Abstract
Peptide:N-glycanase (PNGase) cleaves oligosaccharide chains from glycopeptides and glycoproteins. Recently the deduced amino acid sequence of a cytoplasmic PNGase has been identified in various eukaryotes ranging from yeast to mammals, suggesting that deglycosylation may play a central role in some catabolic process. Several lines of evidence indicate that the cytoplasmic enzyme is involved in the quality control system for newly synthesized glycoproteins. Two-hybrid library screening by using mouse PNGase as the target yielded several PNGase-interacting proteins that previously had been implicated in proteasome-dependent protein degradation: mHR23B, ubiquitin, a regulatory subunit of the 19S proteasome, as well as a protein containing an ubiquitin regulatory motif (UBX) and an ubiquitin-associated motif (UBA). These findings by using the two-hybrid system were further confirmed either by in vitro binding assays or size fractionation assays. These results suggest that PNGase may be required for efficient proteasome-mediated degradation of misfolded glycoproteins in mammalian cells.
In eukaryotes, the endoplasmic reticulum-associated degradation (ERAD) process functions to proteolyze unfolded proteins by using the ubiquitin-proteasome machinery (1–6). In mammalian cells, de-N-glycosylated intermediates in the overall degradation process have been detected in the presence of proteasome inhibitors (7–17). The formation of de-N-glycosylated intermediates is catalyzed by peptide:N-glycanase (PNGase), a deglycosylation activity demonstrated in mammalian cells (18, 19), hen oviduct (20), and in the budding yeast Saccharomyces cerevisiae (21). Recently, the gene encoding for the soluble PNGase (PNG1) implicated in the proteasomal degradation has been identified and characterized in S. cerevisiae (22). A comparison of the sequence of yeast Png1p (yPng1p) with available EST clones revealed that PNGase is highly conserved among various eukaryotes including plant, worms, insects, and mammals, suggesting that it is functionally important (22). Very recently, we have identified yeast Rad23p as a Png1p-interacting protein and showed that Rad23p serves to link Png1p to the proteasome (23). However, except for a decrease in the rate of degradation of a misfolded yeast secretory protein (22), no phenotype has been detected in the png1-null strain. Given this finding, we have concentrated our recent efforts on higher eukaryotes, particularly mouse Png1p (mPng1p), with the goal being to elucidate the function of Png1p.
mPng1p exhibits several structural features that differentiate it from yPng1p. First, mPng1p has extended domains at its N and C termini that result in the mPng1p having a mass almost two times greater than that of yPng1p. Second, the N-terminal domain of mPng1p is conserved among many eukaryotic species, implying its importance for some unknown function (T.S., H.P., and W.J.L., unpublished data). Therefore, it was of special interest to understand the difference between mammalian PNGase and yeast PNGase. To study which protein(s) can associate with PNGase in mammalian organisms, we carried out two-hybrid library screening by using mPng1p as the target, and, in sharp contrast to yPng1p, a number of proteins were identified to interact specifically with mPng1p. These mPng1p-interacting proteins included ubiquitin, one of the 19S proteasome regulatory subunits, the mouse homolog of yeast Rad23p, as well as an uncharacterized protein containing an ubiquitin regulatory motif (UBX) and an ubiquitin-associated motif (UBA). These associations were confirmed by biochemical methods. These findings suggest that there is a close relationship between PNGase and the 26S proteasome machinery.
Materials and Methods
Construction of Plasmids Used for the Two-Hybrid Screening.
By using EST clone 948982, which contains the full-length ORF for mPng1p, as a template, PCR reactions were performed to generate various fragments (amino acid 1–171, amino acid 1–471, amino acid 1–651, amino acid 171–471, amino acid 171–651, and amino acid 471–651) having a 5′-SmaI site and a 3′-SalI site. PCR products were purified by using Quiaquick PCR purification kit (Qiagen, Chatsworth, CA). Purified PCR products were digested with SmaI and SalI and subcloned into pBTM116-ADE2 at the same sites to generate pLexA-mPng1N terminus, pLexA-mPng1ΔC, pLexA-mPng1, pLexA-mPng1 M (middle domain), pLexA-mPng1ΔN, and pLexA-mPng1C terminus.
Construction of Plasmids Used for the Escherichia coli Expression.
