Abstract
Although genome analyses have suggested parallels between archaeal and eukaryotic replication systems, little is known about the DNA replication mechanism in Archaea. By two-dimensional gel electrophoreses we positioned a replication origin (oriC) within 1 kb in the chromosomal DNA of Pyrococcus abyssi, an anaerobic hyperthermophile, and demonstrated that the oriC is physically linked to the cdc6 gene. Our chromatin immunoprecipitation assays indicated that P. abyssi Cdc6 and minichromosome maintenance (MCM) proteins bind preferentially to the oriC region in the exponentially growing cells. Whereas the oriC association of MCM was specifically inhibited by stopping DNA replication with puromycin treatment, Cdc6 protein stayed bound to the replication origin after de novo protein synthesis was inhibited. Our data suggest that archaeal and eukaryotic Cdc6 and MCM proteins function similarly in replication initiation and imply that an oriC association of MCM could be regulated by an unknown mechanism in Archaea.
Keywords: Archaea, minichromosome maintenance helicase, origin association, initiator protein
The initiation step of chromosomal DNA replication is of fundamental importance for the inheritance of genetic material and cell cycle regulation and therefore has attracted considerable experimental interest over the years. Detailed studies of DNA replication initiation in Eukarya and Bacteria have identified multistep processes leading to the unwinding of duplex DNA at a replication origin (recently reviewed in refs. 1 and 2). In comparison with the relatively detailed knowledge obtained for Bacteria and Eukarya with regard to their replication initiation, until recently, no data were available on the replication mechanism in Archaea, the third domain of life. Because many eukaryotic replication proteins, including the initiator proteins Orc1, Cdc6, and minichromosome maintenance (MCM), have homologs in Archaea but not in Bacteria (3–6), studies of the archaeal replication system have the potential to elucidate hitherto unresolved questions with regard to the eukaryotic replication mechanism. In the absence of biochemical or genetic in vivo data, whether the archaeal initiator proteins are functionally analogous to their eukaryotic counterparts has remained elusive. Nevertheless, the recent demonstration that the single MCM protein of the archaeon Methanobacterium thermoautotrophicum forms a double-homohexamer complex with a processive 3′ to 5′ ATP-dependent DNA helicase activity in vitro (7–9) has illustrated how identifying the properties of an archaeal replication protein can aid in understanding processes leading to unwinding of the replication origin by eukaryotic replication proteins.
For a long time, the study of archaeal DNA replication in vivo and in vitro has been hampered by the absence of information about the nature of the replication origins in these atypical prokaryotes. Complete archaeal genome sequences have made it possible to predict the location of a single replication origin in M. thermoautotrophicum and Pyrococcus sp. genomes on the basis of asymmetry in base composition between leading and lagging strands (10, 11). In Pyrococcus abyssi, the location of the predicted origin coincides with an early replicating chromosomal segment of 80 kb identified by radioactive labeling of chromosomal DNA in cultures released from a replication block (12). Our in silico and labeling data also allowed us to conclude that the hyperthermophilic archaeon P. abyssi uses a single bidirectional origin to replicate its genome. We proposed that this origin would correspond to the long intergenic region conserved in all three known Pyrococcus sp. genomes directly 5′ upstream of cdc6, which contains sequence elements similar to those found in the other chromosomal replication origins. Our data also indicated that the replication speed in Archaea is similar to that in Bacteria and up to 10 times higher than in Eukarya. Therefore, despite their very different replication proteins, in terms of replication speed and origin utilization, the two prokaryotic domains (i.e., Bacteria and Archaea) share a similar replication mode, whereas Eukarya use multiple replication origins to accurately replicate their much larger linear chromosomes. How subsets of archaeal replication proteins, either similar to eukaryotic proteins or limited to Archaea, like, e.g., a novel type II DNA topoisomerase (13) and a family D DNA polymerase (14), are used for replication in Archaea remains unclear. Thus, in addition to the bacterial features described above, the archaeal replication mechanism could consist of eukaryotic and/or yet to be discovered unique archaeal features.
