Abstract
Streptococcus suis serotype 2 is an important swine pathogen and an emerging zoonotic agent that causes severe infections. Recent studies have reported a eukaryotic-like Ser/Thr protein kinase (STK) gene and characterized its role in the growth and virulence of different S. suis 2 strains. In the present study, phosphoproteomic analysis was adopted to identify substrates of the STK protein. Seven proteins that were annotated to participate in different cell processes were identified as potential substrates, which suggests the pleiotropic effects of stk on S. suis 2 by targeting multiple pathways. Among them, a protein characterized as cell division initiation protein (DivIVA) was further investigated. In vitro analysis demonstrated that the recombinant STK protein directly phosphorylates threonine at amino acid position 199 (Thr-199) of DivIVA. This effect could be completely abolished by the T199A mutation. To determine the specific role of DivIVA in growth and division, a divIVA mutant was constructed. The ΔdivIVA strain exhibited impaired growth and division, including lower viability, enlarged cell mass, asymmetrical division caused by aberrant septum, and extremely weak pathogenicity in a mouse infection model. Collectively, our results reveal that STK regulates the cell growth and virulence of S. suis 2 by targeting substrates that are involved in different biological pathways. The inactivation of DivIVA leads to severe defects in cell division and strongly attenuates pathogenicity, thereby indicating its potential as a molecular drug target against S. suis.
Keywords: eukaryote-like Ser/Thr kinase, DivIVA, Streptococcus suis serotype 2, cell division, virulence
Introduction
Streptococcus suis (S. suis) is an important swine pathogen that causes a wide range of diseases, including arthritis, endocarditis, meningitis, pneumonia, and septicemia (Staats et al., 1997). S. suis strains have been previously classified into 35 serotypes (serotypes 1–34 and serotype 1/2) based on the antigenicity of their surface capsular polysaccharides (CPSs) (Staats et al., 1997; Gottschalk et al., 2007). However, recent genetic studies have suggested that serotypes 20, 22, 26, 32, 33, and 34 should not be attributed to S. suis (Hill et al., 2005; Goyette-Desjardins et al., 2014; Nomoto et al., 2015). Furthermore, among the remaining serotypes, S. suis 2 is considered to be the most virulent and it is frequently isolated from clinically diseased piglets (Lun et al., 2007). S. suis 2 caused two large outbreaks of human infection in China, which resulted in 38 deaths out of 204 reported cases and 14 deaths out of 25 cases in 2005 and 1998, respectively (Tang et al., 2006). Over the past several decades, more than 20 virulence-associated factors of S. suis have been identified (Vecht et al., 1991; Jacobs et al., 1996; Smith et al., 1996, 1999; de Greeff et al., 2002; Gruening et al., 2006; Feng et al., 2009; Zhang et al., 2009, 2014; Bonifait et al., 2011; Shao et al., 2011, 2013; de Buhr et al., 2014; Li et al., 2015). The expression levels of these virulence factors are affected by a series of important transcription factors, various two-component signal transduction (TCS) systems and orphan regulators (Li et al., 2008, 2011; Pan et al., 2009; Aranda et al., 2010; Fulde et al., 2011; Willenborg et al., 2011; Zheng et al., 2011; Tang et al., 2012; Wang et al., 2012).
Post-translational modifications play important roles in the modulation of protein function, signaling transduction, and various important cellular physiological processes (Michard and Doublet, 2015). Reversible protein phosphorylation caused by protein kinases and cognate phosphatases is an important post-translational modification (Ravikumar et al., 2015). In bacteria, a group of Ser/Thr kinases (STK) that is homologous to those of eukaryotic STK has been defined as eukaryote-like STK (eSTK) (Pereira et al., 2011). The bacterial eSTK is first identified from the gram-negative soil microorganism Myxococcus xanthus (Munoz-Dorado et al., 1991) and it has been proven to be widespread in different bacteria, including Pseudomonas aeruginosa, Enterococcus faecalis, Staphylococcus aureus, Mycobacterium tuberculosis, Streptococcus pneumoniae, Streptococcus agalactiae, and Streptococcus pyogenes (Wang et al., 1998; Av-Gay and Everett, 2000; Rajagopal et al., 2003; Echenique et al., 2004; Mougous et al., 2007; Burnside et al., 2010; Molle and Kremer, 2010; Ohlsen and Donat, 2010; Agarwal et al., 2011; Bugrysheva et al., 2011; Cameron et al., 2012; Hall et al., 2013).
Extensive studies involving phosphoproteomic profile analysis have revealed that the eSTK substrates are quite pleiotropic in nature. This property of eSTKs makes it possible to regulate numerous protein substrates for a wide range of biological processes, including transcription and translation, cell wall synthesis, and cell shape/division, sporulation, stress response, bacterial persistence, and the expression of virulence factors (Liu et al., 2011; Mata-Cabana et al., 2012; Ruggiero et al., 2012). In S. pneumoniae, StkP regulates cell division and morphogenesis by phosphorylating the cell initiation division protein DivIVA (Giefing et al., 2008; Nováková et al., 2010). Abolishing the phosphorylation ability of DivIVA by a threonine to alanine mutation at position 201 results in an elongated and bulged cell phenotype (Fleurie et al., 2012).
The DivIVA protein functions as a scaffold for the recruitment of other cell division-related proteins to the septum and pointed poles, and this protein is highly conserved in a wide range of gram-positive bacteria (Kaval and Halbedel, 2012; Laloux and Jacobs-Wagner, 2014). However, the biological functions of DivIVA vary among different species. For example, in S. aureus (Pinho and Errington, 2004), the presence or absence of the DivIVA protein does not affect cell morphology and division, whereas in Bacillus subtilis (Perry and Edwards, 2004), E. faecalis (Ramirez-Arcos et al., 2005), and S. pneumoniae (Nováková et al., 2010), it is essential for natural cell growth and division. In Listeria monocytogenes (Halbedel et al., 2012), DivIVA also influences the activity of the accessory secretion motor ATPase SecA2, which is involved in bacterial virulence factor secretion.
Recently, a STK protein was identified from S. suis 2. Our research group (Du et al., 2014) and other investigators (Zhu et al., 2014; Zhang et al., 2017) have revealed that the inactivation of the stk gene causes severe phenotype alterations, including a deficiency in cell division, decreased adhesion to host cells, greater sensitivity to H2O2, and lower pathogenicity in mice. However, the underlying molecular mechanisms remain unclear. Furthermore, while changes in the phosphorylation state after stk deletion in several proteins were observed (Zhang et al., 2017), whether these were directly targeted by STK and the function of these potential targets in S. suis 2 remain unclear. In the present study, multiple substrates of S. suis 2 STK were identified by phosphoproteomic and mass spectrometric analyses with a specific antibody against phosphothreonine (pThr). Furthermore, DivIVA phosphorylation by STK was validated using an in vitro assay. Site-directed mutagenesis showed that Thr-199 of DivIVA was essential for STK phosphorylation. To further understand the mechanism of STK in regulating cell division and growth in S. suis, we constructed a divIVA-deletion mutant and investigated its morphology and virulence alterations. Our findings indicate that STK controls cell division in S. suis 2 by phosphorylating the cell division initiation protein, DivIVA.
Results
STK deletion in S. suis 2 alters Thr phosphorylation levels
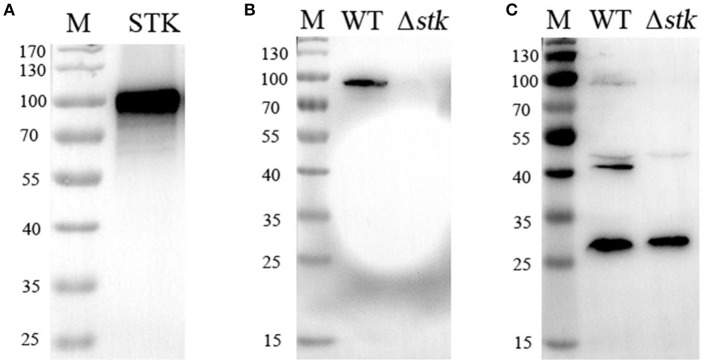
To investigate the effect of the stk deletion on intracellular Thr phosphorylation levels, we first prepared polyclonal antibodies against STK by immunizing mice with purified recombinant STK protein. The results of the Western blot assay showed that the antibody could react specifically with the recombinant STK (Figure 1A). Then, Western blot analyses with the obtained anti-STK antibody were performed to verify the deficient expression of the STK protein in the stk mutant strain, and its normal expression in the WT strain (Figure 1B). Subsequently, Western blotting was performed on the total proteins extracted from the stk mutant and WT strain, respectively, with an anti-phosphothreonine polyclonal antibody. A comparison of the phosphorylated proteins in the cell lysates of the WT strain (05ZYH33) to the isogenic mutant (Δstk) revealed at least four major phosphorylated protein bands in the WT strain, whereas only two of them were detected in the Δstk strain (Figure 1C). This result preliminarily indicated that the deletion of the stk gene in S. suis partially altered the Thr phosphorylation level of its proteins.
Figure 1.
Comparison of phosphorylation levels between the WT and Δstk strains. (A) Detection of the specificity of anti-STK polyclonal antibodies by Western blotting. (B) Assessment of the protein expression levels of Δstk. (C) Thr phosphorylation levels were immunodetected in whole protein lysates of the wild-type strain and the Δstk mutant strain.