The same PCR products used for the two-hybrid vectors were subcloned into pET-28a to generate following plasmids: pET-mPng1N terminus, pET-mPng1ΔC, pET-mPng1, pET-mPng1 M, pET-mPng1ΔN, and mPng1C terminus.
Yeast Strains/Two-Hybrid System.
The two-hybrid experiments were carried out as previously described (24). Strain L4O (MATa leu2 his3 trp1 ade2 URA3∷lexAop-lacZ LYS2∷lexAop-HIS3) was transformed with pLexA-mPng1 (target plasmid; see below) and a two-hybrid cDNA library (mouse testis pACT library, CLONTECH). After screening 3 × 106 transformants per screen, candidates showing reproducible β-galactosidase+ and His+ phenotype in a mPNG1-specific manner were further pursued. After curing pBTM116-ADE2-mPng1 from candidate transformants (L4O), these GAD-cDNA library-harboring yeast were mated with AMR70 strains (MATα leu2 his3 trp1 ade2 URA3∷lexAop-lacZ) transformed with each of several lexA plasmids, including the original target lexA-mPNG1 plasmid. For the candidates that lacked interaction with lexA-lamin or lexA-SIR4c, but had interaction with original lexA-PNG1, the GAD-cDNAs were isolated and sequenced.
PNGase Assay.
PNGase activity assay was performed by using a fetuin-derived asialoglycopeptide I ([14C]CH3)2Leu-Asn(GlcNAc5Man3Gal3)-Asp-Ser-Arg) as substrate as previously described (20, 21). Radioactivity was monitored on a PhosphorImager (Molecular Dynamics) and quantitated by using imagequant (ver. 1.2, Molecular Dynamics).
Production of Anti-mPng1p Antibody.
pET-mPng1N terminus was used to generate the N-terminal domain (amino acid residues 1–171) of mPng1p. The plasmid was transformed in E. coli BL21(DE3), and the protein was expressed. The overexpressed, prepared N-terminal domain of mPng1p then was sliced from a gel after 8% SDS/PAGE and injected into New Zealand white rabbits to generate polyclonal antibody (Pocono Rabbit Farm, Canadensis, PA).
Western Blot Analysis and Antibodies.
Western Blot analysis was carried out by using a 1:1,000 dilution of rabbit anti-mPng1p antibody, a 1:1,000 dilution of anti-hRpt2p (S4) antibody (Affiniti, Nottingham, U.K.), or a 1:4,000 dilution of anti-green fluorescent protein (GFP) antibody (JL-8; CLONTECH). For the secondary antibody, anti-rabbit IgG horseradish peroxidase-conjugated (Roche) (for anti-mPng1p antibody and anti-hRpt2p antibody), or anti-rabbit IgG horse radish peroxidase-conjugated (Roche) (for anti-GFP antibody) was used at 1:2,000. Analysis was performed on 9% SDS/PAGE gels and subsequently visualized by using chemiluminescence (Kirkegaard & Perry Laboratories) on x-ray film (Fuji).
Construction of mPng1-GFP Fusion Plasmid.
To construct a mPng1-GFP fusion plasmid for expression in mammalian cell culture, the full-length mPng1p sequence was amplified by using PCR to introduce a 5′-EcoRI site and a 3′-SalI site. After digestion and purification, full-length mPNG1 was subcloned into a pEGFP-C1 plasmid (CLONTECH) to generate a C-terminal GFP fusion (pmPng1-GFP).
Mammalian Cell Culture/Transfection/Fluorescent Microscopy.
COS-1 and NIH 3T3 cell lines were cultured in DMEM supplemented with 10% FBS, 10 mM l-glutamine, and 100 μg each of penicillin and streptomycin/ml, all from GIBCO/BRL. Transfection of plasmid DNAs was performed by using FuGENE 6 (Roche) according to the manufacturer's protocol. After transfection, cells were washed with cold PBS and analyzed by using a fluorescent microscope to visualize GFP-fusion proteins.
In Vitro Binding Assay.
