TRPML1 [32] (Figure 4). Indeed, enhancing TRPML1 activity either by overexpression or by pharmacological stimulation increases autophagic flux [103].
Figure 4. Cellular effects of TFEB that might be of therapeutic benefit in lysosomal storage disorders.
Lysosomal Ca2+ efflux through TRPML1 activates the Ca2+-dependent phosphatase calcineurin, which mediates dephosphorylation-dependent nuclear translocation of TFEB. Nuclear TFEB up-regulates the transcription of genes involved in lysosome biogenesis and autophagy, thereby enhancing autophagic flux. In addition, Ca2+ efflux from peripheral lysosomes promotes lysosomal exocytosis and the secretion of non-degraded materials.
Several models of MLIV have highlighted a defect in autophagy. Accumulation of lysosomes, autophagosomes and autophagy substrates including polyubiquitinated protein aggregates was reported in patient-derived primary fibroblasts, in Mcoln1-deficient mice, and in Caenorhabditis elegans and Drosophila models [104–108]. These studies also implicated a role for wild-type TRPML1 in regulating lysosomal acidification and autophagosome maturation (alkalinization of lysosomes, for instance by inhibiting the vacuolar H+ATPase, inhibits hydrolase function). Furthermore, defects in CMA have been reported in MLIV, possibly through the interaction between TRPML1 with the molecular chaperones Hsc70 and Hsc40 that are required for protein translocation to the lysosome during CMA [109].
Glycogenoses
The glycogenoses are LSDs with profound autophagy defects that particularly affect skeletal and cardiac muscle [110,111]. As such, these diseases are often referred to as autophagic vacuolar myopathies (AVMs), although neuropathology is also present in some cases [112]. Diseases that lead to AVM are linked to genes involved in lysosome acidification, glycogen hydrolysis, and autophagosome maturation and fusion with lysosomes [113]. They include Pompe disease, Danon disease and X-linked myopathy with excessive autophagy (XMEA).
Pompe disease
Pompe disease is caused by mutations in acid α-glucosidase (GAA), a lysosomal glycogen hydrolase [114]. Enzyme activity is completely absent in Pompe disease and consequently the accumulation of intra-lysosomal glycogen, as well as of autophagosomes, is a hallmark of the disease [115]. Primary myoblasts from Gaa-deficient mice have enlarged endosomes, lysosomes and autophagic vacuoles, in addition to delayed endosomal acidification and mobility [116]. These features have been attributed to a block in autophagic flux since elevated levels of the autophagosomal marker LC3-II and autophagic substrate p62 were observed in Gaa-deficient mice and in the myotubes of infantile-onset patients [117,118]. Interestingly, autophagy has been shown to play a role in the maturation of GAA [117] and the clearance of glycogen [119,120], and thus, defective autophagy is implicated as a disease mechanism in Pompe disease.
Danon disease
Danon disease is an X-linked disorder caused by mutations in the LAMP2 gene, which encodes a lysosomal membrane protein [121]. While LAMP2a is required for protein translocation in CMA [122], mutations in the LAMP2b isoform are associated with Danon disease [121]. Various tissues including liver, skeletal and heart muscle from Lamp2-deficient mice displayed accumulation of autophagic compartments, along with a decline in the lysosomal degradation of long-lived proteins in the hepatocytes that are unresponsive to amino acid starvation [123,124]. Muscle biopsies from patients also exhibit accumulation of LC3-II+ autophagic vesicles and large p62 aggregates [125]. These studies point to a likely block in flux affecting the late stage of autophagy. Consistent with this scenario, impaired autophagy associated with defective mitochondrial clearance (mitophagy) was found in cardiomyocytes differentiated from patient-specific iPSC cells with LAMP2 mutations as well as in Lamp2-deficient mice [126].
X-linked myopathy with excessive autophagy
X-linked myopathy with excessive autophagy (XMEA) is characterized by progressive vacuolation and atrophy of skeletal muscle, and is caused by mutations in the VMA21 gene [127]. The VMA21 protein regulates the assembly of the vacuolar ATPase (v-ATPase) required for lysosomal acidification, and thus XMEA is associated with a reduction in the activity of lysosomal hydrolases [128]. Skeletal muscle biopsies show diminished lysosomal degradation and accumulation of autophagic compartments, suggesting a likely block in autophagic flux [128,129].
Why is defective autophagy relevant for LSD pathology?
Deregulation of autophagy has been observed in myriad human diseases including many forms of cancer and neurodegeneration [10,45,47]. Despite appearing at first glance to have diverse changes in both the cell biology and the genetics underpinning them, deregulated autophagy has been reported in most (or possibly all) LSDs where investigated (Table 1) [14,15]. Potentially, dysfunctional autophagy is the common link between the LSDs but why is impaired autophagy such an important component of disease and how does it contribute to cell death? Clues have emerged from in vivo experiments aimed at depleting basal autophagy in a tissue-specific manner in mice. These result in degeneration and dysfunction of the targeted organ, indicating an essential role for autophagy in maintaining tissue homeostasis [130]. For example, brain-specific deletion of Atg5 or Atg7 in mice results in a neurodegenerative phenotype in the absence of any disease-causing, aggregation-prone proteins [131,132]. These genetic studies suggest that the decline in autophagic capability in the LSDs is likely a major contributory factor to the neuropathology that characterizes many of these disorders. Secondly, some of the non-degraded materials in LSDs such as lipids, cholesterol and glycogen are cleared by autophagy [85,119,120]. Hence, deficient autophagy will contribute to the build-up of these components with ever increasing impact on lysosomal function and autophagic flux. For example, as discussed in NPC1 disease where lipids and cholesterol are the primary non-degraded materials, impairment in autophagy will lead to a further accumulation of these macromolecules by reducing lipophagy [73]. This is because lipophagy is retarded due to loss of autophagy, as evident in autophagy-deficient mouse models [85,133]. Finally, impairment in functional autophagic flux can retard the clearance of autophagic cargo including damaged mitochondria and other undesirable materials, which leads to enhanced oxidative damage of lipids and proteins within the lysosomal membrane and a further reduction in efficiency of clearance [134–137]. Therefore, targeting this common pathway underlying several LSDs could be a promising therapeutic intervention.