Genome-wide screening of putative STK substrates
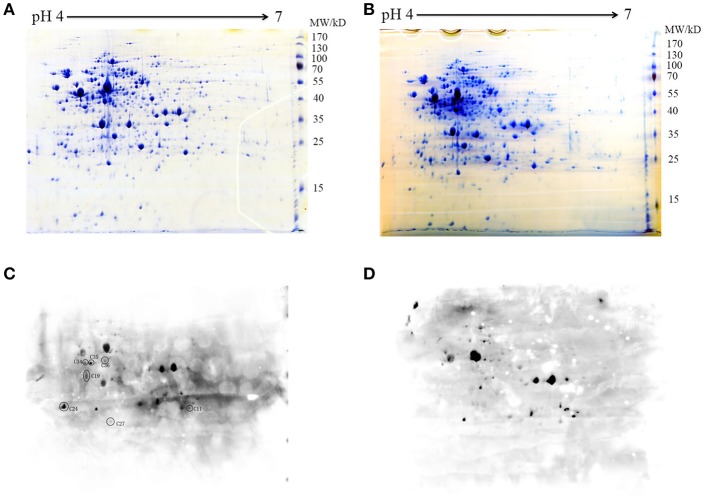
To further explore the potential substrates of STK, the total proteins synthesized by the WT and Δstk strains were separated by two-dimensional SDS-polyacrylamide gel electrophoresis (PAGE). Each of the samples was analyzed by two gels with the same parameters. One was subjected to Coomassie blue staining (Figures 2A,B), while the other was used for detecting the Thr phosphorylation level with a specific anti-phosphothreonine polyclonal antibody (Figures 2C,D). The phosphorylation spots of the WT and Δstk strains were compared, and seven WT-specific phosphorylated spots were identified, namely C11, C19, C24, C27, C34, C35, and C36 (Figure 2C). These spots at the corresponding coordinates in the Coomassie blue-stained gel were extracted for further investigation using mass spectrometry. The results annotated these proteins as ABC-type branched-chain amino acid transporter ATPase, foldase protein (PrsA), DivIVA, manganese-dependent superoxide dismutase (SodA), elongation factor Ts (EF-Ts), glucose-6-phosphate isomerase (PGI), and phosphoglycerate kinase (PGK), respectively (Table 1).
Figure 2.
Two-dimensional map of the whole proteins of (A) WT and (B) Δstk. The gels were stained with Coomassie blue (A,B) or electroblotted and probed with an anti-pThr antibody (C,D). Protein spots C11 to C36 corresponding to phosphorylated proteins were excised and analyzed by MALDI-FTMS. Molecular weights are indicated on the right.
Table 1.
Mass spectrometric identification of STK protein substrates.
| Spot | Accession | Locus | Protein name | pI | Sequence coverage (%) | Mascot score |
|---|---|---|---|---|---|---|
| C11 | ABP90509 | SSU05_1543 | ABC-type branched-chain amino acid transport system | 5.59 | 55 | 318 |
| C19 | ABP90204 | SSU05_1238 | Foldase protein PrsA | 5.01 | 42 | 224 |
| C24 | ABP89453 | SSU05_0487 | Cell division initiation protein | 4.54 | 63 | 328 |
| C27 | ABP90505 | SSU05_1539 | Manganese-dependent superoxide dismutase A | 5.06 | 25 | 86 |
| C34 | ABP90945 | SSU05_1979 | Elongation factor Ts | 4.74 | 56 | 280 |
| C35 | ABP91017 | SSU05_2051 | Glucose-6-phosphate isomerase | 4.7 | 33 | 198 |
| C36 | ABP89127 | SSU05_0157 | Phosphoglycerate kinase | 4.85 | 55 | 542 |
To evaluate the distribution and conservation of STK and its substrates among different S. suis strains, we surveyed the eight proteins in the 34 completely sequenced S. suis genomes (Table S1). The results revealed that STK and the seven potential substrates are widespread and highly conserved in all of the surveyed genomes, with each genome containing one copy of those genes. The average amino acid identity for the eight proteins from the 34 S. suis genomes are: 98.73 (STK), 99.25 (DivIVA), 99.47 (ATPase), 96.08 (PrsA), 99.11 (SodA), 99.21 (EF-Ts), 99.55 (PGK), and 99.47 (PGI), suggesting very low amino acid substitution among orthologous proteins.
DivIVA is phosphorylated by STK in vitro
Among the seven identified proteins that show the presence/absence pattern in phosphothreonine, the C24 spot was annotated as the putative initial cell division protein DivIVA (the product of CDS 05SSU0487). Sequence analysis revealed that the protein is composed of 229 amino acid residues, with an β-helical N-terminal domain that contains a distinct coiled coil region (3–144) and an unstable C-terminal domain (144–229) (Figure S1A). Multiple sequence alignment indicates that the amino acid sequence exhibits 67, 62, 60, and 55% amino acid sequence identity with the DivIVA of S. pneumoniae, S. pyogenes, S. agalactiae, and S. mutans, respectively (Figure S1B).
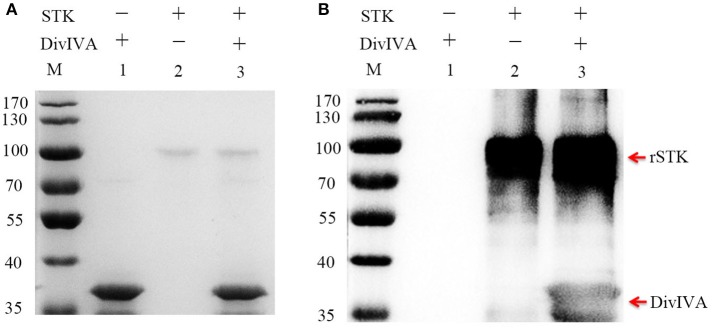
In several bacteria, DivIVA is a substrate that is directly phosphorylated by the STK protein. To test whether DivIVA is also phosphorylated by STK in S. suis 2, the recombinant DivIVA was expressed, purified, and subjected to in vitro phosphorylation by STK. First, the kinase activity of rSTK was tested by incubating with myelin basic protein (MBP), a positive control substance. The kinase assay demonstrated that rSTK is not only capable of phosphorylating MBP, but it also possesses autophosphorylation activity (Figure S2). The phosphorylation of purified rDivIVA by rSTK was then examined by immunodetection using an anti-phosphothreonine polyclonal antibody. The results show that the DivIVA protein was phosphorylated by STK, thereby suggesting that it is a genuine substrate of STK (Figure 3).
Figure 3.
Phosphorylating of DivIVA by STK in vitro. (A) rSTK and DivIVA were incubated together, separated by SDS-PAGE, and stained with Coomassie blue. (B) rSTK and DivIVA were incubated together, separated by SDS-PAGE, electroblotted, and then probed with an anti-pThr antibody.
Determination of phosphorylation sites of DivIVA
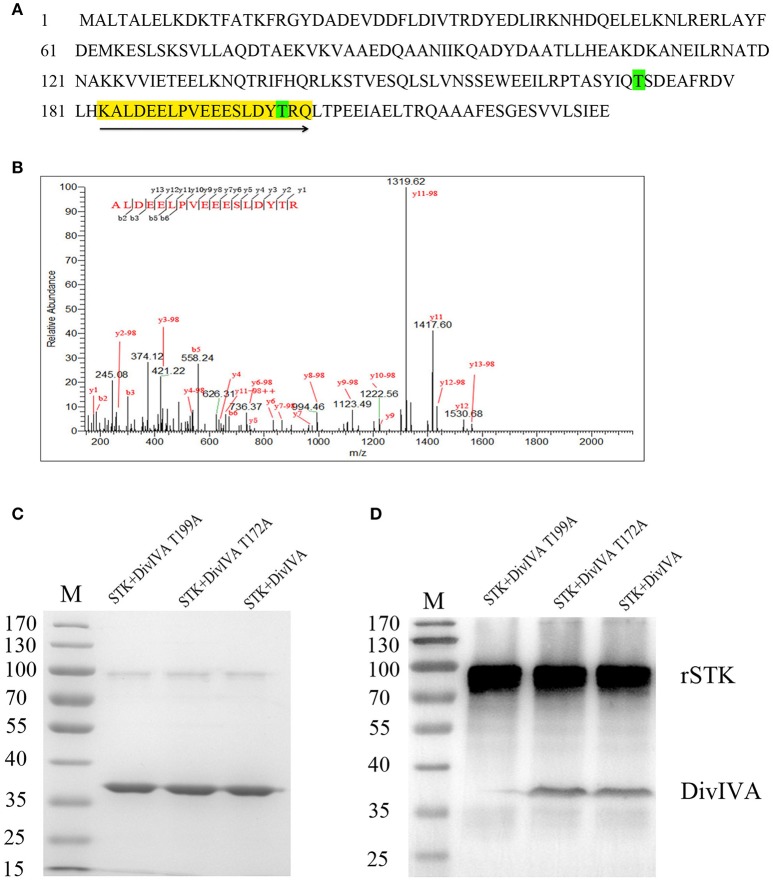
To identify the potential phosphorylation sites in DivIVA, its amino acid sequence was submitted to a kinase-specific phosphorylation site prediction online tool KinasePhos1 (http://kinasephos2.mbc.nctu.edu.tw/index.html), which predicted that the Thr-172 residue is a phosphorylation site. To further test the predictions of KinasePhos1, the in vitro rSTK-phosphorylated rDivIVA was subjected to trypsin digestion, followed by analysis of the digested peptides by mass spectrometry. The results showed that the DivIVA-derived peptide fragment KALDEELPVEEESLDYPTRQ was phosphorylated at position 17, which corresponds to T199 in the DivIVA protein sequence (Figure 4). To further validate the mass spectrometry and online prediction results, Thr-172 and Thr-199 of DivIVA were, respectively, replaced by alanine by site-directed mutagenesis. Western blot analysis revealed that the phosphorylation activity of the DivIVA-T199A protein by rSTK was completely abolished, while that of the mutant protein DivIVA-T172A did not change (Figure 4). The results suggest that STK phosphorylates DivIVA at the Thr-199 residue in S. suis 2.
Figure 4.
Identification of DivIVA phosphorylated sites. (A) Protein sequence of DivIVA: The peptide containing a phosphorylated signal is highlighted in yellow and labeled with arrows. The threonine residues introduced by mutagenesis are labeled in green. (B) Mass spectra showing that DivIVA is phosphorylated at threonine 199. Recombinant DivIVA was incubated with rSTK and then subjected to trypsin digestion. (C) rSTK incubated with mutant DivIVA proteins were separated by SDS-PAGE and then stained with Coomassie blue. (D) An anti-pThr antibody was used to assess the effect of mutant DivIVA proteins.