In vitro binding assay were done as previously described (25). A GST-mHR23B fusion plasmid was constructed to have the C-terminal 173 aa of mHR23B fused to glutathione S-transferase (GST). GST-fusion plasmids were transformed into DH5α cells, and the pET-mPNG1 plasmid was transformed into BL21(DE3) cells. GST-fusion protein expressions were induced by 0.1 mM isopropyl β-d-thiogalactoside whereas the pET vector-driven protein expression was induced by addition of 1 mM isopropyl β-d-thiogalactoside. After induction at 37°C for 3 h with shaking, cells were harvested and lysed by sonication. The lysates were spun for 1 min in a microfuge, and the supernatants containing soluble mPng1p or GST-mHR23 were collected for further experiments. The recombinant mPng1p-containing extract was added to glutathione agarose beads containing GST or GST-mHR23B protein bound to them. After overnight incubation at 4°C, the beads were pelleted and washed three times with PBS. The final pellet in SDS sample buffer was boiled for 5 min. The beads were spun down and the supernatant electrophoresed on SDS/PAGE. The gel was subjected to Western blotting by using an anti-mPng1p antibody.
Gel Filtration.
Gel filtration was performed as previously described with slight modifications (23). Mammalian cell lysates were prepared in a lysis buffer in the presence of 1 mM PMSF and a protease mixture (final concentration: 1 μg/ml leupeptin/2 μg/ml antipain/10 μg/ml benzamide/1 μg/ml chymostatin/1 μg/ml pepstatin). The cytosolic fraction (0.5 ml) was loaded on a Sephacryl S-300 or Sephacryl S-400 column (Amersham Pharmacia; 1.5 × 50 cm) equilibrated with elution buffer (20 mM Hepes, pH 6.8/5 mM magnesium acetate/1 mM DTT/2 mM ATP/150 mM NaCl,/0.4 M sorbitol with protease inhibitors), and fractions of 0.9 ml were collected. Fractions were assayed for proteasome activity, and 0.8 ml of each fraction was precipitated with trichloroacetic acid (10%) and analyzed by SDS/PAGE and Western blotting.
Proteasome Activity Assay.
The peptidase activity of mammalian cell lysates toward the fluorogenic peptide, suc-LLVY-MCA (Sigma), was measured by incubation in 50 mM Tris⋅HCl (pH 7.8), 20 mM KCl, 5 mM MgOAc, and 0.5 mM DTT with 200 μM suc-LLVY-MCA for 1 h at 37°C. The reaction was stopped by addition of an equal volume of 10% SDS and 20 vol. of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). The fluorescence was determined at 380 nm excitation/440 nm emission, by using a LS-3B fluorescence spectrometer (Perkin–Elmer).
Results
Mouse PNGase Contains 651-aa Residues, and the Central Domain Has Strong Homology to yPng1p.
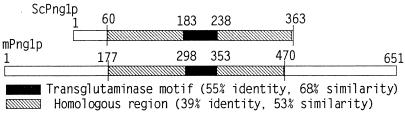
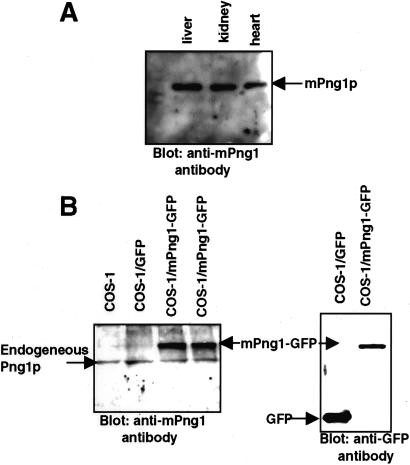
Sequence comparison of mPng1p with yPng1p revealed that the central domain of mPng1p was homologous to yPng1p (53% similarity, 39% identity) (Fig. 1) and that mPng1p contained extended N- and C-terminal domains that were absent in yPng1p. Within the conserved domains in both mouse and yPng1 proteins, a transglutaminase-like domain was found (see below). Northern blot analysis to measure the level of mRNA expression in various mouse tissues showed that mPng1p was ubiquitously expressed, with the highest level being found in testis (data not shown). Western blot analysis by using anti-mPng1p antibody detected a single protein band at 65 kDa from several mouse tissues (Fig. 2A). To document the specificity of the antibody toward mammalian Png1p, COS-1 cells were transfected with a mPng1-GFP fusion plasmid. Western blot results showed that anti-mPng1p antibody recognized both the endogenous mammalian Png1p and the transfected mPng1p-GFP fusion protein (Fig. 2B). Similar results were obtained by using NIH 3T3 cells transfected with a mPng1-GFP fusion plasmid (data not shown).
Figure 1.
Comparison of the deduced amino acid sequences of yPng1p and mPng1p. yPng1p has a 363-aa ORF whereas mPng1p contains a 651-aa ORF. The conserved middle domains contain unique transglutaminase-like motif (26).