Autophagy as a potential therapeutic intervention for LSDs
Autophagy has been exploited for therapeutic benefits in diverse transgenic models of human diseases, including several neurodegenerative disorders, certain liver diseases, myopathies and infectious diseases [34,40,41,138,139]. In most of the conditions studied, induction of autophagy ameliorated the disease phenotypes and, in many cases, improved organismal longevity [140,141]. Multiple lines of evidence suggest that autophagy could also be targeted in LSDs as a treatment strategy [14,15] (Table 2). The most suitable disease contexts for this approach are likely to be LSDs where the lysosomal hydrolytic function is not overtly compromized (e.g., by direct mutation of an enzyme) so that the accumulated autophagic cargo can still be digested efficiently if autophagic flux is stimulated. Some of the mechanisms of how autophagy might be beneficial in LSDs are discussed below.
Bypassing stalled autophagosome maturation by small-molecule autophagy inducers
The dysregulated autophagy underpinning many of the LSDs occurs primarily due to failure of autophagosome maturation, which stalls autophagic flux (Table 1). This phenotype probably arises due to a block in the multi-step route of autophagosome maturation. However, this block may not be absolute because we have recently characterized a bypass mechanism in NPC1 disease models which restores functional autophagic flux [73]. We have shown that induction of autophagy facilitates direct autophagosome–lysosome fusion without requiring the formation of amphisomes in the multi-step route (Figure 3). Restoration of autophagic flux enables cargo clearance, which correlates with improved cell viability in primary mouse neurons with Npc1 knockdown and in NPC1 patient iPSC-derived neuronal and hepatic cells [73,74]. Interestingly, we observed cell-type specificity of small molecule autophagy enhancers in these patient-derived cells: rapamycin and carbamazepine rescued cell death and defective autophagy in both iPSC-derived neuronal and hepatic cells but other autophagy inducers including trehalose, verapamil, BRD2716, BRD5631 and BRD34009 were effective only in neurons [74,142]. Strikingly, enhanced autophagic flux did not reduce the cholesterol load in NPC1 mutant cells, meaning a combination strategy coupling autophagy induction with low doses of a cholesterol-depletion agent such as 2-hydroxypropyl-β-cyclodexterin (HPβCD) may be especially effective for NPC1 disease [73,74,143].
Treatment with autophagy inducers has also been found to be beneficial in other LSDs, including CLN3 disease (Batten disease). Administration of trehalose, an mTOR-independent autophagy inducer [44], in a mouse model of CLN3 disease promoted cellular clearance of undegraded materials, attenuated the neuropathology and extended lifespan [144]. Interestingly, although the autophagy-inducing property of trehalose has been recently attributed to the inhibition of the SLC2A family of glucose transporters [145], trehalose was found to activate TFEB by inhibiting Akt independently of mTORC1 [144]. Furthermore, inositol monophosphatase (IMPase) inhibitors such as lithium and L-690,330, which induce mTOR-independent autophagy by reducing inositol and IP3 levels [35], rescued autophagic flux and improved cell viability in mutant Cln3 cerebellar cells [146]. However, further studies are required to determine whether the bypass mechanism of restoring autophagic flux is mediating the beneficial effects of autophagy induction in these contexts.
Induction of autophagy via the classical mTORC1 pathway also had some beneficial effects in LSD models. Although TORC1 activity is dysregulated in a Drosophila model of Gaucher disease [97], the mTORC1 inhibitor rapamycin [42] partially corrects the shortened lifespan and locomotor defects [97], whereas reactivation of dysregulated mTORC1 by knockdown of the Rheb inhibitor, TSC2, in a mouse model of Pompe disease rescued the muscular atrophy and autophagy defects [147]. Rapamycin, while commonly used in research contexts, particular in cell-based assays in vitro, is unlikely to be useful clinically in all LSDs, in part because its efficacy appears to be context-dependent in vivo and subject to feedback loops. Even in vitro rapamycin has differential effects. For example, it reduces mitochondrial ROS and lipid droplets and induces lysosomal exocytosis in Niemann–Pick type B lymphocytes [148], and improves autophagic flux and GAA maturation in Pompe disease patient myotubes [117], but is toxic in neuronal cells differentiated from Gaucher disease patient-derived iPSCs [91]. Since mTOR governs critical cellular function like cell growth and translation [38], the use of mTOR inhibitors for induction of autophagy can produce potential side-effects [149]. Consequently, the mTOR-independent autophagy inducers are considered more desirable for clinical applications in patients [34,40,41,138].
TFEB promotes lysosomal biogenesis and exocytosis
Genetic up-regulation of autophagy via overexpression of TFEB has shown potential as a therapeutic strategy. Overexpression of TFEB enhances the clearance of proteinaceous aggregates in transgenic models of late-onset neurodegeneration disorders [150–152], and improves autophagy and lysosomal defects while rescuing disease severity in some LSDs [153,154]. Apart from mediating the transcription of genes required for autophagy and lysosomal biogenesis [30,31], TFEB also facilitates lysosomal exocytosis via the recruitment and fusion of the lysosomes to the plasma membrane by increasing local Ca2+ concentrations through its target, the TRPML1/MCOLN1 cation channel (Figure 4) [32,155]. This mechanism may underpin some of the beneficial effects of overexpression because enhanced secretion may enable the clearance of pathologically enlarged lysosomes and undegraded materials associated with LSDs that have an impact on autophagic flux.