Phenotypic differences between WT and isogenic mutant strains
To investigate the role of DivIVA in the cell cycle regulation of S. suis 2, an isogenic divIVA was constructed through homologous recombination (Figure S3). Screening more than 100 Spcr transformants identified one putative mutant. The double-crossover event was confirmed by combined PCR and RT-PCR analyses (Figure S3). The identified mutant strain was then renamed ΔdivIVA. The growth of the ΔdivIVA strain was evaluated. Compared to the wild-type strain 05ZYH33, the growth kinetics of the ΔdivIVA strain, which was monitored by either assessing the OD600-values or counting the viable bacteria amounts during the initial 10–11 h of growth, was relatively slow. At that time, both the WT strain and ΔdivIVA had already reached the post-exponential-growth phase (Figure 5).
Figure 5.
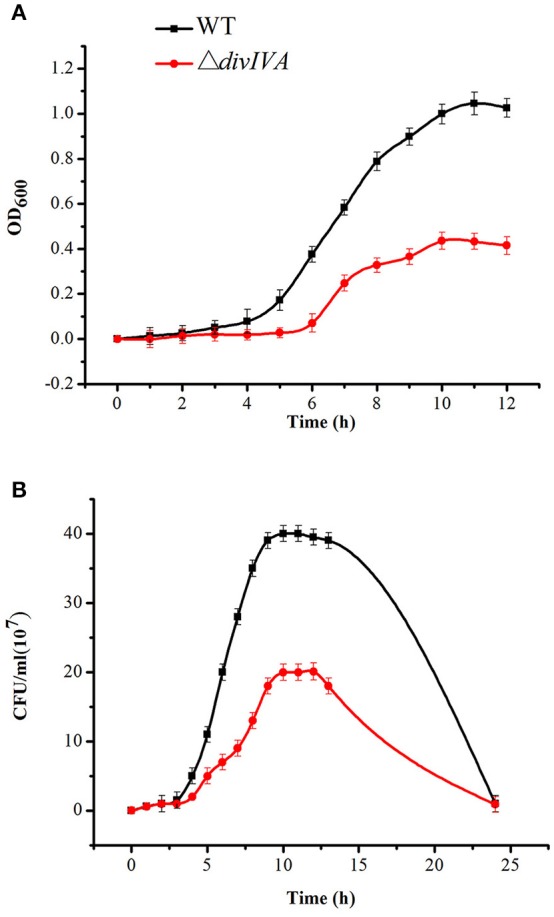
Growth characteristics of the WT and ΔdivIVA mutant strains. (A) Cell density was measured spectrophotometrically at a wavelength of 600 nm, and the data were collected at the indicated time points. (B) Separate aliquots of the bacterial suspensions were serially diluted and plated to determine the CFU numbers per milliliter.
The morphology of the WT and ΔdivIVA strains was first examined by gram staining, which revealed that the inactivation of divIVA results in cell aggregation, forming a giant cell mass (Figure 6A). The SEM results show that the ΔdivIVA strain developed into plump, sausage-shaped cells that were less ovoid than the WT cells. Some cells were unable to divide and separate, and they had lost the typical shape of S. suis (Figure 6B). Additionally, the results of TEM analysis reveal an irregular or disrupted cell septum in the ΔdivIVA cells (Figure 6C). The ΔdivIVA cells also exhibited cell division defects, including abnormal septum position and asymmetrical division. These findings indicate that DivIVA is associated with cell division and suggests that DivIVA plays a crucial role in the selection of the septum position.
Figure 6.
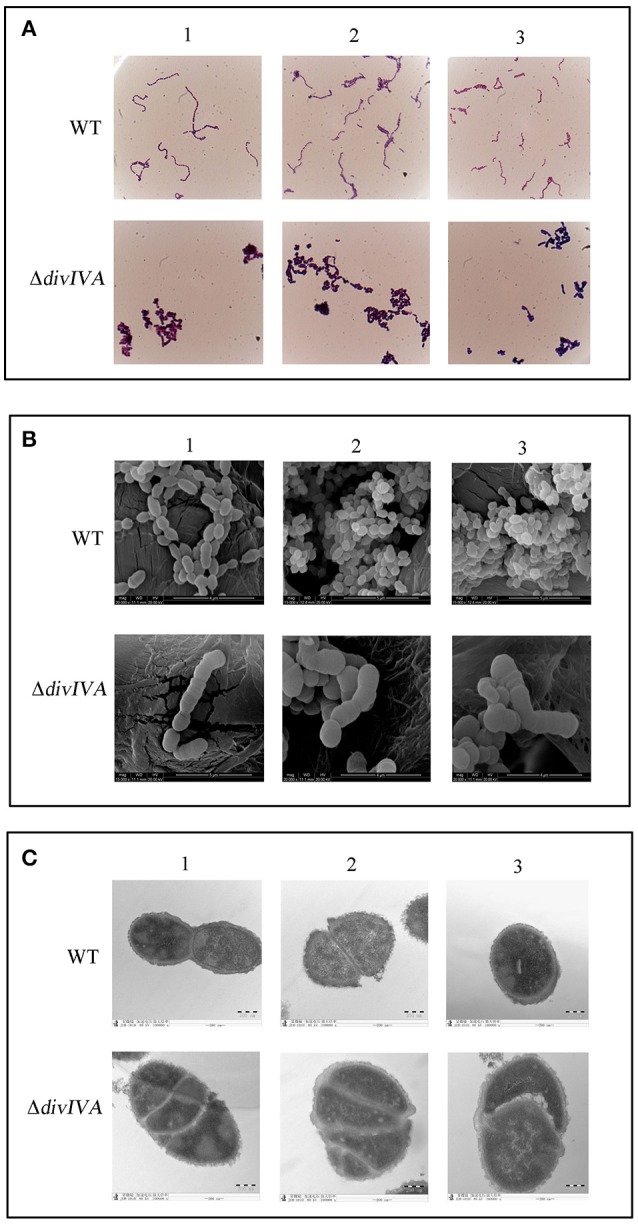
Cell morphology of the wild-type strain and the ΔdivIVA mutant strain. (A) Assessment of the cellular morphology of S. suis using gram staining and light microscopy. (B) Scanning electron micrographs of the strains. (C) Transmission electron microscopy of the strains. The bar indicates the magnification. Each analysis is represented by three pictures captured from different fields of view.
Evaluation of the pathogenicity of the divIVA mutant
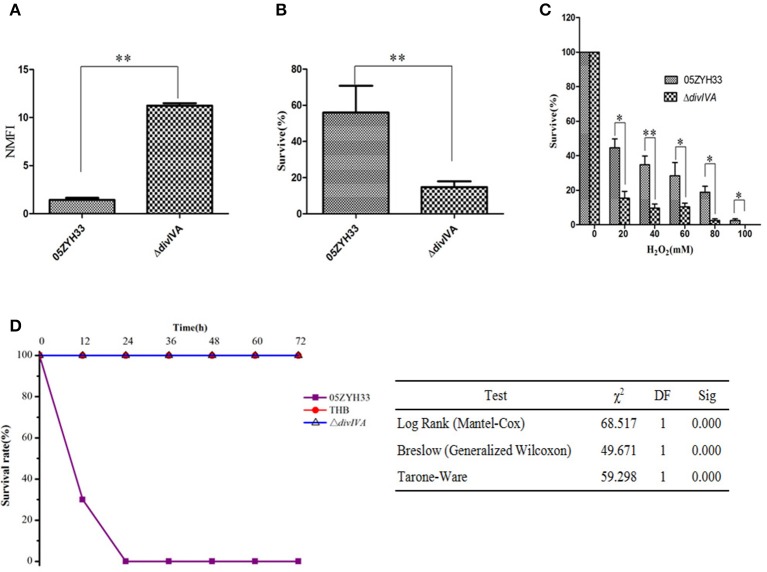
To investigate the differences between the WT and ΔdivIVA mutant strains during the infection of host cells, we measured their anti-phagocytosis ability against mouse macrophagocytes. Fluorescently labeled WT and ΔdivIVA mutant strains were co-cultured with RAW264.7 cells, and then fluorescence signals in the cells were measured. The NMFI of the cells incubated with ΔdivIVA reached 11.24 ± 0.251, which is approximately 8-fold higher than the cells incubated with WT only (1.43 ± 0.215, Figure 7A). These findings indicate that the anti-phagocytosis ability of the ΔdivIVA strain in mouse macrophagocytes significantly decreased in vitro. Furthermore, in the polymorphonuclear cell (PMN)-mediated killing assays, the ΔdivIVA showed a lower survival rate (14.76 ± 3.17%) compared to the WT (55.96 ± 14.89%) (P < 0.01) (Figure 7B). These results indicate that ΔdivIVA is more sensitive to phagocytosis and killing by PMNs. The oxidative stress test showed that the ΔdivIVA strain is more sensitive to H2O2 than the WT strain (Figure 7C).
Figure 7.
Virulence attenuation of the ΔdivIVA mutant strain. (A) Evaluation the anti-phagocytotic ability of S. suis strains in macrophage RAW264.7 cells. (B) The ΔdivIVA strain showing decreased resistance to phagocytosis and killing by neutrophils. The 106 CFU of the wild-type and mutant strains was incubated with PMNs at a bacteria-to-cell ratio of 10:1. The cells were lysed after 1 h of incubation, and the survival percentage of each strain was calculated as follows: (CFUPMN+/CFUPMN) × 100%. The data are expressed as the mean and standard deviation of three independent experiments (**P < 0.01). (C) H2O2 survival test of the WT and the ΔdivIVA mutant. S. suis 2 cultures at the mid-exponential phase (106 CFU) were incubated with increasing concentrations of H2O2 at room temperature for 15 min, and viable counts were monitored. The assay was performed in triplicate. A statistically significant killing effect of H2O2 on the WT05ZYH33 and ΔdivIVA mutant was observed (*P < 0.05; **P < 0.01). (D) Survival curves of mice infected with S. suis 2 strains. Four-week-old BALB/c mice were challenged intraperitoneally with 108 CFU bacteria, and the survival time was monitored. Kaplan-Meier survival analysis was performed with three different tests to evaluate the significant difference of the survival rates among the three groups.