Figure 2.
Anti-mPng1p antibody specifically recognizes mammalian PNGase. (A) Western blot analysis of mouse tissue extracts by using a polyclonal anti-mPng1p antibody. Mouse liver, kidney, and heart were homogenized in B88 buffer and subjected to Western blot analysis. Each mouse tissue revealed the presence of an ≈65-kDa band reacting with the anti-mPng1p antibody. (B) Western blot analysis of COS-1 cells expressing mPng1-GFP fusion protein. Lysates of COS-1 cells were analyzed for expression of mPng1-GFP. (B Left) Western blot using anti-mPng1p antibody against COS-1 lysates (untransfected, GFP-transfected, and mPng1-GFP transfected). Each lane shows endogenous COS-1 Png1p with an approximate Mr of 65-kDa and two right lanes show that transfected mPng1p-GFP is expressed (≈80-kDa band). (B Right) Western blot using an anti-GFP mAb. GFP (24 kDa) and mPng1-GFP (80-kDa) bands are detected in COS1/GFP and COS-1/mPng1-GFP, respectively.
In Vitro Assays Show that E. coli-Expressed mPng1p Has PNGase Activity.
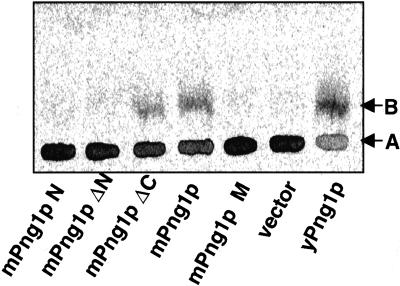
In vitro PNGase assays were performed to determine whether recombinant mPng1p had enzyme activity. E. coli BL21(DE3) lysates expressing mPng1ΔC (amino acid 1–471) or full-length mPng1p showed PNGase activity, whereas mPng1pN terminus, mPng1pΔN, and mPng1 pM showed no detectable activity, indicating the N-terminal domain and the middle domain of mPng1p are important for the observed PNGase activity (Fig. 3). Enzyme assays with an S. cerevisiae Δpng1 strain expressing various mPng1p constructs confirmed the results obtained by using E. coli recombinant mPng1p (data not shown).
Figure 3.
PNGase assay on various mPng1p fragments expressed in E. coli demonstrates that the N-terminal domain and the middle domain of mPng1p are necessary for the enzyme activity. E. coli BL21(DE3) lysates expressing various fragment were tested for PNGase enzyme activity. Each reaction was subjected to a paper chromatography as described (21). A, The starting substrate ([14C]-Leu-Asn(GlcNac5Man3Gal3)-Asn-Ser-Arg); B, the de-N-glycosylated product ([14C]-Leu-Asp-Asn-Ser-Arg).
Localization Study of mPng1-GFP Fusion Protein in Mammalian Cells Indicates Most of the PNGase Is Present in the Cytoplasm.
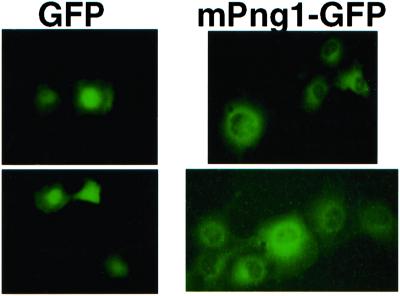
mPng1-GFP fusion plasmids were transfected into COS-1 cells to determine the localization of mPng1p. In the case of yeast, Png1p was found in the cytosol, as well as in the nucleus (23). In COS-1 cells, transfected mPng1p-GFP was found predominately in the cytosol, with a very low level associated with the nucleus. The mPng1p-GFP detected in the cytosol seemed to be concentrated around the nuclear envelope (Fig. 4).
Figure 4.
Localization of mPng1-GFP in COS-1 cells. COS-1 cells transfected with pEGFP and pmPng1-GFP were subjected to a fluorescent microscope to detect expression of GFP proteins. Left, The expression of control GFP protein (GFP localized mostly at nucleus). Right, The expression pattern of mPng1-GFP. mPng1-GFP appeared to localize mostly at the cytoplasm, around the nuclear envelope.
Two-Hybrid Library Screening Revealed That Many Proteins Interact with mPng1p.