Experimentally, overexpression of TFEB reduces glycogen load and the size of lysosomes, and promotes autophagosome maturation and lysosomal exocytosis in immortalized myogenic cells from Pompe disease patients [156] and rescues tissue pathology in vivo in autophagy-dependent manner [155,156]. In addition, overexpression of TFEB together with GAA cooperatively facilitated glycogen clearance and improved pathology in skeletal muscles differentiated from Pompe disease patient-derived iPSCs [157]. Similarly, lysosomal exocytosis and reduction in cellular vacuolization occur after TFEB overexpression in various cell and mouse models of LSDs, including multiple sulfatase deficiency (MSD) and mucopolysaccharidosis type IIIA (MPS-IIIA) where a block in autophagy was previously reported [155,158]. Overexpression of TFEB also causes reduction in substrate accumulation in various cell models of Pompe disease, MSD, MPS-II, MPS-IIIA and Batten disease [150,155,156], although it did not rescue the lysosomal depletion phenotype in neuronal cells differentiated from Gaucher disease patient-derived iPSCs [91]. However, TFEB did enhance lysosomal biogenesis in the presence of recombinant GCase in these cells [91]. Taken together, these studies highlight TFEB as a promising therapeutic target with the potential to be manipulated in multiple LSDs.
Combining ERT with autophagy induction
Enzyme replacement therapy (ERT) to replace the absent or non-functional hydrolytic enzyme is approved for patients with Gaucher, Pompe and Fabry diseases and some mucopolysaccharidoses, including MPS type I [159,160]. For Gaucher disease, ERT for the GCase enzyme is a relatively mature therapy with several drugs on the market, including imiglucerase, velaglucerase alfa and taliglucerase alfa [161–163], along with two substrate reduction therapies, miglustat and eliglustat [164,165]. Recent data demonstrate that ERT for the GAA enzyme deficient in Pompe disease improves muscle mass and autophagic flux in muscle biopsies of some patients after treatment [117,166]. As with the Gaucher disease experiments described above [91], combination strategies that include induction of autophagy may prove more effective in Pompe disease and similar LSDs than ERT alone, in this case possibly by stimulating clearance of the accumulating glycogen and facilitating GAA maturation [117,157]. Indeed, experiments with a Pompe mouse model in vivo demonstrated increasing autophagic flux with rapamycin or its analogue CCI-779 alongside ERT for the GAA enzyme reduced muscle glycogen levels compared with either treatment alone [167]. These emerging data highlight the potential benefits of combinatorial treatment strategies in the LSDs.
Conclusions
The LSDs are characterized by accumulation of undegraded macromolecules due to defective or absent lysosomal hydrolases, or because of dysregulated endosomal–lysosomal processes, including altered vesicular trafficking, autophagosome maturation, lysosomal acidification or transport of molecules across the lysosomal membrane. The separate classes of LSDs represent several different defects in lysosomal biology and to some extent require tailored therapies to overcome them. However, one feature apparent in many of the LSDs is dysregulated autophagy (Table 1), and this may be the common therapeutic target that can be exploited similarly in different disorders. Based on the common blockage of autophagic flux, therapies designed to stimulate autophagy and alleviate the block are a logical option to trial. While autophagy inducers have already shown benefit in a few LSDs (Table 2), a broader evaluation of autophagy modulation in the LSDs is urgently required. Autophagy induction shows particular promise in combination with existing therapeutic options such as ERT. However, ERT is applicable to only a subset of the LSDs and the treatment costs are high, and therefore, identifying novel therapeutic interventions for the LSDs that are cheaper and more widely useful is essential. Ultimately, different combinations of therapies are likely to be required for each of the LSDs but autophagy regulators may play a significant role in many.
Summary
Autophagy is a vital cellular process requiring the degradative function of lysosomes.
Abnormal lysosomal function leading to accumulation of undegraded metabolites occurs in the LSDs.
Defects in autophagy are emerging to be a common disease mechanism underlying LSDs.
Stimulation of autophagy is a potential therapeutic intervention in LSDs.
Abbreviations
- ERT
enzyme replacement therapy
- GAA
acid α-glucosidase
- GCase
glucocerebrosidase
- iPSC
induced pluripotent stem cells
- LAMP2
lysosome-associated membrane protein 2
- LSD
Lysosomal storage disorder
- MLIV
mucolipidosis type IV
- MPS
mucopolysaccharidosis
- MSD
multiple sulfatase deficiency
- mTORC1
mechanistic target of rapamycin complex 1
- NCL
neuronal ceroid lipofuscinosis
- NPC1
Niemann–Pick type C1 disease
- ROS
reactive oxygen species
- SNARE
N-ethylmaleimide-sensitive factor-attachment protein receptors
- TFEB
transcription factor EB
- XMEA
X-linked myopathy with excessive autophagy
Funding
The authors thank the funding agencies for supporting our research. S.S. is funded by Wellcome Trust Seed Award [109626/Z/15/Z], UKIERI (UK-India Education and Research Initiative) DST Thematic Partnership Award [2016-17-0087] and Birmingham Fellowship. R.I.T. is funded by a New Investigator Award from the Biotechnology and Biological Sciences Research Council [BBSRC; BB/N008472/1]. T.B. is funded by the Medical Research Council (MRC) Biomedical Catalyst: Developmental Pathway Funding Scheme (DPFS) Award [MR/P007732/1]. T.R.R. is funded by Fundação de Amparo à Pesquisa do Estado de São Paulo [FAPESP; 2015/02041-1], Fundação de Apoio à Pesquisa da Faculdade de Ciências Médicas da Santa Casa de São Paulo (FAP FCMSCSP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)/Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). S.S. is also a Former Fellow for life at Hughes Hall, University of Cambridge, U.K.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Luzio J.P., Pryor P.R. and Bright N.A. (2007) Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–632 [DOI] [PubMed] [Google Scholar]