Mouse infection assays were further adopted to address the contribution of the divIVA gene to bacterial pathogenicity. The results demonstrate that all of the mice infected with the ΔdivIVA mutant had no detectable clinical symptoms 2 days post-infection and at later time points (Figure 7D). The ΔdivIVA mutant may possibly be avirulent in the animal model, which coincides with the findings of the cell line-based experiments. Thus, we concluded that the inactivation of DivIVA significantly attenuates the pathogenicity of S. suis, suggesting that the protein may be potentially used as an antibacterial target.
Discussion
S. suis 2 STK substrates are involved in different biological processes of the bacterium
eSTK regulates various cellular processes in bacteria, including cell growth and division, metabolism, stress response, and virulence. The results from the present and two previous studies (Zhu et al., 2014; Zhang et al., 2017) suggest that numerous biological activities of the stk mutants of S. suis 2 are also affected to different extents such as delayed growth rate, abnormal cell division, sensitivity to oxidative stress, and attenuated virulence. However, the underlining mechanisms of these phenotypic changes have not been well-elucidated. By performing a comparative phosphoproteomic analysis of Δstk and the WT strains, we detected seven potential substrate proteins of STK that show the presence/absence of Thr phosphorylation, including PGK, PGI, SodA, EF-Ts, PrsA, DivIVA, and an amino acid transport-related protein. Literature-based function analysis revealed that the seven proteins are involved in cell division, oxidative stress, glucose metabolism, translation regulation, and protein transport process, thereby suggesting that the STK in S. suis 2 is equipped with a complex regulatory mechanism.
PGI catalyzes the reversible isomerization of glucose-6-phosphate (G6P) and fructose-6-phosphate (F6P) and it is involved in the regeneration of G6P molecules in the glycolytic and oxidized pentose phosphate pathway (OPPP) (Tran et al., 2009). PGK plays an important role in glycolysis and gluconeogenesis (Wang et al., 2014). The phosphorylation at threonine has been detected for PGK and PGI in B. subtilis (Macek et al., 2007) and Escherichia coli (Macek et al., 2008), suggesting a regulatory role of phosphorylation on their function. EF-Ts plays a central role in protein biosynthesis. EF-Ts regulates the abundance and stability of the EF-Tu·GTP·aminoacyl-tRNA ternary complex, which contributes to protein synthesis (Burnett et al., 2013). The threonine of EF-Ts is phosphorylated in B. subtilis and L. monocytogenes (Misra et al., 2011), suggesting that the translation process is also regulated by phosphorylation. The deficiency in threonine phosphorylation in these proteins may disrupt its function and therefore affect basic bacterial metabolism, which in turn may contribute to the delay in the growth of stk mutant strains.
Superoxide dismutase (SOD) are metalloproteins that convert superoxide anions to molecular oxygen and hydrogen peroxide. Manganese-dependent superoxide dismutase (SodA) is a class of SOD (McMillan et al., 2004). S. suis 2 utilizes SodA for survival not only by scavenging ROS but also by alleviating the host autophagic responses due to ROS stimulation (Fang et al., 2015). The inactivation of a serine/threonine phosphatase (stp1) increases S. suis 2 resistance to oxidative stress and survival in macrophage cells (Fang et al., 2017). In the present study, the phosphorylation of SodA was abolished in the stk mutant. The present study showed that Δstk has decreased tolerance to H2O2 and lower survival in the anti-phagocytic experiment, which is similar to that observed in a previous study (Zhu et al., 2014). Taken together, the following scenario is proposed. Phosphorylated SodA removes ROS, which contributes to S. suis 2 resistance to macrophage cells. stk mutants have unphosphorylated SodA that is unable to remove ROS in time, thereby leading to Δstk tolerance to H2O2 and anti-phagocytic decline. In contrast, when stp1 is mutated, SodA remains phosphorylated and it is more capable of removing ROS, which in turn enhances Δstp anti-phagocytosis.
S. suis 2 STK regulates cell division by directly phosphorylating DivIVA protein
In S. pneumoniae (Fleurie et al., 2012) and S. agalactiae (Rajagopal et al., 2003), stk mutants exhibit disruptions in cell shape and size. DivIVA has been identified as a conserved target of STK in these bacteria, wherein its absence or phosphorylation deficiency significantly disrupts cell morphology and division. Our study observed a decrease in the growth rate in the Δstk mutants. STK deficiency also caused an increase in the chain length and blockage of daughter cell segregation. Both our study and a previous study (Zhang et al., 2017) revealed that DivIVA phosphorylation is abolished in stk mutants, thereby suggesting that DivIVA is a potential target of STK in S. suis.
To clarify whether DivIVA plays a similar role in regulating S. suis cell division, a DivIVA mutant was constructed in this study. The disruption of S. suis DivIVA significantly affected bacterial phenotypes and growth characteristics. Similar changes in the growth rate have also been observed in E. faecalis, in which the turbidity and colony count measurements of the divIVA gene mutant strain were significantly decreased (Ramirez-Arcos et al., 2005). The absence of DivIVA in S. suis 2 inhibited normal cell division, which resulted in abnormal cell clusters possessing an enlarged cell mass instead of typical short chains. Furthermore, the lower viability of the divIVA mutant might be due to improper septa segregation within numerous cells that were generated in each cluster. These findings suggest that divIVA is an essential gene for the growth and maintenance of normal cell morphology in S. suis.
Pneumococcus cells expressing non-phosphorylatable DivIVA-T201A possess an elongated shape with a polar bulge and aberrant spatial organization of nascent peptidoglycan (Fleurie et al., 2012). DivIVA is also a key phosphorylation substrate of STK in S. agalactiae (Silvestroni et al., 2009), in which DivIVA reversible phosphorylation is important for normal cell division. In our study, the direct phosphorylation of DivIVA by STK was clearly detected in an in vitro experiment. Furthermore, we also determined that STK phosphorylates DivIVA at the Thr199 residue. The determination of the phosphorylation sites provides an important basis for further investigation on DivIVA in S. suis 2. In summary, our findings provide strong evidence that DivIVA is a direct substrate of STK in S. suis 2 that is involved in normal cell division.
A superposition effect involving multiple pathways contributes to virulence attenuation in stk mutants
Apart from affecting cell morphology and division, several studies have described the involvement of stk in regulating bacterial virulence. In the present study and other previous investigations (Zhang et al., 2017), the attenuation of virulence was also observed in various S. suis 2 strains in different mouse models. The identification of various STK targets and direct evaluation of the function of DivIVA suggest that the attenuation of virulence in the stk mutant of S. suis 2 involves a superposition that is due to the disruption of the normal functions of target proteins from multiple pathways. The inactivation of each of the proteins may contribute to the observed decrease in virulence. For example, as detected in this study, the deletion of the single target gene divIVA in S. suis 2 significantly delayed cell growth, affected cell division, and attenuated virulence. Such attenuation of virulence in ΔdivIVA is not surprising because the normal growth of bacterial cells was significantly delayed.
Similarly, three other proteins, including PGK, PGI, and EF-Ts, also play important roles in basic bacterial metabolism. Functional deficiencies in these proteins due to a disruption in phosphorylation in stk mutants may also contribute to decreased virulence. Unlike proteins involved in basic metabolism, SodA is a STK substrate protein that directly affects the survival of bacteria in the host environment. A recent study showed that SodA plays a role in the anti-autophagic response by scavenging ROS in infected macrophages (Fang et al., 2015). Our results suggest that the unphosphorylated state of the stk mutants may affect their normal function and ultimately contribute to decreased virulence. Taken together, we concluded that stk affects S. suis 2 virulence by directly modulating the phosphorylation state of some virulence factors, such as SodA. Meanwhile, the phosphorylation deficiency of STK substrates that are involved in basic cell metabolism and division may also contribute to a decrease in S. suis 2 pathogenicity.
Materials and methods
Bacterial strains, plasmids, cells, and growth conditions
The bacterial strains and plasmids used in the present study are listed in Table S2. The virulent strain S. suis 05ZYH33 (wild-type, WT) was isolated from an infected patient during the 2005 outbreak in Sichuan, China (Tang et al., 2006; Chen et al., 2007). The WT strain was grown in Todd-Hewitt broth (THB; Difco Laboratories, Detroit, MI, USA) liquid medium or plated on THB agar plates containing 5% (v/v) sheep blood at 37°C. Spectinomycin (100 μg/mL) was used to screen the S. suis mutant strain. The E. coli strains used in the cloning and expression of the recombinant proteins were purchased from Transgen Co. (Beijing, China). The E. coli DH5α, E. coli TOP10, and E. coli BL21 were maintained in Luria-Bertani (LB) broth liquid medium or plated on LB agar at 37°C. E. coli strain DH5α was used as host strain for cloning, while BL21 and TOP10 were used in the expression of the fusion proteins. Ampicillin or kanamycin (100 μg/ml) was added as needed.
DNA extraction and manipulation
Genomic DNA was extracted from the WT strain using a bacterial genomic DNA extraction kit (Omega Bio-tek Co., Ohio State, USA), according to the manufacturer's instructions. The mini-preparation of recombinant plasmids and transformation of E. coli were performed according to standard procedures. The restriction enzymes, DNA modifying enzymes, and Taq DNA polymerases were purchased from Takara Bio Co. (Dalian, China) and used according to the manufacturer's instructions. Oligonucleotide primers were obtained from Invitrogen (Shanghai, China).