Previously, Rad23p was identified in yeast as a Png1p-interacting protein and found to provide a link to the 26S proteasome (23). Because mPng1p has extended N- and C-terminal domains, we searched for any other proteins interacted with the mammalian Png1p. Two-hybrid library screening by using mPng1p as a target was performed by using a mouse cDNA library prepared from testis (CLONTECH). All of the candidates (total of 15; six of them are listed in Table 1) were identified based on the fact that they showed interaction with mPng1p but not with other unrelated proteins, i.e., lamin, Sir4p, and Top1p (data not shown), which were used to eliminate so called “false positives” (see ref. 24).
Table 1.
mPng1p-interacting proteins detected using the two-hybrid system
| Name | Features |
|---|---|
| mHR23B (416-aa ORF; P54728) | Yeast Rad23p homolog with 2 UBA and 1 UbL motifs |
| Present in nucleotide excision repair complex (XPC) | |
| Interacts with the 26S proteasome | |
| C-terminal 173 aa required for mPng1p binding | |
| Mouse S4 (NP_032973) | Yeast Rpt2 homolog, a 19S protesome subunit (ATPase) |
| Insert starts from 44th amino acid (440-aa ORF) | |
| Ubiquitin | A 76-aa molecule involved in the modification of protein substrate for the proteasome degradation pathway |
| Mouse homolog of human Y33K protein (297-aa ORF) | A hypothetical protein containing 1 UBA and 1 UBX motif |
| Similar to ubiquitin isopeptidase T (≈35% identity) in terms of UBA and UBX motif organization | |
| mPng1p-interacting domain includes the UBX motif | |
| Insert starts at the 52nd amino acid | |
| Mouse autocrine motility factor receptor (AMFR) (643-aa ORF; NP_035917) | A multi-transmembrane protein (cytosolic C-terminal domain) |
| Involved in the metastasis | |
| C-terminal domain of AMFR has RING finger motif and CUE motif | |
| Insert starts from 371st amino acid | |
| Mouse importin α2 (529-aa ORF; NP_034785.1) | Functions as NLS receptor |
| C-terminal 116 aa are sufficient to interact with mPng1p |
The Mouse Homolog of Rad23p and S4, a 19S Proteasome Subunit, Interacts with mPng1p.
Several of the mouse protein candidates detected in the two-hybrid screening were found to be mHR23B, a homolog of yeast Rad23p. In vitro binding assays were performed to directly test whether mPng1p interacts with mHR23B. In the experiment shown in Fig. 5, GST-mHR23B bound to glutathione agarose beads was shown to bind specifically mPng1p, establishing that a direct physical interaction between these two proteins occurs.
Figure 5.
In vitro binding assay for mPng1p-GST-mHR23B interaction. E. coli DH5α cells expressing GST or GST-mHR23B (amino acid 244–416) were lysed and subjected to glutathione-agarose binding. E. coli BL21(DE3) cell lysates expressing full-length mPng1p was used for in vitro binding. Lane 1, GST + mPng1p; lanes 2 and 3, GST-mHR23B + mPng1p, and lane 4, mPng1p only (input). For lanes 1–3, glutathione agarose-bound GST or GST-mHR23B was incubated with BL21(DE3) lysates expressing mPng1p and beads were washed and subjected to SDS/PAGE. For lane 4, BL21(DE3) lysates expressing mPng1p was used.