- 2.Perera R.M. and Zoncu R. (2016) The lysosome as a regulatory hub. Annu. Rev. Cell Dev. Biol. 32, 223–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H. and Ren D. (2015) Lysosomal physiology. Annu. Rev. Physiol. 77, 57–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Futerman A.H. and van Meer G. (2004) The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 5, 554–565 [DOI] [PubMed] [Google Scholar]
- 5.Platt F.M., Boland B. and van der Spoel A.C. (2012) The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J. Cell Biol. 199, 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parenti G., Andria G. and Ballabio A. (2015) Lysosomal storage diseases: from pathophysiology to therapy. Annu. Rev. Med. 66, 471–486 [DOI] [PubMed] [Google Scholar]
- 7.Boustany R.M. (2013) Lysosomal storage diseases–the horizon expands. Nat. Rev. Neurol. 9, 583–598 [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N., Levine B., Cuervo A.M. and Klionsky D.J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravikumar B., Sarkar S., Davies J.E., Futter M., Garcia-Arencibia M., Green-Thompson Z.W. et al. (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 90, 1383–1435 [DOI] [PubMed] [Google Scholar]
- 10.Levine B. and Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stolz A., Ernst A. and Dikic I. (2014) Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16, 495–501 [DOI] [PubMed] [Google Scholar]
- 12.Boya P., Reggiori F. and Codogno P. (2013) Emerging regulation and functions of autophagy. Nat. Cell Biol. 15, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J., Tan S.H., Nicolas V., Bauvy C., Yang N.D., Zhang J. et al. (2013) Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res. 23, 508–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman A.P., Puertollano R., Raben N., Slaugenhaupt S., Walkley S.U. and Ballabio A. (2012) Autophagy in lysosomal storage disorders. Autophagy 8, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward C., Martinez-Lopez N., Otten E.G., Carroll B., Maetzel D., Singh R. et al. (2016) Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim. Biophys. Acta 1861, 269–284 [DOI] [PubMed] [Google Scholar]
- 16.Klionsky D.J. and Schulman B.A. (2014) Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 21, 336–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D. et al. (1998) A protein conjugation system essential for autophagy. Nature 395, 395–398 [DOI] [PubMed] [Google Scholar]
- 18.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T. et al. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima N. (2007) Autophagy: process and function. Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 20.Ganley I.G. (2013) Autophagosome maturation and lysosomal fusion. Essays Biochem. 55, 65–78 [DOI] [PubMed] [Google Scholar]
- 21.Saftig P. and Klumperman J. (2009) Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 [DOI] [PubMed] [Google Scholar]
- 22.Reggiori F. and Ungermann C. (2017) Autophagosome maturation and fusion. J. Mol. Biol. 429, 486–496 [DOI] [PubMed] [Google Scholar]
- 23.Levine B., Liu R., Dong X. and Zhong Q. (2015) Beclin orthologs: integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol. 25, 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen T.N., Padman B.S., Usher J., Oorschot V., Ramm G. and Lazarou M. (2016) Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 215, 857–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D. et al. (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 57, 39–54 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Li L., Hou C., Lai Y., Long J., Liu J. et al. (2016) SNARE-mediated membrane fusion in autophagy. Semin. Cell Dev. Biol. 60, 97–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itakura E., Kishi-Itakura C. and Mizushima N. (2012) The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 [DOI] [PubMed] [Google Scholar]
- 28.Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y. et al. (2015) ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520, 563–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuboyama K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H. and Mizushima N. (2016) The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041 [DOI] [PubMed] [Google Scholar]
- 30.Settembre C., Di Malta C., Polito V.A., Arencibia M.G., Vetrini F., Erdin S. et al. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Settembre C., Fraldi A., Medina D.L. and Ballabio A. (2013) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R. et al. (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He C. and Klionsky D.J. (2009) Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar S. (2013) Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem. Soc. Trans. 41, 1103–1130 [DOI] [PubMed] [Google Scholar]
- 35.Sarkar S., Floto R.A., Berger Z., Imarisio S., Cordenier A., Pasco M. et al. (2005) Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 170, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H. et al. (2008) Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 4, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y.C. and Guan K.L. (2015) mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 125, 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxton R.A. and Sabatini D.M. (2017) mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar S. (2013) Chemical screening platforms for autophagy drug discovery to identify therapeutic candidates for Huntington’s disease and other neurodegenerative disorders. Drug Discover. Today 10, e137–e144 [DOI] [PubMed] [Google Scholar]
- 40.Levine B., Packer M. and Codogno P. (2015) Development of autophagy inducers in clinical medicine. J. Clin. Invest. 125, 14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubinsztein D.C., Codogno P. and Levine B. (2012) Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discover. 11, 709–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blommaart E.F.C., Luiken J.J.F.P., Blommaart P.J.E., van Woerkom G.M. and Meijer A.J. (1995) Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270, 2320–2326 [DOI] [PubMed] [Google Scholar]
- 43.Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y. et al. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkar S., Davies J.E., Huang Z., Tunnacliffe A. and Rubinsztein D.C. (2007) Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α-synuclein. J. Biol. Chem. 282, 5641–5652 [DOI] [PubMed] [Google Scholar]