Construction of a divIVA knockout mutant
A previously constructed stk mutant (Du et al., 2014) was used in this study. A similar method was used to construct the divIVA mutant strain of S. suis 05ZYH33. Briefly, the spectinomycin resistance (Spcr) gene was amplified from pSET2, and then it was inserted into a pUC18 vector (Takara, Japan) to create the recombinant plasmid pUC18-Spc. Two DNA fragments (LA and RA) flanking the divIVA gene were amplified from 05ZYH33 genomic DNA and cloned into pUC18-Spc to generate the knockout plasmid pUC::divIVA. The pUC::divIVA plasmid was then used for the transformation of the 05ZYH33 competent cells. The obtained transformants were confirmed by multiplex-PCR using a series of specific primers (Table 2) and reverse transcription-PCR (RT-PCR). Finally, the direct DNA sequencing of a PCR product amplified by primers Check Out1 and Check Out2 from the mutant strain was performed to confirm the successful deletion of the divIVA gene.
Table 2.
Primers used for PCR amplification and detection.
| Primers | Sequences (5′-3′)a | Restriction enzyme | Amplification target |
|---|---|---|---|
| Div-F1 | GGATCCATGGCACTTACAGCATT | BamHI | divIVA gene |
| Div-R1 | CTCGAGTTCTTCTATTGAAAGTACTAC | XhoI | divIVA gene |
| STK-F1 | GGATCCATGATTCAAATCGGTAAG | NdeI | stk gene |
| STK-R1 | CTCGAGTTGTCCGCTACCTGTTG | HindIII | stk gene |
| Div-T172A-F1 | CAACAGCAAGTTACATTCAAGCAAGTGACGAAG | Mutant divIVA gene | |
| Div-T172A-F1 | CTTGAATGTAACTTGCTGTTGGGCGGAGGAT | Mutant divIVA gene | |
| Div-T199A-F1 | AAAGTTTGGATTATACGCATCAATTGACACCAGAAG | Mutant divIVA gene | |
| Div-T199A-F1 | TTTCAAACCTAATATGCGTAGTTAACTGTGGTCTTC | Mutant divIVA gene | |
| L1 | CGAATTCGCTTTGCTAAGT TGGTTT | EcoRI | Upstream border of divIVA |
| L2 | CGGATCCCTTTCCTCCTAAGTTTTAAC | BamHI | Upstream border of divIVA |
| Spc1 | GGATCCGTTCGTGAATACATGTTATA | BamHI | SpcR gene |
| Spc2 | GTCGACGTTTTCTAAAATCTGAT | SalI | SpcR gene |
| R1 | CGTCGACATTTTAAGCGAGTAGGAG | SalI | Downstream border of divIVA |
| R2 | GCATGCAGACTTGCTCAATAGGA | Sph1 | Downstream border of divIVA |
| Check In1 | GACGCAGATGAAGTTGATGACTT | Internal region of divIVA | |
| Check In2 | TGAATGTAACTTGCTGTTGGGC | Internal region of divIVA | |
| Check Out1 | ATATGTCGGAGCAACAAGCAAGAC | For combined PCR detection | |
| Check Out2 | TTCGATTTCAGCTTCTGCAAGG | For combined PCR detection |
The underlined sequences are the restriction sites or nucleotides involved in site directed mutation.
Assessment of the growth characteristics and genetic stability of the mutant strains
The ΔdivIVA was passaged more than 50 times and genetic stability was analyzed by PCR using the primers Spc1/Spc2. The WT and ΔdivIVA strains were separately inoculated in flasks containing 100 mL Spc-free THB media and incubated at 37°C. The absorbance of the cultures was monitored at 1 h intervals using a spectrophotometer (Bio-Rad, USA) at a wavelength of 600 nm, and sterile THB media was used as the blank. Simultaneously, the cell cultures were also monitored using a viable bacterial count method.
Microscopic evaluation of morphology
The WT and ΔdivIVA strains were grown in THY broth and harvested at the mid-log phase, then washed twice with ddH2O. Each sample was fixed on glass slides by flaming. Gram staining was conducted according to the instructions provided in the gram staining kit (Jiancheng, China). The stained samples were observed under a light microscope.
Transmission electron microscope (TEM) and scanning electron microscope (SEM) observation were performed as previously described (Pan et al., 2009). For TEM evaluation, S. suis cells (WT and mutants) were harvested at an optical density at the wavelength of 600 nm (OD600) of 0.8 and fixed in 5% glutaraldehyde for 2 h, followed by washing with PBS. Post fixation with 1% osmium tetroxide in cacodylate buffer was conducted for 1 h at room temperature in the dark. Then, the cells were dehydrated for 20 min in each ethanol gradient and then embedded in Epon-812 epoxy resin. Ultra-thin sections were post-stained with uranyl acetate and lead citrate and observed in a JEM-1010 TEM (JEOL, Ltd., Tokyo, Japan) at an accelerating voltage of 100 kV.
For SEM analysis, all of the samples were grown in THY broth and harvested at the OD600 of 0.8. The cells were spotted on polylysine coverslips followed by washing with PBS (Fleurie et al., 2012). The cells were fixed in 0.18 M cacodylate buffer (pH 7.6) containing 2% glutaraldehyde. This was followed by dehydration across an ethanol gradient, passage in 1,1,1,3,3,3-hexamethyldisilazane (HMDS), and then air-drying. The dried samples were covered with a 10 nm-thick gold/platinum layer. Samples were then observed with a Quanta200 (FEI Co. Ltd., Oregon, USA) SEM.
Oxidative stress assays
To compare H2O2 sensitivity between the WT strain and the ΔdivIVA strain, the bacteria were grown in THB to a logarithmic phase (OD600 ≈ 0.6), and 106 CFU cells were used in each oxidative stress assay. The WT and mutant cells were treated with 0, 20, 40, 60, 80, and 100 mM H2O2 and incubated at 37°C for 15 min. Survival rates were calculated using the following equation:
by obtaining CFU counts from dilution plating after 48 h incubation.
Assays for cell phagocytosis
For phagocytosis, murine macrophage Raw 264.7 cells were cultured in RPMI 1640 (Invitrogen, USA) supplemented with 10% fetal calf serum (FCS) and maintained at 37°C with 5% CO2. Confluent monolayers of Raw 264.7 cells grown in 24-well plates were, respectively, infected with the ΔdivIVA and WT strains that were labeled by carboxyfluorescein diacetate, succinimidyl ester (CFSE) at a ratio of 100:1. The mixtures were incubated at 37°C with gentle agitation for 2 h, followed by penicillin and streptomycin treatment for 1 h in the dark. The Raw 264.7 and bacterial cells were pelleted, washed twice with PBS, and then fixed with 4% (wt/vol) paraformaldehyde for flow cytometry (BD) analysis. The anti-phagocytotic ability of bacteria was assessed based on the NMFI-values.
Cell-killing experiments
Polymorphonuclear neutrophils (PMNs) were isolated from the heparinized venous blood of healthy human volunteers using the dextran sedimentation method as described by Chabot-Roy et al. (Chabot-Roy et al., 2006). Bacteria were opsonized in 10% normal human serum and incubated with human PMNs at a ratio of 10:1 for 1 h. The controls used in testing each strain were samples containing PMNs and heat-inactivated sera and samples lacking PMNs but containing human sera. The cells were lysed with 0.1% saponin (20 min on ice), and serial dilutions of the lysates were plated on THB agar. The colonies were counted, and the percentage of S. suis 2 bacteria that survived was determined as follows: (CFUPMN+/CFUPMN−) × 100% (Kobayashi et al., 2003). Each assay was performed in triplicate.
Experimental infection of mice
Randomized groups of 10 4-week-old female BALB/c mice were challenged intraperitoneally with the WT, Δstk, or ΔdivIVA strains at a dose of 108 CFU/mouse. THB was used as the negative control. A total of three groups was used, each consisting of 10 mice. The infected mice were monitored in terms of clinical symptoms for 2 weeks. Deaths were recorded and moribund animals were humanely killed. The animal experiments were carried out in accordance with the recommendations of the laboratory animal administration rules, State Scientific and Technological Commission. The protocol was approved by the Ethics Committee of Hua Dong Research Institute for Medicine and Biotechnics.
Preparation of S. suis protein lysate and total protein
The WT and Δstk strains were grown at 37°C in THB liquid medium until the desired optical densities was, respectively, reached. The cultures were harvested by centrifugation at 8,000 rpm for 10 min at 4°C. The S. suis cells were suspended in lysis buffer containing a protease inhibitor cocktail (Sigma, USA), respectively. The cells were disrupted by sonication to obtain crude extracts of the S. suis strains. The supernatant was collected by centrifugation at 12,000 rpm for 20 min at 4°C. The proteins were precipitated with acetone, and the total protein of the S. suis strains was collected by centrifugation. Protein concentrations were determined using a bicinchoninic acid (BCA) protein estimation kit (Pierce, USA). The protein samples were stored at −80°C.