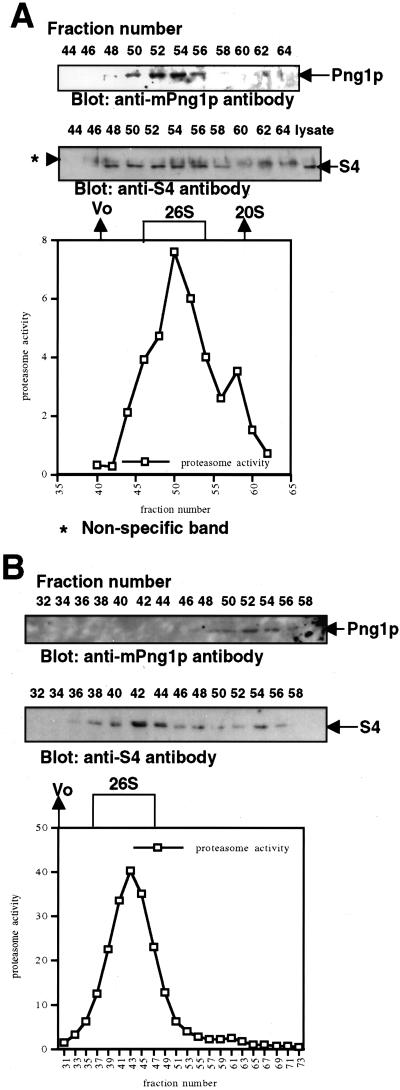
As shown in Table 1, S4, a subunit of the 19S proteasome, also was found to specifically interact with mPng1p in the two-hybrid library screening. To demonstrate an S4-mPng1p interaction in vivo, gel filtration was performed on various mammalian cell lysates. COS-1 cells were lysed in a lysis buffer containing ATP, Mg2+, and DTT to maintain integrity of the 26S proteasome and mammalian Png1p. When the lysate was fractionated on a Sephacryl 400 column most of the mPng1p was co-eluted in the same fractions that contained proteasome activity (Fig. 6A). As expected, the mammalian Png1p-rich fractions were found to contain S4 by Western blot by using anti-S4 antibody (Fig. 6A). Similar results were obtained when a Sepharcyl 300 column was used (data not shown). When the gel filtration was carried out in the absence of ATP, mPng1p was found only in fractions of lower molecular weight (Fig. 6B), indicating that the binding of mPng1p to the proteasome is ATP-dependent.
Figure 6.
Gel filtration assays on mammalian cell lysates show that mammalian Png1p co-migrates with the proteasome. (A) Sephacryl 400 column for COS-1 lysates. (B) Sephacryl 400 column for NIH 3T3 lysates in the absence of ATP. Mammalian cell lysates in 0.5-ml lysis buffer were applied on Sephacryl 400 column with or without ATP. Proteasome assays were done in the presence of ATP (A and B). Nonspecific bands in A (for anti-S4 antibody) were sometimes seen but clearly distinct from real S4 bands.
Discussion
A Comparison of yPng1p and mPng1p.
Previously, the existence of mPng1p was identified by a homology search of a mouse EST database by using yPng1p sequence as the template. A candidate mouse EST clone was sequenced and named mPNG1 (22). In this study, we first tested whether mPng1p has PNGase enzyme activity. Enzyme assay of the bacterially expressed mPng1p revealed that mPng1p, like yPng1p, has PNGase activity in vitro. Therefore, both yPng1p and mPng1p possess PNGase activity in vitro and contain a highly conserved central domain. The conserved regions of yPng1p and mPng1p were found to contain a transglutaminase-like motif (Fig. 1; ref. 26), which in yeast has been hypothesized to be essential for the catalytic activity of Png1p. It is interesting to note, however, that the mass of mPng1p is approximately twice that of yPng1p because of the presence of a 171-aa residue extension at the N terminus and a 181-aa extension at the C terminus (Fig. 1). Because mPng1p and yPng1p have marked structural differences, we searched for differences in terms of protein–protein interactions. From an extensive two-hybrid screening by using yPng1p as a target, we had previously identified yeast Rad23p, a protein which is involved in nucleotide excision repair and also known to be associated with the proteasome (27), as a yPng1p-interacting protein (23). So far, Rad23p is the only protein that we have detected to interact specifically with yPng1p. mPng1p, however, was found to interact with various proteins (Table 1). In most cases, these interactions required the N-terminal domain as well as the conserved middle domain, but not the C-terminal domain of mPng1p, suggesting the importance of the N-terminal domain of mPng1p for protein–protein interaction.
mPng1p Interacts with mHR23B.
It has been previously shown that S5a, one of the regulatory subunits of the 19S proteasome, interacts with hHR23B and hHR23A, two human homologs of Rad23p (27), providing the first evidence that the proteasome interacts with Rad23p homologs in mammalian cells. Consistent with the earlier observation that yPng1p interacts with Rad23p and this interaction provides a link to the 26S proteasome (23), we found that several fragments of a mouse homolog of Rad23p (mHR23B), interacted with mPng1p via a two-hybrid library screening. As shown in Table 1, one of the mHR23B candidates lacking the N-terminal domain, but containing the C-terminal domain, still interacted with mPng1p. In vitro binding assays confirmed that mHR23B-mPng1p interaction occurs directly (Fig. 5) and suggested that mHR23B can also provide a physical link to the 26S proteasome like yeast Rad23p.
mPng1p Interacts with S4.