Pyrococcus species are unique among those Archaea for which full-length genome sequences are available, in that they contain only a single gene similar to eukaryotic Cdc6 and the Orc1 subunit of the origin recognition complex (ORC) (Cdc6/Orc1, referred to hereafter as Cdc6 for simplicity) as well as the MCM proteins. These proteins are the only known candidate initiator proteins present in these species. With the use of neutral/neutral two-dimensional (N/N 2D) agarose gel electrophoreses, the defined origin was found in a ≈10-kb window in the vicinity of genes encoding several replication proteins [i.e., the Cdc6, two subunits of the family D DNA polymerase, and two subunits of a clamp loader (RF-C)], in accordance with our earlier proposal for the origin location in P. abyssi (12). With the use of in vivo chemical cross-linking assays we also have investigated the specific origin association of P. abyssi Cdc6 and MCM. These studies revealed that P. abyssi Cdc6 and MCM proteins associate specifically with origin DNA in exponential phase cultures. Whereas origin association of P. abyssi Cdc6 was maintained, association of MCM was abolished in cells not replicating their DNA. These data are experimental proof that, as in Eukarya, Cdc6 and MCM helicase form a core complex required for replication initiation in Archaea.
Materials and Methods
Cell Culture Techniques.
P. abyssi GE5 (strain Orsay) was cultivated anaerobically in liquid medium at 95°C with enriched medium as described (12). Cultures were inoculated with 1/20th of the final culture volume with freshly obtained inoculate. Cells were harvested after 5–6 h of growth at approximate cell densities of 5 × 107 (for N/N 2D gel electrophoresis) or 1–2 × 107 (for other purposes) cells per milliliter. The absence of chromosomal rearrangements in the P. abyssi genome was confirmed by contour-clamped homogeneous electric field electrophoreses (data not shown).
N/N 2D Agarose Gel Electrophoresis.
P. abyssi cells from asynchronous cultures were washed and resuspended under aerobic conditions with buffer A (250 mM NaCl/25 mM Tris⋅Cl, pH 8.0/10 mM EDTA). Cells were suspended at 1010 cells per milliliter and encapsulated with an equal volume of 0.8% low-gelling-temperature agarose (Type VII; Sigma) in buffer A. DNA was prepared from agarose plugs after their overnight treatment in 1 mg/ml proteinase K, 10 mM Tris⋅Cl (pH 9.1), 0.5 M EDTA at 37°C. Obtained plugs, which were washed extensively with buffer A and TE (10 mM Tris/1 mM EDTA, pH 8.0) before further manipulations, contained ≈20–30 μg of DNA/ml.
Intact DNA was digested in agarose plugs with the use of the indicated restriction enzymes, following the manufacturer's recommendations for pulse-field electrophoresis qualified restriction enzymes (Promega or New England Biolabs), including 2 units of β-agarose (Sigma) per 100-μl plug. After the digestions were completed, the plugs were extracted with phenol and chloroform, followed by isopropanol precipitation of the DNA samples and two washes with 70% ethanol at room temperature. Ten to 15 micrograms of digested DNA was run on N/N 2D gels (15) with 1× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) in a cold room (first dimension: 0.4% agarose, 0.7 V/cm, ≈40 h; second dimension: 1% agarose, 3 V/cm, 13–15 h). Southern blots using Hybond-N (Amersham Pharmacia) filters and probe preparations were performed as described (16). All radioactive signals were revealed by exposing the filters to PhosphorImager screens for up to 72 h, and signals were analyzed with imagequant software (Molecular Dynamics).
Chromatin Binding and Detection of Association Between the P. abyssi oriC and Archaeal Initiator Proteins.
Purification of P. furiosus Cdc6 and MCM proteins after their production in Escherichia coli and polyclonal antibodies raised against these proteins has yet to be reported (Y.I., S. Ishino, M. Nakae, and K. Komori, unpublished data). Antisera thus obtained recognized one major chromatin-associated band of approximately expected molecular weight in cell-free extracts of P. furiosus and were shown to cross-react with the corresponding P. abyssi proteins. For chromatin association studies, P. abyssi GE5 cells were harvested in the late exponential phase, 5–6 h after inoculation. Stationary phase cultures were collected after overnight cultivation. Samples were washed once with washing buffer (25 mM Hepes, pH 7.0/1 M sorbitol). Cell suspensions (≈109 cells per milliliter) were disrupted in extraction buffer (25 mM Hepes, pH 7.0/15 mM MgCl2/100 mM NaCl/0.4 M sorbitol/0.5% Triton X-100/1 mM PMSF/0.5 μg/ml leupeptin/1 μg/ml pepstatin A). After 10 min on ice, soluble proteins (supernatant) and chromosomal DNA-enriched insoluble fraction (pellet) were separated by centrifugation at 14,000 × g for 20 min. The insoluble fraction, containing DNA and chromatin-associated proteins, was washed once and dissolved in an equal volume of extraction buffer by brief sonication (45 W, four times for 15 s each), yielding DNA fragments of 500–1,000 bp. The obtained fractions were analyzed by immunoblotting with polyclonal rabbit antiserums. An enhanced chemiluminescence kit (Amersham Pharmacia) was used to detect immunocomplexes as recommended. In each case, several protein dilutions and exposures were used to ensure the linearity of analyzed signals. Dot plot and quantitative Western blot analyses were performed with the use of purified proteins as controls.