Identification of endogenous substrate by two-dimensional SDS-PAGE and western blotting
To further detect the phosphorylated STK protein in the WT and Δstk strains, two-dimensional SDS-PAGE and Western blotting were performed. Initially, the protein extracts of the WT and stk mutant strains were, respectively, dissolved in a rehydration/sample buffer. Proteins were adsorbed onto an 11-cm Immobiline DryStrip (IPG, pH range 4–7, BioRad), and rehydrated overnight. For the first dimension, isoelectric focusing (IEF, BioRad) was performed using a voltage that was increased up to the steady state under the following conditions: S1, 300 V 30 min; S2, 700 V 30 min; S3, 1,500 V 1.5 h; S4, 9,000 V 3 h; and S5, 9,000 V 4 h. Prior to the second dimension, the strips were equilibrated for 2 × 15 min in equilibration buffer (6 M urea, 2% SDS, 30% glycerol, and 75 mM Tris-HCl pH 8.8, bromophenol blue), containing 2% DTT and 2.5% iodoacetamide, respectively. The proteins were separated in a 12.5% polyacrylamide gel without a spacer gel. Electrophoresis was performed at 5 mA per gel for 30 min and then at 10 mA per gel until the tracking dye reached the bottom of the gel. Each of the samples was analyzed by two gels with the same parameters. One of them was subjected to Coomassie blue staining, and the other one was detected by a specific anti-phosphothreonine polyclonal antibody. The phosphoproteins were detected with an anti-phosphothreonine polyclonal antibody at a dilution of 1:1,000, and a goat anti-rabbit secondary antibody HRP conjugate (Bio-Rad, USA) was used at a dilution of 1:8,000. STK was identified with a STK-mouse polyclonal antibody at a dilution of 1:1,000. Three replicates were run for each samples of each strain and then analyzed using the software Image Master Platinum 7.0 (GE Healthcare, New Jersey, USA). The normalized protein amount for each protein spot was calculated as the ratio of that spot's volume to the total spot's volume on the gel. A student's t-test (P < 0.05) and a threshold of 1.5-fold change in the expression spot were utilized in the data analysis.
Mass spectrometry analysis of protein and database searches
TYPHOON SCANNE and ImageMaster 2D platinum 5.0 were employed to collect the protein point position coordinates and expression data. The phosphoprotein spots of the WT and Δstk were compared, and the WT-specific spots were identified as possible substrates of STK. Furthermore, the spots of interest were visually confirmed by three independent readers prior to mass spectroscopy. Coomassie Blue-stained protein spots of interest were cut out of the gels and sent to the Guangzhou Fit Gene Biological Technology Co. Ltd. (Guangzhou, China) for trypsin in-gel digestion and Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF-MS) analysis. Protein spots with low Mascot scores were analyzed by MALDI-TOF/TOF-MS. Data from MALDI-TOF-MS and MALDI-TOF/TOF-MS analyses were used against the National Center for Biotechnology Information (NCBI) nr protein database using MASCOT (http://www.matrixscience.com), with the parameter settings of trypsin digestion, a maximum of one missed cleavage, variable modification of oxidation (M), and a peptide mass tolerance for monoisotopic data of 50 ppm. The sequences matching the peptide mass fingerprinting (PMF) data in MASCOT were downloaded. The probability score for the match, molecular weight (MW), isoelectric point (pI), number of peptide matches, and percentage of the total translated open reading frame (ORF) sequence covered by the peptides were analyzed for confident spot identification.
Protein expression and purification
The primers used in the amplification of target DNA fragments in the present study are listed in Table 2. The coding sequence of stk and divIVA was amplified using the primers STK-F1/STK-R1 and Div-F1/Div-R1 from the 05ZYH33 genome, respectively. The PCR products of stk and divIVA were, respectively, restricted with NdeI/HindIII or BamHI/XhoI, respectively, and then inserted into the digested pET-30a and pET-28a vector to generate the recombinant plasmids pET-stk and pET-divIVA, which were then transformed into E. coli TOP10 and E. coli BL21 (DE3) cells, respectively. When the OD600 of the cells reached around 0.8, the expression was induced by adding 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Sigma, USA) and incubating at 16°C for 12 h. His-tagged rSTK and rDivIVA were purified using Ni-NTA columns (GE Healthcare, Sweden) according to the manufacturer's recommendations. The purified protein was identified by Western blot analysis using a corresponding peroxidase-conjugated anti-polyhistidine monoclonal antibody (Sigma, USA).
Production of antibodies against the rSTK and rDivIVA proteins
Adult female New Zealand white rabbits were used for the production of antibodies. Approximately 400 μg of rDivIVA was emulsified in an equal volume of complete Freund's adjuvant (CFA, Sigma-Aldrich, USA) and injected into the white rabbits intradermally. Rabbits were given two booster injections of 200 μg of protein with incomplete Freund's adjuvant (IFA, Sigma-Aldrich, USA) every 2 weeks. The rabbits received the final immunization consisting of 400 μg of purified rDivIVA protein without complete/incomplete Freund's adjuvant. Seven days later, the rabbits were bled, and the sera were separate and assayed for antibodies against rDivIVA. A similar approach was used in the production of the rSTK polyclonal antibodies using Balb/c mice. Western blotting was performed to assess the specificity of the polyclonal antibodies produced by two different animals.
In vitro protein phosphorylation
Approximately 0.34 μg of rSTK was incubated with the substrate protein (1.0 μg−1.2 μg) in a kinase buffer containing 50 mM Tris-HCl (pH 7.5), 25 mM NaCl, 10 mM MnCl2, 1 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA, and 10 mM ATP for 30 min at 37°C. The reaction was stopped by adding 5 × SDS sample buffer. The proteins were separated by 10% SDS-PAGE and transferred onto a PVDF membrane. The proteins were detected with an anti-phosphothreonine polyclonal antibody. For substrate phosphorylation, MBP (Sigma, USA) was used as the positive control in testing for kinase activity.
Determination of phosphorylation sites in DivIVA
First, the DivIVA amino acid sequence was submitted to a kinase-specific phosphorylation site prediction online tool, KinasePhos1 (http://kinasephos2.mbc.nctu.edu.tw/index.html) to predict the phosphorylated residues. Then, rDivIVA was phosphorylated by rSTK in vitro and then separated on a 10% SDS polyacrylamide gel. The band corresponding to rDivIVA was excised and subjected to trypsin digestion, and then the digested peptides were analyzed by mass spectrometry. Based on the results of network analysis and amino acid substitution predictions, we then conducted site-directed mutagenesis. First, PCR was performed using pET28a-divIVA as a template with the primer pairs DivIVA-T172A-F1/DivIVA-T172A-R1 and DivIVA-T199A-F1/DivIVA-T199A-R1 to generate pET28a-divIVAT172A and pET28a-divIVAT199A. Second, the two plasmids containing the Ala mutation at position 172 or 199 were digested by the DpnI restriction enzyme and then transformed into the E. coli competent cells, respectively. The resulting constructs were then verified by DNA sequencing. Finally, the plasmids pET28a-divIVAT172A and pET28a-divIVAT199A were extracted from the E. coli cells, respectively, and then transformed into E. coli BL21 cells. The site-directed mutated protein was overexpressed, purified, and subjected to phosphorylation assays as earlier described earlier.
Statistical analysis
All of the assays were performed in triplicate at least three times. Statistical analysis was performed using a student's t-test. Differences were considered to be statistically significant when the calculated P-value was < 0.05.
Author contributions
XP, Z-QS, and XC conceived and designed the project. HN and WF performed most experiments and data analysis. CL, QW, HH, DH, FZ, XZ, and CW participated in some experiments and data analysis. XP, Z-QS, HN, and WF wrote the paper. All authors contributed to discussion of the results, reviewed the manuscript, and approved the final article.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Doctor Daisuke Takamatsu at the National Institute of Animal Health of Japan for kindly providing the pSET2 E. coli-S. suis shuttle vector. We would also like to thank LetPub for its linguistic assistance during the preparation of this manuscript. This work was supported by the National Natural Science Foundation of China (No.81571965, No.81471920, and No.81501725), Natural Science Foundation of Jiangsu Province (BK20151091), and the Fundamental Research Funds for the Central Universities (020814380084).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00085/full#supplementary-material
Sequence analysis of S. suis DivIVA. (A) Schema of S. suis DivIVA protein. N terminal coiled-coil DivIVA domain and C terminal unstable region, respectively. (B) Multiple sequence alignment of S. suis DivIVA with related homologous proteins at the amino acid level.
Kinase activity of STK in vitro. (A) After incubating rSTK with MBP as a positive substrate control, the protein was separated by SDS-PAGE and stained with Coomassie blue. (B) After incubating rSTK with MBP, the proteins were separated by SDS-PAGE, electroblotted, and then probed with an anti-pThr antibody.
Construction of an isogenic divIVA mutant of S. suis 05ZYH33. (A) Diagram of gene divIVA knockout. The divIVA gene represented by a thick orange arrow was replaced by the spectinomycin resistance gene, as indicated by a yellow arrow. The thin arrows indicate the primer positions used in the construction and identification of the divIVA knockout mutant (ΔdivIVA). (B) Confirmatory PCRs of the ΔdivIVA mutant. The primer pairs and templates used in the PCR analysis are indicated above the lanes. The WT and Δdiv represent genome DNA of the wild-type strain 05ZYH33 and mutant strain, respectively. (C) Extraction of total RNAs. Total RNAs were extracted from the wild-type strain 05ZYH33 and the ΔdivIVA mutant. (D) RT-PCR analysis of the divIVA gene transcripts. The transcripts of the spc and divIVA genes were detected using reverse transcription (RT-PCR) analysis with cDNA, cDNA reaction mixtures without reverse transcriptase (cDNA-RT), or genomic DNA (gDNA) as templates. The RT-PCR products were analyzed by electrophoresis on a 1.0% agarose gel (lanes 1, 2, and 3 represent the application using gDNA, cDNA-RT, and cDNA from the mutant ΔdivIVA strain as template, respectively, and Spc1/Spc2 primers; lanes 4, 5, and 6 represent the application using gDNA, cDNA-RT, and cDNA from the mutant ΔdivIVA strain as a template, respectively, and In1/In2 primers; and lanes 7, 8, and 9 represent the application using gDNA, cDNA-RT, and cDNA from the wild-type strain 05ZYH33 as a template respectively, and In1/In2 primers).
Genomes used in this study.
Bacterial strains and plasmids used in this study.