- 45.Jiang P. and Mizushima N. (2014) Autophagy and human diseases. Cell Res. 24, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubinsztein D.C. (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 47.Nixon R.A. (2013) The role of autophagy in neurodegenerative disease. Nat. Med. 19, 983–997 [DOI] [PubMed] [Google Scholar]
- 48.Menzies F.M., Moreau K. and Rubinsztein D.C. (2011) Protein misfolding disorders and macroautophagy. Curr. Opin. Cell Biol. 23, 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nita D.A., Mole S.E. and Minassian B.A. (2016) Neuronal ceroid lipofuscinoses. Epileptic Disord. 18, 73–88 [DOI] [PubMed] [Google Scholar]
- 50.Palmer D.N., Barry L.A., Tyynela J. and Cooper J.D. (2013) NCL disease mechanisms. Biochim. Biophys. Acta 1832, 1882–1893 [DOI] [PubMed] [Google Scholar]
- 51.Mole S.E. and Cotman S.L. (2015) Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim. Biophys. Acta 1852, 2237–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonati A., Pezzini F., Moro F. and Santorelli F.M. (2014) Neuronal ceroid lipofuscinosis: the increasing spectrum of an old disease. Curr. Mol. Med. 14, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 53.Cárcel-Trullols J., Kovács A.D. and Pearce D.A. (2015) Cell biology of the NCL proteins: What they do and don’t do. Biochim. Biophys. Acta. 1852, 2242–2255 [DOI] [PubMed] [Google Scholar]
- 54.Brandenstein L., Schweizer M., Sedlacik J., Fiehler J. and Storch S. (2016) Lysosomal dysfunction and impaired autophagy in a novel mouse model deficient for the lysosomal membrane protein Cln7. Hum. Mol. Genet. 25, 777–791 [DOI] [PubMed] [Google Scholar]
- 55.Thelen M., Daμμe M., Schweizer M., Hagel C., Wong A.M.S., Cooper J.D. et al. (2012) Disruption of the autophagy-lysosome pathway is involved in neuropathology of the nclf mouse model of neuronal ceroid lipofuscinosis. PLoS One 7, e35493, 10.1371/journal.pone.0035493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leinonen H., Keksa-Goldsteine V., Ragauskas S., Kohlmann P., Singh Y., Savchenko E. et al. (2017) Retinal degeneration in a mouse model Of CLN5 disease is associated with compromised autophagy. Sci. Rep. 7, 1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cannelli N., Garavaglia B., Simonati A., Aiello C., Barzaghi C., Pezzini F. et al. (2009) Variant late infantile ceroid lipofuscinoses associated with novel mutations in CLN6. Biochem. Biophys. Res. Commun. 379, 892–897 [DOI] [PubMed] [Google Scholar]
- 58.Cao Y., Espinola J.A., Fossale E., Massey A.C., Cuervo A.M., MacDonald M.E. et al. (2006) Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J. Biol. Chem. 281, 20483–20493 [DOI] [PubMed] [Google Scholar]
- 59.Lojewski X., Staropoli J.F., Biswas-Legrand S., Simas A.M., Haliw L., Selig M.K. et al. (2014) Human iPSC models of neuronal ceroid lipofuscinosis capture distinct effects of TPP1 and CLN3 mutations on the endocytic pathway. Hum. Mol. Genet. 23, 2005–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vidal-Donet J.M., Cárcel-Trullols J., Casanova B., Aquado C. and Knecht E. (2013) Alterations in ROS activity and lysosomal pH account for distinct patterns of macroautophagy in LINCL and JNCL fibroblasts. PLoS One 8, e55526, 10.1371/journal.pone.0055526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chandrachud U., Walker M.W., Simas A.M., Heetveld S., Petcherski A., Klein M. et al. (2015) Unbiased cell-based screening in a neuronal cell model of batten disease highlights an interaction between Ca2+ homeostasis, autophagy, and CLN3 Protein function. J. Biol. Chem. 290, 14361–14380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schultz M.L., Tecedor L., Stein C.S., Stamnes M.A. and Davidson B.L. (2014) CLN3 Deficient cells display defects in the ARF1-Cdc42 pathway and actin-dependent events. PLoS One 9, e96647, 10.1371/journal.pone.0096647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shacka J.J. and Roth K.A. (2005) Cathepsin deficiency as a model for neuronal ceroid lipofuscinoses. Am. J. Pathol. 167, 1473–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koike M., Nakanishi H., Saftig P., Ezaki J., Isahara K., Ohsawa Y. et al. (2000) Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J. Neurosci. 20, 6898–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koike M., Shibata M., Waguri S., Yoshimura K., Tanida I., Kominami E. et al. (2005) Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am. J. Pathol. 167, 1713–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shacka J.J., Klocke B.J., Young C., Shibata M., Olney J.W., Uchiyama Y. et al. (2007) Cathepsin D deficiency induces persistent neurodegeneration in the absence of Bax-dependent apoptosis. J. Neurosci. 27, 2081–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olsen A.S.B. and Færgeman N.J. (2017) Sphingolipids: membrane microdomains in brain development, function and neurological diseases. Open Biol. 7, 10.1098/rsob.170069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Platt F.M. (2014) Sphingolipid lysosomal storage disorders. Nature 510, 68–75 [DOI] [PubMed] [Google Scholar]
- 69.Sandhoff K. and Kolter T. (2003) Biosynthesis and degradation of mammalian glycosphingolipids. Philos. Trans. R. Soc. B Biol. Sci. 358, 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schuchman E.H. and Desnick R.J. (2017) Types A and B Niemann-Pick disease. Mol. Genet. Metab. 120, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vanier M.T. (2010) Niemann-Pick disease type C. Orphanet J. Rare Dis. 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Storch J. and Xu Z. (2009) Niemann–Pick C2 (NPC2) and intracellular cholesterol trafficking. Biochim. Biophys. Acta 1791, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarkar S., Carroll B., Buganim Y., Maetzel D., Ng A.H.M., Cassady J.P. et al. (2013) Impaired autophagy in the lipid-storage disorder Niemann-Pick Type C1 disease. Cell Rep. 5, 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maetzel D., Sarkar S., Wang H., Abi-Mosleh L., Xu P., Cheng A.W. et al. (2014) Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Rep. 2, 866–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boland B., Smith D.A., Mooney D., Jung S.S., Walsh D.M. and Platt F.M. (2010) Macroautophagy is not directly involved in the metabolism of amyloid precursor protein. J. Biol. Chem. 285, 37415–37426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liao G., Yao Y., Liu J., Yu Z., Cheung S., Xie A. et al. (2007) Cholesterol accumulation is associated with lysosomal dysfunction and autophagic stress in Npc1− /- mouse brain. Am. J. Pathol. 171, 962–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pacheco C.D., Kunkel R. and Lieberman A.P. (2007) Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum. Mol. Genet. 16, 1495–1503 [DOI] [PubMed] [Google Scholar]