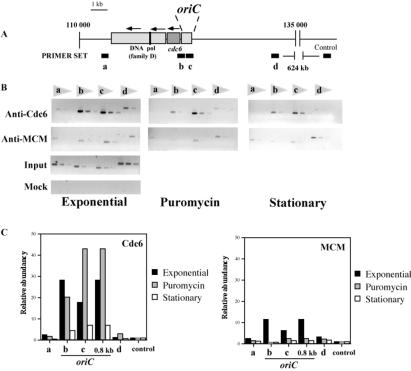
For chromatin immunoprecipitation assays, P. abyssi cells obtained as described above were quickly cooled down to room temperature, followed by fixation with 1% formaldehyde for 15 min under aerobic conditions with gentle agitation. Samples for immunoprecipitation were prepared by sonication in extraction buffer followed by centrifugation as above. Supernatant (0.5 ml) was incubated with 5 μl of anti-Cdc6 and anti-MCM sera for 4 h after pretreatment with protein A-Sepharose. Immunocomplexes were collected by adding 150 μl of protein A-Sepharose with gentle shaking for 2 h, followed by extensive washes with extraction buffer. Precipitated DNA was extracted from immunocomplexes as described (17). The obtained DNA was directly used as a template in PCR reactions [1.25 units of Pfu DNA polymerase (Promega), 20 cycles under conditions recommended by the manufacturer]. Six different primer sets (primer sequences are available on request; see also Fig. 4) with three different template dilutions were used to ensure the linearity of the assays, addressing the specific association of Cdc6 and MCM with P. abyssi oriC DNA. Quantifications described in the text were obtained by determining relative amounts of amplified PCR products with the use of a PCR target located 624 kb from the mapped P. abyssi origin as the control locus. The intensities of each PCR product stained by ethidium bromide were determined from digitized gel images with National Institutes of Health image software, accounting for differences in amplification efficiency observed for different primer sets with the use of input DNA. In each case, values obtained for the control locus were set to one arbitrary unit. All results shown are averages of 2–3 independent experiments. All signals were dependent on the antisera used. In the absence of genetic tools for P. abyssi, we have not been able to directly address whether signals detected at the distant control locus are due to antiserum-specific background.
Figure 4.
In vivo association of Cdc6 and MCM with the P. abyssi replication origin detected by formaldehyde cross-linking. (A) The locations of oriC and cdc6 in P. abyssi sequence coordinates, as well as five primer sets used for PCR amplification (a, 113,000; b, oriC (Left); c, oriC (Right); d, 133,000; control, 745,000) within a genome segment (110,000–135,000). (B) Specific association of Cdc6 and MCM proteins with chromatin was addressed with specific antisera and primer sets a–d, normalized against signals observed for the control locus 624 kb from the origin. 1/240, 1/960, and 1/3,840 of total precipitated DNA were used as PCR templates (loaded from left to right). Input and Mock refer to reactions performed with total sonicated input DNA and negative control reactions performed without antiserum, respectively. Analyses were performed with exponential-phase (with and without puromycin treatment) and stationary-phase cultures. (C) The relative abundance of each PCR product obtained under various conditions is indicated; 0.8 kb refers to data obtained with the use of an 800-nt-long oriC-specific PCR target (data not shown).
Results
An Active Replication Origin Is Located in the Vicinity of the P. abyssi cdc6 Gene.