S4, one of the 19S regulatory ATPase subunits, also was identified as mPng1p-interacting protein in the two-hybrid screen (Table 1). This interaction required full-length mPng1p or mPng1p ΔC, suggesting that the N-terminal domain of mPng1p is required for the interaction (the middle domain alone cannot interact with S4) (data not shown). Gel filtration analyses established that fractions showing proteasome activity co-eluted with the fractions containing mPng1p fractions when a Sephacryl 400 column was used. This was further confirmed by Western blot analysis by using an anti-S4 antibody. Strikingly, this proteasome-mPng1p interaction was found to require ATP (see Fig. 6). Overall, these results differ from those obtained in yeast where it was found that only a small fraction of yPng1p co-migrated with the proteasome activity (23). Also, mPng1p-GFP expressed in COS-1 cells showed a greater localization around the nuclear periphery (Fig. 5) than that earlier observed in yeast (22). It remains to be determined by in vitro experiment whether S4 and mPng1p can directly interact with each other without mHR23B. However, because it is clear that the 19S proteasome subunit S4 interacts with mPng1p in the two-hybrid assay, it is likely that a link between the deglycosylating event and the process of unwinding protein substrates for proteasome-mediated degradation exists. Rpt2p, the S4 homolog in yeast, has been found to mediate axial channel opening (gating) of the proteasome via its ATPase domain (28). It will be of interest to determine the in vivo role of mPng1p in proteasome-mediated degradation of (glyco)proteins. In this context it is interesting that it has been found that a Caenorhabditis elegans strain mutated in Png1p has been identified in a screen for mutations that affect axon development and branching by an unknown mechanism (A. Colavita and M. Tessier-Lavigne, personal communication).
Other Proteins That Interact with mPng1p.
The two-hybrid analysis (Table 1) revealed that ubiquitin and proteins containing other ubiquitin-related motifs (e.g., UBX or UBA) also interact with mPng1p and, in case of ubiquitin, only the N-terminal domain of mPng1p was responsible for the interaction. A previously uncharacterized protein containing both UBA and UBX domains also was isolated from two-hybrid screening (Table 1). The mouse homolog of human Y33K protein, which has been reported as a hypothetical protein, has an N-terminal UBA (ubiquitin associated) and a C-terminal UBX motif. The UBA domain was originally identified as a motif present in multiple enzymes in the ubiquitin pathway in organisms ranging from yeast to human (29) and was found to be essential for interaction of ubiquitin with yeast Rad23p and Ddi1p (30). The UBX domain is an 80-aa sequence and its three-dimensional structure suggests that it has distant homology with ubiquitin and may be involved in ubiquitin-related processes (31). Interestingly, the presence of an N-terminal UBA and a C-terminal UBX domain in the same protein is evolutionary conserved. This domain pattern also is found in ubiquitin hydrolases, ubiquitin conjugating enzymes, and ubiquitin ligases (31), which strongly suggests that the UBX domain is involved in the ubiquitination processes. This finding also suggested a possible connection between mammalian PNGase and ubiquitin-related metabolism.
The cytoplasmic tail of autocrine motility factor receptor (AMFR) also was identified as a mPng1p-interacting molecule (Table 1). AMFR is a seven transmembrane protein that has been implicated in various cancers and the full-length cDNAs for human and mouse were recently cloned (32). The mPng1p-interacting cytoplasmic domain of AMFR contains two unique domains: a CUE motif and a RING finger motif (33). The CUE motif was originally identified in yeast Cue1p, which recruits an ubituitin-conjugating enzyme to the endoplasmic reticulum (ER) membrane and is found to be one of a large family of putative scaffolding domain-containing proteins. Interestingly, the structural arrangement of AMFR (RING finger motif and five transmembrane domains) was found to be identical with the architecture of yeast Der3p/Hrd1p, whose RING finger motif has been found to be essential for endoplasmic reticulum associated degradation (ERAD) (34). It is also interesting to note that Der3p/Hrd1p was recently identified as an ER membrane-anchored ubiquitin ligase (34–36). Another candidate that was found to interact with mPng1p in the two-hybrid screen was the α subunit of importin. Yeast importin α, Srp1p, has been recently shown to have two distinct functions: nuclear localization signal motif binding in nuclear import and in regulation of protein degradation via the ubiquitin-proteasome pathway (37). Because in our study mPng1-GFP was found predominantly in the cytoplasm (Fig. 4), the interaction of importin α and mPng1p may regulate proteasome-dependent protein degradation.