- 78.Meske V., Erz J., Priesnitz T. and Ohm T.G. (2014) The autophagic defect in Niemann-Pick disease type C neurons differs from somatic cells and reduces neuronal viability. Neurobiol. Dis. 64, 88–97 [DOI] [PubMed] [Google Scholar]
- 79.Ordonez M.P., Roberts E.A., Kidwell C.U., Yuan S.H., Plaisted W.C. and Goldstein L.S. (2012) Disruption and therapeutic rescue of autophagy in a human neuronal model of Niemann Pick type C1. Hum. Mol. Genet. 21, 2651–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee H., Lee J.K., Park M.H., Hong Y.R., Marti H.H., Kim H. et al. (2014) Pathological roles of the VEGF/SphK pathway in Niemann-Pick type C neurons. Nat. Commun. 5, 5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fraldi A., Annunziata F., Lombardi A., Kaiser H.J., Medina D.L., Spampanato C. et al. (2010) Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO 29, 3607–3620 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Lloyd-Evans E., Morgan A.J., He X., Smith D.A., Elliot-Smith E., Sillence D.J. et al. (2008) Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat. Med. 14, 1247–1255 [DOI] [PubMed] [Google Scholar]
- 83.Kiselyov K., Yamaguchi S., Lyons C.W. and Muallem S. (2010) Aberrant Ca2+ handling in lysosomal storage disorders. Cell Calcium 47, 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guo H., Zhao M., Qiu X., Deis J.A., Huang H., Tang Q.Q. et al. (2016) Niemann-Pick type C2 deficiency impairs autophagy-lysosomal activity, mitochondrial function, and TLR signaling in adipocytes. J. Lipid Res. 57, 1644–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M. et al. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mistry P.K., Lopez G., Schiffmann R., Barton N.W., Weinreb N.J. and Sidransky E. (2017) Gaucher disease: progress and ongoing challenges. Mol. Genet. Metab. 120, 8–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hruska K.S., LaMarca M.E., Scott C.R. and Sidransky E. (2008) Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA). Hum. Mutat. 29, 567–583 [DOI] [PubMed] [Google Scholar]
- 88.Wong K., Sidransky E., Verma A., Mixon T., Sandberg G.D., Wakefield L.K. et al. (2004) Neuropathology provides clues to the pathophysiology of Gaucher disease. Mol. Genet. Metab. 82, 192–207 [DOI] [PubMed] [Google Scholar]
- 89.Vaccaro A.M., Motta M., Tatti M., Scarpa S., Masuelli L., Bhat M. et al. (2010) Saposin C mutations in Gaucher disease patients resulting in lysosomal lipid accumulation, saposin C deficiency, but normal prosaposin processing and sorting. Hum. Mol. Genet. 19, 2987–2997 [DOI] [PubMed] [Google Scholar]
- 90.Tamargo R.J., Velayati A., Goldin E. and Sidransky E. (2012) The role of saposin C in Gaucher disease. Mol. Genet. Metab. 106, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Awad O., Sarkar C., Panicker L.M., Miller D., Zeng X., Sgambato J.A. et al. (2015) Altered TFEB-mediated lysosomal biogenesis in Gaucher disease iPSC-derived neuronal cells. Hum. Mol. Genet. 24, 5775–5788 [DOI] [PubMed] [Google Scholar]
- 92.Tatti M., Motta M., Di Bartolomeo S., Scarpa S., Cianfanelli V., Cecconi F. et al. (2012) Reduced cathepsins B and D cause impaired autophagic degradation that can be almost completely restored by overexpression of these two proteases in Sap C-deficient fibroblasts. Hum. Mol. Genet. 21, 5159–5173 [DOI] [PubMed] [Google Scholar]
- 93.Sun Y., Liou B., Ran H., Skelton M.R., Williams M.T., Vorhees C.V. et al. (2010) Neuronopathic Gaucher disease in the mouse: viable combined selective saposin C deficiency and mutant glucocerebrosidase (V394L) mice with glucosylsphingosine and glucosylceramide accumulation and progressive neurological deficits. Hum. Mol. Genet. 19, 1088–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Osellame L.D., Rahim A.A., Hargreaves I.P., Gegg M.E., Richard-Londt A., Brandner S. et al. (2013) Mitochondria and quality control defects in a mouse model of gaucher disease—links to Parkinson’s disease. Cell Metab. 17, 941–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Y-h Xu, K Xu, Sun Y., Liou B., Quinn B., R-h Li et al. (2014) Multiple pathogenic proteins implicated in neuronopathic Gaucher disease mice. Hum. Mol. Genet. 23, 3943–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Farfel-Becker T., Vitner E.B., Kelly S.L., Bame J.R., Duan J., Shinder V. et al. (2014) Neuronal accumulation of glucosylceramide in a mouse model of neuronopathic Gaucher disease leads to neurodegeneration. Hum. Mol. Genet. 23, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kinghorn K.J., Grönke S., Castillo-Quan J.I., Woodling N.S., Li L., Sirka E. et al. (2016) A Drosophila model of neuronopathic gaucher disease demonstrates lysosomal-autophagic defects and altered mTOR Signalling and is functionally rescued by rapamycin. J. Neurosci. 36, 11654–11670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bassi M.T., Manzoni M., Monti E., Pizzo M.T., Ballabio A. and Borsani G. (2000) Cloning of the gene encoding a novel integral membrane protein, mucolipidin—and identification of the two major founder mutations causing mucolipidosis Type IV. Am. J. Hum. Genet. 67, 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bargal R., Avidan N., Ben-Asher E., Olender Z., Zeigler M., Frumkin A. et al. (2000) Identification of the gene causing mucolipidosis type IV. Nat. Genet. 26, 118–123 [DOI] [PubMed] [Google Scholar]
- 100.LaPlante J.M., Ye C.P., Quinn S.J., Goldin E., Brown E.M., Slaugenhaupt S.A. et al. (2004) Functional links between mucolipin-1 and Ca2+-dependent membrane trafficking in mucolipidosis IV. Biochem. Biophys. Res. Commun. 322, 1384–1391 [DOI] [PubMed] [Google Scholar]