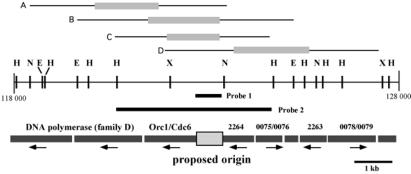
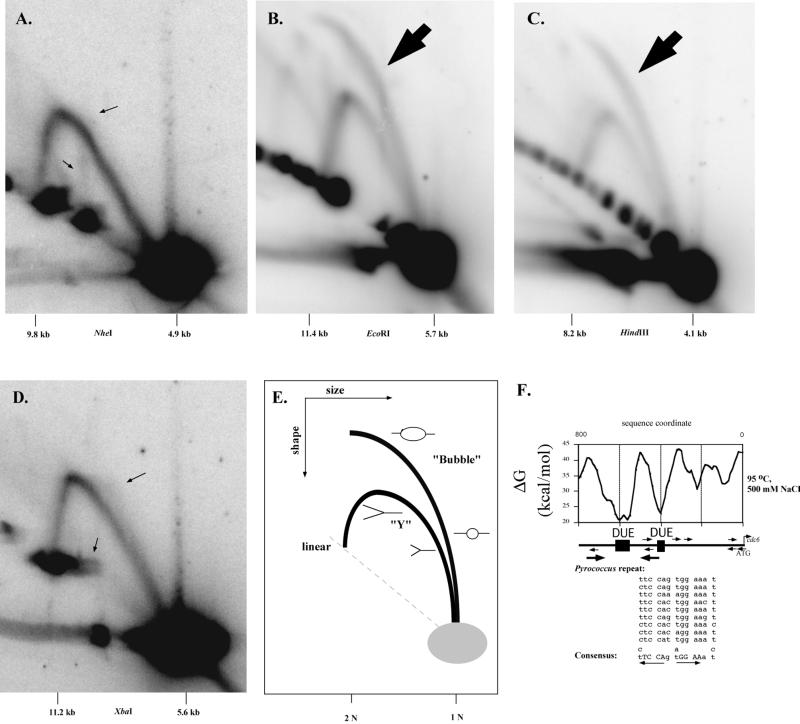
Because our earlier observations have suggested a physical linkage between a replication origin and the P. abyssi cdc6 gene, we investigated how a 10-kb chromosomal segment centered around the cdc6 gene (P. abyssi sequence coordinates 118,000 to 128,000; Fig. 1) is replicated in vivo. A replication pattern of this region was revealed by using N/N 2D agarose gel electrophoreses (15), which rely on electrophoretic separation of the different classes of replication intermediates (Fig. 2E). Total intact DNA, thus including all replication intermediates, was gently prepared from P. abyssi exponential growth phase cells embedded in agarose plugs. After N/N 2D gel electrophoreses, the replication mode of four restriction fragments covering the aforementioned P. abyssi chromosomal region was investigated with the use of two specific radioactive probes (Fig. 1). These experiments with probe 1 (Figs. 1 and 2) indicated that the fragments A and D, which are located distally to the cdc6 [4.9-kb NheI fragment (Fig. 2A); 5.6-kb XbaI fragment (Fig. 2D)], produced a strong “Y” arc; hence, they are replicated with one fork traveling from one end to the other. In contrast, the 5.7-kb EcoRI (Fig. 2B) and 4.1-kb HindIII (Fig. 2C) fragments produced a composite pattern consisting of a well-defined “bubble” arc (indicated by filled arrows) and a typical “Y” arc. As bubble arcs can be detected reliably only when a replication origin is located in the central third of a restriction fragment (18) and cdc6 is centrally located only within the latter fragments, these results demonstrate that the P. abyssi chromosomal replication origin is located in a ≈1-kb window either just upstream from or partially overlapping the cdc6 locus. The structure of the oriC region, together with the identification of two duplex unwinding elements (Fig. 2F), as well as conserved nucleotide repeats (indicated by arrows), further supports the notion that an active replication origin is located within 800 nt immediately 5′ upstream of P. abyssi cdc6. No obvious replication pause sites, manifesting themselves as dark spots along Y arcs or bubble arcs, originated from this 10-kb chromosomal region. Therefore, considering our earlier data showing that Pyrococcus DNA replication proceeds bidirectionally (12), the P. abyssi origin studied here fires symmetrically in two directions within the analyzed chromosomal segment.
Figure 1.
Physical map of a P. abyssi chromosomal segment (sequence coordinates 118,000–128,000) carrying the predicted replication origin (12). Locations of several restriction sites (H, HindIII; N, NheI; E, EcoRI; X, XbaI), probes used for N/N 2D analyses are indicated. Fragments A (4.9-kb NheI fragment), B (5.7-kb EcoRI fragment), C (4.1-kb HindIII fragment), and D (5.6-kb XbaI fragment) correspond to those analyzed in Fig. 2. The central third of the each fragment [a bubble detection zone (18)] is indicated by gray shading. A ≈1-kb boxed region upstream of, and partially overlapping with, the P. abyssi cdc6 indicates the region containing an active replication origin as demonstrated in Fig. 2.
Figure 2.
N/N 2D gel analyses of replication intermediates from the predicted replication origin region of P. abyssi. Replication intermediates were prepared from asynchronous P. abyssi cultures by an agarose plug method. A–D correspond to the analysis of restriction fragments A to D with the use of probe 1 (Fig. 1). (E) The expected pattern for fragments containing either one replication fork (Y arc) or two replication forks with an internal initiation site (bubble arc). The filled arrows indicate the bubble arcs observed for fragments B and C by probe 1 (similar results for fragments B and C were obtained when probe 2 was used). In addition to a well-defined Y arc, fragments A and D show weak signals (small arrows) detected by probe 1. These signals are consistent with an asymmetrically located replication origin within the central third of the fragments B and C. The mobilities of the dark spots on the line traced by linear fragments are consistent with impartial digests. (F) Structure of the oriC region with the identification of the two duplex unwinding elements (DUE) (19) and conserved nucleotide repeats (indicated by arrows) (12). Longer arrows indicate 34-bp repeats earlier unnoticed.