References
- Agarwal S., Agarwal S., Pancholi P., Pancholi V. (2011). Role of serine/threonine phosphatase (SP-STP) in Streptococcus pyogenes physiology and virulence. J. Biol. Chem. 286, 41368–41380. 10.1074/jbc.M111.286690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda J., Garrido M. E., Fittipaldi N., Cortés P., Llagostera M., Gottschalk M., et al. (2010). The cation-uptake regulators AdcR and Fur are necessary for full virulence of Streptococcus suis. Vet. Microbiol. 144, 246–249. 10.1016/j.vetmic.2009.12.037 [DOI] [PubMed] [Google Scholar]
- Av-Gay Y., Everett M. (2000). The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8, 238–244. 10.1016/S0966-842X(00)01734-0 [DOI] [PubMed] [Google Scholar]
- Bonifait L., Vaillancourt K., Gottschalk M., Frenette M., Grenier D. (2011). Purification and characterization of the subtilisin-like protease of Streptococcus suis that contributes to its virulence. Vet. Microbiol. 148, 333–340. 10.1016/j.vetmic.2010.09.024 [DOI] [PubMed] [Google Scholar]
- Bugrysheva J., Froehlich B. J., Freiberg J. A., Scott J. R. (2011). Serine/Threonine protein kinase Stk is required for virulence, stress response, and penicillin tolerance in Streptococcus pyogenes. Infect. Immun. 79, 4201–4209. 10.1128/IAI.05360-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett B. J., Altman R. B., Ferrao R., Alejo J. L., Kaur N., Kanji J., et al. (2013). Elongation factor Ts directly facilitates the formation and disassembly of the Escherichia coli elongation factor Tu.GTP.aminoacyl-tRNA ternary complex. J. Biol. Chem. 288, 13917–13928. 10.1074/jbc.M113.460014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside K., Lembo A., de Los Reyes M., Iliuk A., Binhtran N. T., Connelly J. E., et al. (2010). Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS ONE 5:e11071. 10.1371/journal.pone.0011071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D. R., Ward D. V., Kostoulias X., Howden B. P., Moellering R. C., Jr., Eliopoulos G. M., et al. (2012). Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J. Infect. Dis. 205, 1677–1687. 10.1093/infdis/jis252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot-Roy G., Willson P., Segura M., Lacouture S., Gottschalk M. (2006). Phagocytosis and killing of Streptococcus suis by porcine neutrophils. Microb. Pathog. 41, 21–32. 10.1016/j.micpath.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Chen C., Tang J., Dong W., Wang C., Feng Y., Wang J., et al. (2007). A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE 2:e315. 10.1371/journal.pone.0000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Buhr N., Neumann A., Jerjomiceva N., von Köckritz-Blickwede M., Baums C. G. (2014). Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiology 160(Pt 2), 385–395. 10.1099/mic.0.072199-0 [DOI] [PubMed] [Google Scholar]
- de Greeff A., Buys H., Verhaar R., Dijkstra J., van Alphen L., Smith H. E. (2002). Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect. Immun. 70, 1319–1325. 10.1128/IAI.70.3.1319-1325.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X. J., Ni H., Wang L., Zheng F., Hu D., Wang C., et al. (2014). The Effects of serine/threonine kinase of Streptococcus suis type 2 on biological characteristics. Chin. J. Zoonoses 30:3 10.3969/cjz.j.issn.1002-2694.2013.12.007 [DOI] [Google Scholar]
- Echenique J., Kadioglu A., Romao S., Andrew P. W., Trombe M. C. (2004). Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72, 2434–2437. 10.1128/IAI.72.4.2434-2437.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Fan P., Yang Y., Zhou J., Shen H., Fang W. (2017). A serine/threonine phosphatase 1 of Streptococcus suis type 2 is an important virulence factor. J. Vet. Sci. 18, 439–447. 10.4142/jvs.2017.18.4.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Shen H., Tang Y., Fang W. (2015). Superoxide dismutase of Streptococcus suis serotype 2 plays a role in anti-autophagic response by scavenging reactive oxygen species in infected macrophages. Vet. Microbiol. 176, 328–336. 10.1016/j.vetmic.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Feng Y. J., Pan X. Z., Sun W., Wang C. J., Zhang H. M., Li X. F., et al. (2009). Streptococcus suis enolase functions as a protective antigen displayed on the bacterial cell surface. J. Infect. Dis. 200, 1583–1592. 10.1086/644602 [DOI] [PubMed] [Google Scholar]
- Fleurie A., Cluzel C., Guiral S., Freton C., Galisson F., Zanella-Cleon I., et al. (2012). Mutational dissection of the S/T-kinase StkP reveals crucial roles in cell division of Streptococcus pneumoniae. Mol. Microbiol. 83, 746–758. 10.1111/j.1365-2958.2011.07962.x [DOI] [PubMed] [Google Scholar]
- Fulde M., Willenborg J., de Greeff A., Benga L., Smith H. E., Valentin-Weigand P., et al. (2011). ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology 157, 572–582. 10.1099/mic.0.043067-0 [DOI] [PubMed] [Google Scholar]
- Giefing C., Meinke A. L., Hanner M., Henics T., Bui M. D., Gelbmann D., et al. (2008). Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205, 117–131. 10.1084/jem.20071168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk M., Segura M., Xu J. (2007). Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim. Health Res. Rev. 8, 29–45. 10.1017/S1466252307001247 [DOI] [PubMed] [Google Scholar]
- Goyette-Desjardins G., Auger J. P., Xu J., Segura M., Gottschalk M. (2014). Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 3:e45. 10.1038/emi.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruening P., Fulde M., Valentin-Weigand P., Goethe R. (2006). Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 188, 361–369. 10.1128/JB.188.2.361-369.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbedel S., Hahn B., Daniel R. A., Flieger A. (2012). DivIVA affects secretion of virulence-related autolysins in Listeria monocytogenes. Mol. Microbiol. 83, 821–839. 10.1111/j.1365-2958.2012.07969.x [DOI] [PubMed] [Google Scholar]
- Hall C. L., Tschannen M., Worthey E. A., Kristich C. J. (2013). IreB, a Ser/Thr kinase substrate, influences antimicrobial resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 57, 6179–6186. 10.1128/AAC.01472-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Gottschalk M., Brousseau R., Harel J., Hemmingsen S. M., Goh S. H. (2005). Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 107, 63–69. 10.1016/j.vetmic.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Jacobs A. A., van den Berg A. J., Loeffen P. L. (1996). Protection of experimentally infected pigs by suilysin, the thiol-activated haemolysin of Streptococcus suis. Vet. Rec. 139, 225–228. 10.1136/vr.139.10.225 [DOI] [PubMed] [Google Scholar]
- Kaval K. G., Halbedel S. (2012). Architecturally the same, but playing a different game: the diverse species-specific roles of DivIVA proteins. Virulence 3, 406–407. 10.4161/viru.20747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S. D., Braughton K. R., Whitney A. R., Voyich J. M., Schwan T. G., Musser J. M., et al. (2003). Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc. Natl. Acad. Sci. U.S.A. 100, 10948–10953. 10.1073/pnas.1833375100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux G., Jacobs-Wagner C. (2014). How do bacteria localize proteins to the cell pole? J. Cell Sci. 127(Pt 1), 11–19. 10.1242/jcs.138628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tan C., Zhou Y., Fu S., Hu L., Hu J., et al. (2011). The two-component regulatory system CiaRH contributes to the virulence of Streptococcus suis 2. Vet. Microbiol. 148, 99–104. 10.1016/j.vetmic.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Li M., Shao Z. Q., Guo Y. Q., Wang L., Hou T. Q., Hu D., et al. (2015). The type II histidine triad protein HtpsC is a novel adhesion with the involvement of Streptococcus suis virulence. Virulence 6, 643–653. 10.1080/21505594.2015.1056971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wang C. J., Feng Y. J., Pan X. Z., Cheng G., Wang J., et al. (2008). SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS ONE 3:e2080. 10.1371/journal.pone.0002080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fan J. J., Niu C., Wang D. C., Wang J. P., Wang X., et al. (2011). The eukaryotic-type serine/threonine protein kinase Stk is required for biofilm formation and virulence in Staphylococcus epidermidis. PLoS ONE 6:e25380. 10.1371/journal.pone.0025380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun Z. R., Wang Q. P., Chen X. G., Li A. X., Zhu X. Q. (2007). Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7, 201–209. 10.1016/S1473-3099(07)70001-4 [DOI] [PubMed] [Google Scholar]
- Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., et al. (2008). Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell Proteomics 7, 299–307. 10.1074/mcp.M700311-MCP200 [DOI] [PubMed] [Google Scholar]
- Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., et al. (2007). The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707. 10.1074/mcp.M600464-MCP200 [DOI] [PubMed] [Google Scholar]
- Mata-Cabana A., García-Domínguez M., Florencio F. J., Lindahl M. (2012). Thiol-based redox modulation of a cyanobacterial eukaryotic-type serine/threonine kinase required for oxidative stress tolerance. Antioxid. Redox Signal. 17, 521–533. 10.1089/ars.2011.4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan D. J., Davies M. R., Good M. F., Sriprakash K. S. (2004). Immune response to superoxide dismutase in group A streptococcal infection. FEMS Immunol. Med. Microbiol. 40, 249–256. 10.1016/S0928-8244(04)00003-3 [DOI] [PubMed] [Google Scholar]
- Michard C., Doublet P. (2015). Post-translational modifications are key players of the Legionella pneumophila infection strategy. Front. Microbiol. 6:87. 10.3389/fmicb.2015.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S. K., Milohanic E., Aké F., Mijakovic I., Deutscher J., Monnet V., et al. (2011). Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 11, 4155–4165. 10.1002/pmic.201100259 [DOI] [PubMed] [Google Scholar]
- Molle V., Kremer L. (2010). Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75, 1064–1077. 10.1111/j.1365-2958.2009.07041.x [DOI] [PubMed] [Google Scholar]