- 101.Eichelsdoerfer J.L., Evans J.A., Slaugenhaupt S.A. and Cuajungco M.P. (2010) Zinc dyshomeostasis is linked with the loss of mucolipidosis IV-associated TRPML1 ion channel. J. Biol. Chem. 285, 34304–34308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dong X.-P., Cheng X., Mills E., Delling M., Wang F., Kurz T. et al. (2008) The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 455, 992–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.De Leo M.G., Staiano L., Vicinanza M., Luciani A., Carissimo A., Mutarelli M. et al. (2016) Autophagosome-lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat. Cell Biol. 18, 839–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Micsenyi M.C., Dobrenis K., Stephney G., Pickel J., Vanier M.T., Slaugenhaupt S.A. et al. (2009) Neuropathology of the Mcoln1 −/− knockout mouse model of mucolipidosis Type IV. J. Neuropathol. Exp. Neurol. 68, 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Venkatachalam K., Long A.A., Elsaesser R., Nikolaeva D., Broadie K. and Montell C. (2008) Motor deficit in a Drosophila model of mucolipidosis Type IV due to defective clearance of apoptotic cells. Cell 135, 838–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miedel M.T., Rbaibi Y., Guerriero C.J., Colletti G., Weixel K.M., Weisz O.A. et al. (2008) Membrane traffic and turnover in TRP-ML1–deficient cells: a revised model for mucolipidosis type IV pathogenesis. J. Exp. Med. 205, 1477–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thompson E.G., Schaheen L., Dang H. and Fares H. (2007) Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 8, 54, 10.1186/1471-2121-8-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Curcio-Morelli C., Charles F.A., Micsenyi M.C., Cao Y., Venugopal B., Browning M.F. et al. (2010) Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiol. Dis. 40, 370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Venugopal B., Mesires N.T., Kennedy J.C., Curcio-Morelli C., LaPlante J.M., Dice J.F. et al. (2009) Chaperone-mediated autophagy is defective in mucolipidosis type IV. J. Cell. Physiol. 219, 344–353 [DOI] [PubMed] [Google Scholar]
- 110.Oldfors A. and DiMauro S. (2013) New insights in the field of muscle glycogenoses. Curr. Opin. Neurol. 26, 544–553 [DOI] [PubMed] [Google Scholar]
- 111.Godfrey R. and Quinlivan R. (2016) Skeletal muscle disorders of glycogenolysis and glycolysis. Nat. Rev. Neurol. 12, 393–402 [DOI] [PubMed] [Google Scholar]
- 112.Nishino I. (2003) Autophagic vacuolar myopathies. Curr. Neurol. Neurosci. Rep. 3, 64–69 [DOI] [PubMed] [Google Scholar]
- 113.Malicdan M.C. and Nishino I. (2012) Autophagy in lysosomal myopathies. Brain Pathol. 22, 82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raben N., Plotz P. and Byrne B.J. (2002) Acid α-glucosidase deficiency (glycogenosis Type II, Pompe disease). Curr. Mol. Med. 2, 145–166 [DOI] [PubMed] [Google Scholar]
- 115.Dasouki M., Jawdat O., Almadhoun O., Pasnoor M., McVey A.L., Abuzinadah A. et al. (2014) Pompe disease: literature review and case series. Neurol. Clin. 32, 751–776, ix [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fukuda T., Ewan L., Bauer M., Mattaliano R.J., Zaal K., Ralston E. et al. (2006) Dysfunction of endocytic and autophagic pathways in a lysosomal storage disease. Ann. Neurol. 59, 700–708 [DOI] [PubMed] [Google Scholar]
- 117.Nascimbeni A.C., Fanin M., Masiero E., Angelini C. and Sandri M. (2012) The role of autophagy in the pathogenesis of glycogen storage disease type II (GSDII). Cell Death Differ. 19, 1698–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N. et al. (2008) Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 17, 3897–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zirin J., Nieuwenhuis J. and Perrimon N. (2013) Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol. 11, e1001708, 10.1371/journal.pbio.1001708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kotoulas O.B., Kalamidas S.A. and Kondomerkos D.J. (2004) Glycogen autophagy. Microsc. Res. Tech. 64, 10–20 [DOI] [PubMed] [Google Scholar]
- 121.Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T. et al. (2000) Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406, 906–910 [DOI] [PubMed] [Google Scholar]
- 122.Bandyopadhyay U., Kaushik S., Varticovski L. and Cuervo A.M. (2008) The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell. Biol. 28, 5747–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tanaka Y., Guhde G., Suter A., Eskelinen E.-L., Hartmann D., Lullmann-Rauch R. et al. (2000) Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902–906 [DOI] [PubMed] [Google Scholar]
- 124.Eskelinen E.-L., Illert A.L., Tanaka Y., Schwarzmann G., Blanz J., von Figura K. et al. (2002) Role of LAMP-2 in lysosome biogenesis and autophagy. Mol. Biol. Cell 13, 3355–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nascimbeni A.C., Fanin M., Angelini C. and Sandri M. (2017) Autophagy dysregulation in Danon disease. Cell Death Dis. 8, e2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hashem S.I., Murphy A.N., Divakaruni A.S., Klos M.L., Nelson B.C., Gault E.C. et al. (2017) Impaired mitophagy facilitates mitochondrial damage in Danon disease. J. Mol. Cell Cardiol. 108, 86–94 [DOI] [PubMed] [Google Scholar]
- 127.Dowling J.J., Moore S.A., Kalimo H. and Minassian B.A. (2015) X-linked myopathy with excessive autophagy: a failure of self-eating. Acta Neuropathol. (Berl.) 129, 383–390 [DOI] [PubMed] [Google Scholar]
- 128.Ramachandran N., Munteanu I., Wang P., Ruggieri A., Rilstone J.J., Israelian N. et al. (2013) VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol. (Berl.) 125, 439–457 [DOI] [PubMed] [Google Scholar]
- 129.Munteanu I., Kalimo H., Saraste A., Nishino I. and Minassian B.A. (2017) Cardiac autophagic vacuolation in severe X-linked myopathy with excessive autophagy. Neuromuscul. Disord. 27, 185–187 [DOI] [PubMed] [Google Scholar]
- 130.Mariño G., Madeo F. and Kroemer G. (2011) Autophagy for tissue homeostasis and neuroprotection. Curr. Opin. Cell Biol. 23, 198–206 [DOI] [PubMed] [Google Scholar]
- 131.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I. et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 132.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R. et al. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 133.Singh R. and Cuervo A.M. (2012) Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012, 282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Youle R.J. and Narendra D.P. (2011) Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Green D.R. and Levine B. (2014) To be or not to be? how selective autophagy and cell death govern cell fate Cell 157, 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schneider J.L. and Cuervo A.M. (2014) Autophagy and human disease: emerging themes. Curr. Opin. Genet. Dev. 26, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee J., Giordano S. and Zhang J. (2012) Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 441, 523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sarkar S., Ravikumar B., Floto R.A. and Rubinsztein D.C. (2009) Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 16, 46–56 [DOI] [PubMed] [Google Scholar]