P. abyssi Cdc6 and MCM Are Abundantly Expressed Proteins.
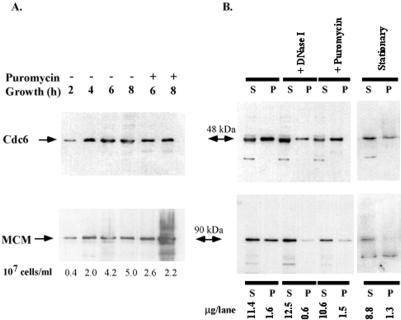
To gain insight into steady-state expression levels of putative archaeal initiator proteins, whole-cell samples of P. abyssi were collected and analyzed by immunoblotting with the use of polyclonal Cdc6- and MCM-specific antisera. As shown in Fig. 3A, the steady-state expression level of P. abyssi Cdc6 (calculated molecular mass of 48 kDa) was relatively high from the early exponential phase to the late stationary phase. A similar constitutive expression pattern was observed for MCM protein (calculated molecular mass of 77 kDa, assuming splicing of two inteins). Dot plot and quantitative Western blot analyses, with purified proteins for calibration, indicated that 5,000–10,000 and 200–400 molecules of Cdc6 and MCM proteins, respectively, were present in a rapidly dividing P. abyssi cell (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). P. abyssi Cdc6 (Fig. 3A) and MCM proteins were still detected after de novo protein synthesis was blocked by puromycin for 2 h, thus inhibiting DNA synthesis in P. abyssi (12). When puromycin treatment was continued for 4 h, some additional bands were detected with MCM-specific antiserum. As the identity of these additional bands is unknown, all experiments described hereafter were performed with shorter (2–3 h) puromycin treatments. The apparent molecular masses of Cdc6 and MCM were not affected by treatment with lambda-phosphatase (data not shown). Furthermore, cell counts of control and puromycin-treated cultures (Fig. 3A) and further microscopic analyses indicated that, as in Sulfolobus acidocaldarius (20), inhibiting DNA and/or protein synthesis resulted in cell division arrest in P. abyssi.
Figure 3.
Chromatin association of P. abyssi Cdc6 (PAB2265) and MCM (PAB2373). (A) Expression patterns of P. abyssi Cdc6 and MCM were investigated by immunoblotting of 10% or 8%, respectively, SDS/PAGE gels after various times after inoculation. Puromycin was added after 4 h of growth. The cell densities of samples (107 cells per milliliter) are indicated below each lane. As whole-cell lysates were directly analyzed, the results should be considered semiquantitative. (B) Chromatin binding of P. abyssi Cdc6 and MCM was investigated by separating cell lysates into soluble (S) and chromatin-enriched (P) fractions. Where indicated, puromycin (200 μg/ml)-treated or stationary phase cells were used for assay. All samples were extracted from a comparable amount of cells. Protein amounts analyzed per lane are indicated below each lane in micrograms.
P. abyssi Cdc6 and MCM Proteins Associate with Chromatin.
To investigate possible chromatin association of P. abyssi Cdc6 and MCM, cell-free extracts of P. abyssi cells in the late exponential phase were prepared by gentle lysis with 0.5% Triton X-100, followed by fractionation into soluble and chromatin-enriched fractions by low-speed centrifugation through sorbitol. Chromatin-enriched fractions of exponential phase cultures contained only 12% of total cellular protein but nearly all of total DNA, thus indicating a high level of enrichment for chromatin-associated proteins. In addition, to confirm chromatin association of archaeal initiator proteins, samples were treated with DNase I before fractionation, and the absence of DNA after DNase I treatment was confirmed by agarose gel electrophoresis. As indicated in Fig. 3B, both Cdc6- and MCM-specific antisera recognized a single major band in chromatin-enriched fractions. Fractions of full-length Cdc6 and MCM proteins (≈50% and ≈42%, respectively) were found in the pellet fraction, pointing out their direct or indirect chromatin association. P. abyssi Cdc6 persisting in puromycin-treated cells was still capable of associating with chromatin (Fig. 3B). On the contrary, the majority of MCM after puromycin treatment is found in the supernatant fraction. Inasmuch as the amounts of MCM protein in pellets of DNase I and puromycin-treated samples were comparable (Fig. 3B, Lower), these data indicate that puromycin treatment arrests P. abyssi DNA replication by inhibiting or preventing a chromatin association of MCM helicase. Finally, Cdc6 and MCM were also present in stationary-phase cultures after overnight incubations at 95°C. However, their steady-state expression levels were slightly lower than those observed during exponential phase, and chromatin association of a full-length MCM protein was abolished.