- Mougous J. D., Gifford C. A., Ramsdell T. L., Mekalanos J. J. (2007). Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9, 797–803. 10.1038/ncb1605 [DOI] [PubMed] [Google Scholar]
- Munoz-Dorado J., Inouye S., Inouye M. (1991). A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell 67, 995–1006. 10.1016/0092-8674(91)90372-6 [DOI] [PubMed] [Google Scholar]
- Nomoto R., Maruyama F., Ishida S., Tohya M., Sekizaki T., Osawa R. (2015). Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22 and 26: Streptococcus parasuis sp. nov. Int. J. Syst. Evol. Microbiol. 65(Pt 2), 438–443. 10.1099/ijs.0.067116-0 [DOI] [PubMed] [Google Scholar]
- Nováková L., Bezousková S., Pompach P., Spidlová P., Sasková L., Weiser J., et al. (2010). Identification of multiple substrates of the StkP Ser/Thr protein kinase in Streptococcus pneumoniae. J. Bacteriol. 192, 3629–3638. 10.1128/JB.01564-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsen K., Donat S. (2010). The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int. J. Med. Microbiol. 300, 137–141. 10.1016/j.ijmm.2009.08.016 [DOI] [PubMed] [Google Scholar]
- Pan X., Ge J., Li M., Wu B., Wang C., Wang J., et al. (2009). The orphan response regulator CovR: a globally negative modulator of virulence in Streptococcus suis serotype 2. J. Bacteriol. 191, 2601–2612. 10.1128/JB.01309-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S. F., Goss L., Dworkin J. (2011). Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 75, 192–212. 10.1128/MMBR.00042-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S. E., Edwards D. H. (2004). Identification of a polar targeting determinant for Bacillus subtilis DivIVA. Mol. Microbiol. 54, 1237–1249. 10.1111/j.1365-2958.2004.04363.x [DOI] [PubMed] [Google Scholar]
- Pinho M. G., Errington J. (2004). A divIVA null mutant of Staphylococcus aureus undergoes normal cell division. FEMS Microbiol. Lett. 240, 145–149. 10.1016/j.femsle.2004.09.038 [DOI] [PubMed] [Google Scholar]
- Rajagopal L., Clancy A., Rubens C. E. (2003). A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 278, 14429–14441. 10.1074/jbc.M212747200 [DOI] [PubMed] [Google Scholar]
- Ramirez-Arcos S., Liao M., Marthaler S., Rigden M., Dillon J. A. (2005). Enterococcus faecalis divIVA: an essential gene involved in cell division, cell growth and chromosome segregation. Microbiology 151(Pt 5), 1381–1393. 10.1099/mic.0.27718-0 [DOI] [PubMed] [Google Scholar]
- Ravikumar V., Jers C., Mijakovic I. (2015). Elucidating host-pathogen interactions based on post-translational modifications using proteomics approaches. Front. Microbiol. 6:1313. 10.3389/fmicb.2015.01312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero A., De Simone P., Smaldone G., Squeglia F., Berisio R. (2012). Bacterial cell division regulation by Ser/Thr kinases: a structural perspective. Curr. Protein Pept. Sci. 13, 756–766. 10.2174/138920312804871201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Pan X., Li X., Liu W., Han M., Wang C., et al. (2011). HtpS, a novel immunogenic cell surface-exposed protein of Streptococcus suis, confers protection in mice. FEMS Microbiol. Lett. 314, 174–182. 10.1111/j.1574-6968.2010.02162.x [DOI] [PubMed] [Google Scholar]
- Shao Z. Q., Zhang Y. M., Pan X. Z., Wang B., Chen J. Q. (2013). Insight into the evolution of the histidine triad protein (HTP) family in Streptococcus. PLoS ONE 8:e60116. 10.1371/journal.pone.0060116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestroni A., Jewell K. A., Lin W. J., Connelly J. E., Ivancic M. M., Tao W. A., et al. (2009). Identification of serine/threonine kinase substrates in the human pathogen group B streptococcus. J. Proteome Res. 8, 2563–2574. 10.1021/pr900069n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. E., Damman M., van der Velde J., Wagenaar F., Wisselink H. J., Stockhofe-Zurwieden N., et al. (1999). Identification and characterization of the CPS locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67, 1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. E., Vecht U., Wisselink H. J., Stockhofe-Zurwieden N., Biermann Y., Smits M. A. (1996). Mutants of Streptococcus suis types 1 and 2 impaired in expression of muramidase-released protein and extracellular protein induce disease in newborn germfree pigs. Infect. Immun. 64, 4409–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats J. J., Feder I., Okwumabua O., Chengappa M. M. (1997). Streptococcus suis: past and present. Vet. Res. Commun. 21, 381–407. 10.1023/A:1005870317757 [DOI] [PubMed] [Google Scholar]
- Tang J., Wang C., Feng Y., Yang W., Song H., Chen Z., et al. (2006). Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e151. 10.1371/journal.pmed.0030151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Wu W., Zhang X., Lu Z., Chen J., Fang W. (2012). Catabolite control protein A of Streptococcus suis type 2 contributes to sugar metabolism and virulence. J. Microbiol. 50, 994–1002. 10.1007/s12275-012-2035-3 [DOI] [PubMed] [Google Scholar]
- Tran S. T., Le D. T., Kim Y. C., Shin M., Choi J. D. (2009). Cloning and characterization of phosphoglucose isomerase from Sphingomonas chungbukensis DJ77. BMB Rep. 42, 172–177. 10.5483/BMBRep.2009.42.3.172 [DOI] [PubMed] [Google Scholar]
- Vecht U., Wisselink H. J., Jellema M. L., Smith H. E. (1991). Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect. Immun. 59, 3156–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Shen X., Zhao Y., Wang M., Zhong Q., Chen T., et al. (2012). Identification and proteome analysis of the two-component VirR/VirS system in epidemic Streptococcus suis serotype 2. FEMS Microbiol. Lett. 333, 160–168. 10.1111/j.1574-6968.2012.02611.x [DOI] [PubMed] [Google Scholar]
- Wang J., Li C., Yang H., Mushegian A., Jin S. (1998). A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J. Bacteriol. 180, 6764–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. T., Huang H. Y., Tsai M. A., Wang P. C., Jiang B. H., Chen S. C. (2014). Phosphoglycerate kinase enhanced immunity of the whole cell of Streptococcus agalactiae in tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 41, 250–259. 10.1016/j.fsi.2014.09.008 [DOI] [PubMed] [Google Scholar]
- Willenborg J., Fulde M., de Greeff A., Rohde M., Smith H. E., Valentin-Weigand P., et al. (2011). Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology 157(Pt 6), 1823–1833. 10.1099/mic.0.046417-0 [DOI] [PubMed] [Google Scholar]
- Zhang C., Sun W., Tan M., Dong M., Liu W., Gao T., et al. (2017). The eukaryote-like serine/threonine kinase STK regulates the growth and metabolism of zoonotic Streptococcus suis. Front. Cell. Infect. Microbiol. 7:66. 10.3389/fcimb.2017.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. H., He K. W., Duan Z. T., Zhou J. M., Yu Z. Y., Ni Y. X., et al. (2009). Identification and characterization of inosine 5-monophosphate dehydrogenase in Streptococcus suis type 2. Microb. Pathog. 47, 267–273. 10.1016/j.micpath.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Zhang Y. M., Shao Z. Q., Wang J., Wang L., Li X., Wang C., et al. (2014). Prevalent distribution and conservation of Streptococcus suis Lmb protein and its protective capacity against the Chinese highly virulent strain infection. Microbiol. Res. 169, 395–401. 10.1016/j.micres.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Zheng F., Ji H., Cao M., Wang C., Feng Y., Li M., et al. (2011). Contribution of the Rgg transcription regulator to metabolism and virulence of Streptococcus suis serotype 2. Infect. Immun. 79, 1319–1328. 10.1128/IAI.00193-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zhou J., Ni Y., Yu Z., Mao A., Hu Y., et al. (2014). Contribution of eukaryotic-type serine/threonine kinase to stress response and virulence of Streptococcus suis. PLoS ONE 9:e91971. 10.1371/journal.pone.0091971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence analysis of S. suis DivIVA. (A) Schema of S. suis DivIVA protein. N terminal coiled-coil DivIVA domain and C terminal unstable region, respectively. (B) Multiple sequence alignment of S. suis DivIVA with related homologous proteins at the amino acid level.
Kinase activity of STK in vitro. (A) After incubating rSTK with MBP as a positive substrate control, the protein was separated by SDS-PAGE and stained with Coomassie blue. (B) After incubating rSTK with MBP, the proteins were separated by SDS-PAGE, electroblotted, and then probed with an anti-pThr antibody.
Construction of an isogenic divIVA mutant of S. suis 05ZYH33. (A) Diagram of gene divIVA knockout. The divIVA gene represented by a thick orange arrow was replaced by the spectinomycin resistance gene, as indicated by a yellow arrow. The thin arrows indicate the primer positions used in the construction and identification of the divIVA knockout mutant (ΔdivIVA). (B) Confirmatory PCRs of the ΔdivIVA mutant. The primer pairs and templates used in the PCR analysis are indicated above the lanes. The WT and Δdiv represent genome DNA of the wild-type strain 05ZYH33 and mutant strain, respectively. (C) Extraction of total RNAs. Total RNAs were extracted from the wild-type strain 05ZYH33 and the ΔdivIVA mutant. (D) RT-PCR analysis of the divIVA gene transcripts. The transcripts of the spc and divIVA genes were detected using reverse transcription (RT-PCR) analysis with cDNA, cDNA reaction mixtures without reverse transcriptase (cDNA-RT), or genomic DNA (gDNA) as templates. The RT-PCR products were analyzed by electrophoresis on a 1.0% agarose gel (lanes 1, 2, and 3 represent the application using gDNA, cDNA-RT, and cDNA from the mutant ΔdivIVA strain as template, respectively, and Spc1/Spc2 primers; lanes 4, 5, and 6 represent the application using gDNA, cDNA-RT, and cDNA from the mutant ΔdivIVA strain as a template, respectively, and In1/In2 primers; and lanes 7, 8, and 9 represent the application using gDNA, cDNA-RT, and cDNA from the wild-type strain 05ZYH33 as a template respectively, and In1/In2 primers).
Genomes used in this study.
Bacterial strains and plasmids used in this study.