- 139.Sarkar S., Perlstein E.O., Imarisio S., Pineau S., Cordenier A., Maglathlin R.L. et al. (2007) Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat. Chem. Biol. 3, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Madeo F., Zimmermann A., Maiuri M.C. and Kroemer G. (2015) Essential role for autophagy in life span extension. J. Clin. Invest. 125, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Morselli E., Galluzzi L., Kepp O., Criollo A., Maiuri M.C., Tavernarakis N. et al. (2009) Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging 1, 961–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuo S.Y., Castoreno A.B., Aldrich L.N., Lassen K.G., Goel G., Dančík V. et al. (2015) Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc. Natl Acad. Sci. U.S.A. 112, E4281–E4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sarkar S., Maetzel D., Korolchuk V.I. and Jaenisch R. (2014) Restarting stalled autophagy a potential therapeutic approach for the lipid storage disorder, Niemann-Pick type C1 disease. Autophagy 10, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Palmieri M., Pal R., Nelvagal H.R., Lotfi P., Stinnett G.R., Seymour M.L. et al. (2017) mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 8, 14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.DeBosch B.J., Heitmeier M.R., Mayer A.L., Higgins C.B., Crowley J.R., Kraft T.E. et al. (2016) Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci. Signal. 9, ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chang J.-W., Choi H., Cotman S.L. and Jung Y.-K. (2011) Lithium rescues the impaired autophagy process in CbCln3 Δex7/8/Δex7/8 cerebellar cells and reduces neuronal vulnerability to cell death via IMPase inhibition. J. Neurochem. 116, 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lim J.-A., Li L., Shirihai O.S., Trudeau K.M., Puertollano R. and Raben N. (2017) Modulation of mTOR signaling as a strategy for the treatment of Pompe disease. EMBO Mol. Med. 9, 353–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Canonico B., Cesarini E., Salucci S., Luchetti F., Falcieri E., Di Sario G. et al. (2016) Defective autophagy, mitochondrial clearance and lipophagy in Niemann-Pick Type B lymphocytes. PLoS One 11, e0165780, 10.1371/journal.pone.0165780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pallet N. and Legendre C. (2013) Adverse events associated with mTOR inhibitors. Expert Opin. Drug Saf. 12, 177–186 [DOI] [PubMed] [Google Scholar]
- 150.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A. et al. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
- 151.Decressac M., Mattsson B., Weikop P., Lundblad M., Jakobsson J. and Björklund A. (2013) TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl Acad. Sci. U.S.A. 110, E1817–E1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Martini-Stoica H., Xu Y., Ballabio A. and Zheng H. (2016) The autophagy–lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci. 39, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sardiello M. (2016) Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases. Ann. N.Y. Acad. Sci. 1371, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Napolitano G. and Ballabio A. (2016) TFEB at a glance. J. Cell Sci. 129, 2475–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Medina Diego L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C. et al. (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 21, 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Spampanato C., Feeney E., Li L., Cardone M., Lim J.-A., Annunziata F. et al. (2013) Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 5, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sato Y., Kobayashi H., Higuchi T., Shimada Y., Ida H. and Ohashi T. (2016) TFEB overexpression promotes glycogen clearance of Pompe disease iPSC-derived skeletal muscle. Mol. Ther.–Methods Clin. Dev. 3, 16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Settembre C., Fraldi A., Jahreiss L., Spampanato C., Venturi C., Medina D. et al. (2008) A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 17, 119–129 [DOI] [PubMed] [Google Scholar]
- 159.Desnick R.J. and Schuchman E.H. (2012) Enzyme replacement therapy for lysosomal diseases: lessons from 20 years of experience and remaining challenges. Annu. Rev. Genomics Hum. Genet. 13, 307–335 [DOI] [PubMed] [Google Scholar]
- 160.Ratko T.A., Marbella A., Godfrey S. and Aronson N. (2013) Enzyme-Replacement Therapies for Lysosomal Storage Diseases. Agency for Healthcare Research and Quality, Comparative Effectiveness Technical Briefs, Rockville (MD) Report No.: 12(13)-EHC154-EF [PubMed] [Google Scholar]
- 161.Cox T.M. (2010) Recommendations for treating patients with Gaucher disease with emerging enzyme products. Blood Cells Mol. Dis. 44, 84–85 [DOI] [PubMed] [Google Scholar]
- 162.Aerts J.M.F.G., Yasothan U. and Kirkpatrick P. (2010) Velaglucerase alfa. Nat. Rev. Drug Discover. 9, 837–838 [DOI] [PubMed] [Google Scholar]
- 163.Hollak C.E. (2012) An evidence-based review of the potential benefits of taliglucerase alfa in the treatment of patients with Gaucher disease. Core Evidence 7, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.McEachern K.A., Fung J., Komarnitsky S., Siegel C.S., Chuang W.-L., Hutto E. et al. (2007) A specific and potent inhibitor of glucosylceramide synthase for substrate inhibition therapy of Gaucher disease. Mol. Genet. Metab. 91, 259–267 [DOI] [PubMed] [Google Scholar]
- 165.Bennett L.L. and Mohan D. (2013) Gaucher disease and its treatment options. Ann. Pharmacother. 47, 1182–1193 [DOI] [PubMed] [Google Scholar]
- 166.Ripolone M., Violano R., Ronchi D., Mondello S., Nascimbeni A., Colombo I. et al. (2017) Effects of short-to-long term enzyme replacement therapy (ERT) on skeletal muscle tissue in late onset Pompe disease (LOPD). Neuropathol. Appl. Neurobiol. doi: 10.1111/nan.12414 [DOI] [PubMed] [Google Scholar]
- 167.Ashe K.M., Taylor K.M., Chu Q., Meyers E., Ellis A., Jingozyan V. et al. (2010) Inhibition of glycogen biosynthesis via mTORC1 suppression as an adjunct therapy for Pompe disease. Mol. Genet. Metab. 100, 309–315 [DOI] [PubMed] [Google Scholar]