Association of the P. abyssi Cdc6 and MCM Proteins with the Replication Origin.
In Archaea, Ccd6/Orc1 and MCM proteins are the only proteins showing sequence similarity to eukaryotic proteins known to interact either directly or indirectly with replication origins, prompting us to investigate their specific association with the oriC of P. abyssi. Cell-free extracts from asynchronous cultures cross-linked with formaldehyde were prepared, and the relative abundance of several chromosomal segments bound to immunoprecipitated Cdc6 or MCM proteins was measured by PCR. Different primer sets amplifying the oriC, two regions located ≈10 kb of the origin, or the control region located 624 kb from the origin was used (Fig. 4A). Quantitative analyses of our data indicate that immunoprecipitation of Cdc6 resulted in a 30- to 40-fold enrichment of origin DNA sequences in comparison with our control when exponential phase cells were used for assays (Fig. 4). In contrast to this high level of enrichment of Cdc6 at the replication origin, chromatin immunoprecipitation assays performed with MCM-specific polyclonal antiserum indicated that chromatin association of MCM is less specific for origin DNA than that observed for Cdc6 (Fig. 4 B and C). To ease comparisons between Cdc6 and MCM chromatin immunoprecipitation assays, quantitative data for Cdc6 and MCM origin association were plotted with the same scale.
Similar chromatin immunoprecipitation assays using puromycin-treated or stationary phase cells suggest that chromatin association of archaeal initiator proteins takes place in a dynamic fashion. In particular, in puromycin-treated cells Cdc6 protein stays bound to the replication origin, whereas chromatin (Fig. 3B) as well as the origin (Fig. 4C) association of MCM protein is somehow prevented under these conditions. This result cannot be simply explained by degradation of MCM, as full-length MCM protein can still be detected in the supernatant fraction of puromycin-treated cultures (Fig. 3B). Our data also suggest that during stationary phase some Cdc6 is still present at the origin, whereas specific origin association of MCM protein at the origin was not detectable after overnight cultivation.
Discussion
Jacob et al. (21) proposed in their replicon model that in bacteria, a trans-acting factor activates a cis-acting DNA element, the replicator, to initiate DNA replication at a specific chromosomal locus. Our analyses of P. abyssi DNA replication in vivo now extend this model to the third domain of life, the Archaea, by showing the association of P. abyssi initiator proteins Cdc6 and MCM with the chromosomal replication origin. We have shown by N/N 2D gel electrophoreses (Fig. 2) that a defined replication origin, physically linked to the cdc6 gene, is present in the P. abyssi chromosomal region predicted earlier by in silico analysis to carry a replication start site (12). The 0.8-kb region immediately upstream of P. abyssi cdc6, corresponding to the mapped origin, contains several conserved repeats and AT-rich regions (Fig. 2F). Whether these repeats are required for interactions of an origin with initiator proteins and origin unwinding, respectively, remains to be determined. Nevertheless, similar features are present in origins we have predicted earlier for P. furiosus, P. horikoshii, Mb. thermoautorophicum, and a plasmid pFZ1 of Mb. thermoformicicum (11), thus suggesting their functional importance. Previous N/N 2D gel data for the chromosomal replication origin of Bacteria Mycoplasma capricolum (22) also have revealed a composite pattern consisting of Y and bubble arcs, similar to what we have now found for the P. abyssi oriC. Thus, these patterns observed for replication origins in Bacteria and Archaea cannot be explained by multiple replication origins as in Eukarya, but possibly by nicking of replication intermediates during sample preparation. Another possibility for these analogous composite patterns is that they are somehow related to larger replicon sizes and/or higher replication speed in Prokaryotes than in Eukarya.
We show that, as the number of chromosome equivalents per Pyrococcus cell during exponential phase is up to 15 (R. Bernander, personal communication), P. abyssi cells from exponential phase cultures harbor up to 300–600 molecules of Cdc6 per replication origin. In addition, P. abyssi Cdc6 remains chromatin bound at the replication origin in puromycin-treated cells (Fig. 4), making it unlikely that Cdc6 would limit DNA replication under these conditions. This observation is different from what has been observed for the unstable Cdc6 protein in Eukarya (23) but analogous to what has been observed for the ORC complex that stays bound at replication origins throughout the eukaryotic cell cycle (24). In agreement with the possibility that chromatin association of archaeal Cdc6 proteins with the replication origin could be direct, the Cdc6 protein from the archaeon Pyrobaculum aerophilum contains a putative DNA binding domain formed by a winged helix-turn-helix motif (25). This domain is also present in other archaeal Cdc6 proteins, and in the Orc1 subunit of eukaryotic ORC directly interacting with replication origins. Thus, regardless of whether the archaeal initiator protein originally dubbed Cdc6 is orthologous to eukaryotic Cdc6 or Orc1, our data uniquely suggest that a single P. abyssi polypeptide codes for the separate functions of the evolutionarily related ORC complexes and Cdc6 proteins in origin recognition (26) and MCM loading (27), respectively. However, on the basis of our current data, we cannot exclude more complicated possibilities (e.g., that unknown novel proteins might assist the P. abyssi Cdc6 either in origin recognition or in MCM loading).
In contrast to Cdc6 immunoprecipitates showing 30- to 40-fold enrichment of P. abyssi origin DNA, the chromatin association of the P. abyssi MCM protein is less specific for oriC (Fig. 4 B and C). This observation suggests that the archaeal MCM helicase (7–9) could be more broadly distributed along the P. abyssi chromosome than it is along the Cdc6 protein. Although more experiments are necessary to address this observation, these data would be consistent with what has been observed in Eukarya (24, 28), where MCM proteins participate in the initiation and elongation stages of DNA replication. This possibility is further supported by our observation that chromatin and origin association of the P. abyssi MCM is abolished by the inhibition of DNA replication or, alternatively, in stationary phase cells not replicating their DNA (Figs. 3 and 4). Indeed, pyromycin treatment prevents origin association of MCM helicase, thus explaining, at least in part, why puromycin treatment stops DNA replication and can synchronize replication forks at the replication origin in Pyrococcus abyssi (12). Its relatively low steady-state expression level also supports the possibility that MCM could limit DNA replication in Archaea.
Our results, when combined with previous computer analyses, indicate that archaeal initiation replication complexes are in some ways analogous to those of Eukarya. However, as many eukaryotic replication proteins are missing in Archaea (29), the archaeal replication initiation system shows some important differences with those of Eukarya (Fig. 5). Although P. abyssi Cdc6 is presumably required for the origin and chromatin association of P. abyssi MCM protein, binding of Cdc6 at the P. abyssi origin alone is not sufficient for origin association of MCM (Fig. 4). We also show that inhibiting P. abyssi protein synthesis and/or DNA replication leads to cell division arrest, indirectly suggesting that chromosome replication and cell division events in P. abyssi and S. acidocaldarius (20) are somehow linked. Strikingly, chromatin association of MCM is inhibited in puromycin-treated and stationary-phase cells, raising the important possibility that loading of MCM helicase onto chromatin could be used to regulate cell cycle events in Archaea. As archaeal genomes lack candidate genes for cyclin as well as for geminin and Cdt1, regulating chromatin loading of MCM in Eukarya (30–33), Archaea must use hitherto uncharacterized factors for chromatin loading of MCM and/or its regulation. Whether this mechanism is unique for Archaea or, alternatively, its functional counterpart has not yet characterized in Eukarya (or Bacteria), remains to be investigated. Note also that Pyrococcus sp. contain many “bacterial” proteins, such as FtsZ and MinD, presumably taking part in cell division, thus suggesting a functional linkage between “eukaryotic” replication and “bacterial” cell division proteins.
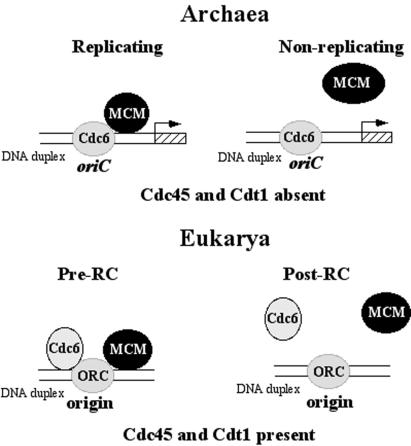
Figure 5.
A schematic model of archaeal initiation complexes at the replication origin in replicating (exponential-phase culture) and nonreplicating (puromycin-treated samples) P. abyssi cells. The shaded box and arrow indicate the P. abyssi cdc6 gene and its transcriptional start in the vicinity of the mapped oriC. Cdc45 and Cdt1 missing in Archaea are essential for replication initiation in Eukarya. RC, replication complex.
In summary, our detailed studies on the archaeal replication mechanism have identified Cdc6 and MCM proteins as components of archaeal replication complexes assembled at the replication origin. We also have revealed a fluctuation of the archaeal MCM helicase in chromatin-bound and soluble forms. To further elucidate how loading of the MCM helicase proceeds and how it is regulated in Archaea requires an oriC-dependent in vitro system for archaeal DNA replication, with the Pyrococcus oriC and purified replication proteins.
Supplementary Material
Acknowledgments
We thank O. Hyrien for help with two-dimensional gel electrophoreses and U. Liebl for helpful comments on the manuscript. We are also grateful to Y. Shimura and K. Morikawa for continuous encouragement. This work was supported by the Association pour la Recherche sur le Cancer, and the Programme de Recherches Fondamentales en Microbiologie, Maladies Infectieuses et Parasitologie of the French Ministry of Science and Education. F.M and H.M. thank the Human Frontier Science Program, European Molecular Biology Organization, and Fondation des Treilles for their financial support.
Abbreviations
- MCM
minichromosome maintenance
- ORC
origin recognition complex
- N/N 2D
neutral/neutral two-dimensional
References
- 1.Kelly T J, Brown G W. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 2.Boye E, Lobner-Olesen A, Skarstad K. EMBO Rep. 2000;1:479–483. doi: 10.1093/embo-reports/kvd116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edgell D R, Doolittle W F. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 4.Forterre P. Mol Microbiol. 1999;33:457–465. doi: 10.1046/j.1365-2958.1999.01497.x. [DOI] [PubMed] [Google Scholar]
- 5.Leipe D D, Aravind L, Koonin E V. Nucleic Acids Res. 1999;27:389–401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishino Y, Cann I K. Genes Genet Syst. 1998;73:323–336. doi: 10.1266/ggs.73.323. [DOI] [PubMed] [Google Scholar]
- 7.Kelman Z, Lee J K, Hurwitz J. Proc Natl Acad Sci USA. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong J P, Hayashi M K, Simon M N, Xu R M, Stillman B. Proc Natl Acad Sci USA. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. . (First published February 4, 2000; 10.1073/pnas.030539597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shechter D F, Ying C Y, Gautier J. J Biol Chem. 2000;275:15049–15059. doi: 10.1074/jbc.M000398200. [DOI] [PubMed] [Google Scholar]
- 10.Grigoriev A. Nucleic Acid Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez P, Philippe H, Myllykallio H, Forterre P. Mol Microbiol. 1999;32:883–886. doi: 10.1046/j.1365-2958.1999.01370.x. [DOI] [PubMed] [Google Scholar]
- 12.Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, Zivanovic Y, Philippe H, Forterre P. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- 13.Bergerat A, de Massy B, Gadelle D, Varoutas P C, Nicolas A, Forterre P. Nature (London) 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 14.Cann I K, Ishino Y. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brewer B J, Fangman W L. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 16.Shinomiya T, Ina S. Nucleic Acids Res. 1991;19:3935–3941. doi: 10.1093/nar/19.14.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlando V, Paro R. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- 18.Linskens M H, Huberman J A. Nucleic Acids Res. 1990;18:647–652. doi: 10.1093/nar/18.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natale D A, Schubert A E, Kowalski D. Proc Natl Acad Sci USA. 1992;89:2654–2658. doi: 10.1073/pnas.89.7.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hjort K, Bernander R. Mol Microbiol. 2001;40:225–234. doi: 10.1046/j.1365-2958.2001.02377.x. [DOI] [PubMed] [Google Scholar]
- 21.Jacob F, Brenner S, Cuzin F. Cold Spring Harbor Symp Quant Biol. 1964;28:329–348. [Google Scholar]
- 22.Miyata M, Fukumura T. Gene. 1997;193:39–47. doi: 10.1016/s0378-1119(97)00075-9. [DOI] [PubMed] [Google Scholar]
- 23.Piatti S, Lengauer C, Nasmyth K. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Smith C L, DeRyckere D, DeAngelis K, Martin G S, Berger J M. Mol Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 26.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;22:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 28.Labib K, Tercero J A, Diffley J F. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 29.Tye B K. Proc Natl Acad Sci USA. 2000;97:2399–2401. doi: 10.1073/pnas.97.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiorano D, Moreau J, Mechali M. Nature (London) 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 31.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. Nature (London) 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 32.Wohlschlegel J A, Dwyer B T, Dhar S K, Cvetic C, Walter J C, Dutta A. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 33.Tada S, Li A, Maiorano D, Mechali M, Blow J J. Nat Cell Biol. 2001;2:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.