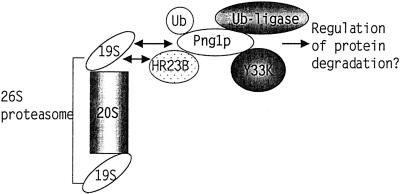
The current study, by using mPng1p as bait in a two-hybrid library screening, identified number of proteins likely to be in the ubiquitin-proteasome pathway. This result is a sharp contrast to the results from the earlier yPng1p two-hybrid library screening study (23), which indicated Rad23p as the sole Png1p-interacting protein. This result is consistent with the finding that only mHR23B was detected when the middle domain of mPng1p, which exhibits high homology to yPng1p, was used as a target for the two-hybrid screening. All of the other candidates were found to require the N-terminal domain for interaction. Thus it seems clear that the N-terminal domain is required not only for PNGase enzymatic activity, but also for an interaction with the other proteins identified from two-hybrid screening. We have found that the N-terminal domain of mPng1p has a unique conserved motif, putatively named as PUB domain (Peptide:N-glycanase/UBA or UBX domain), which is found in several Arabidopsis thaliana genes implicated in the ubiquitin-proteasome pathway (T.S., H.P., and W.J.L., unpublished data). Although, in yeast, Rad23p provides a link between Png1p and the proteasome, the relative amount of Png1p associated with the proteasome is minor. However, in mammalian cell lysates, mammalian Png1p interacts more extensively with the proteasome, probably via an ATP-dependent interaction with S4, a 19S proteasome subunit. Perhaps, as outlined in Fig. 7, mammalian Png1p recruits these ubiquitin pathway-related proteins to efficiently degrade unfolded proteins and glycoproteins exported out of the ER and regulates the ubiqutin-proteasome machinery. For example, mPng1p interacts with ubiquitin, a 19S proteasome subunit, Rad23p homolog, Y33K protein (which has an UBA and an UBX domain), and a putative ubiquitin ligase (AMFR). Thus, PNGase may provide a platform to recruit these ubiquitin-proteasome pathway proteins as well as proteins interacting with them and link this protein complex to the 26S proteasome to efficiently degrade unfolded glycoproteins. Although many details are yet to be clarified, this study has provided evidence for a tightly knitted relationship between PNGase and the proteasome pathway, and it will be of great interest to determine how mutation of this enzyme affects growth and development.
Figure 7.
Model for protein–protein interactions of mPng1p. mPng1p might act as a “platform” to recruit various ubiquitin pathway-related proteins and present them to the 26S proteasome.
Acknowledgments
We thank members of Drs. Michael Hayman's and Deborah Brown's laboratories for mammalian cell lines and Dr. Chong Yon Park for helping with mammalian cell culture. We thank Drs. Bernadette Holdener and Liqun Zhang for providing mouse tissues, members of Dr. Dafna Bar-Sagi laboratory for the use of fluorescent microscope, and Lennarz laboratory members for critical comments on manuscript. We also thank Drs. Marc Tessier-Lavigne and Antonio Colavita of University of California San Francisco for communicating valuable information and Dr. Rolf Sternglanz for providing valuable suggestions. This work was supported by National Institutes of Health Grant GM 33814 (to W.J.L.).
Abbreviations
- PNGase
Peptide:N-glycanase
- AMFR
autocrine motility factor receptor
- PNGase
mPng1p, mouse Png1p
- yPng1p
yeast Png1p
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- GST
glutathione S-transferase
References
- 1.Kopito R R. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 2.Cresswell P, Hughes E A. Curr Biol. 1997;7:R552–R555. doi: 10.1016/s0960-9822(06)00279-x. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki T, Yan Q, Lennarz W J. J Biol Chem. 1998;273:10083–10086. doi: 10.1074/jbc.273.17.10083. [DOI] [PubMed] [Google Scholar]
- 4.Brodsky J L, McCracken A A. Semin Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- 5.Plemper R K, Wolf D H. Mol Biol Rep. 1999;26:125–130. doi: 10.1023/a:1006913215484. [DOI] [PubMed] [Google Scholar]
- 6.Römish K. J Cell Sci. 1999;112:4185–4191. doi: 10.1242/jcs.112.23.4185. [DOI] [PubMed] [Google Scholar]
- 7.Wiertz E J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Nature (London) 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 8.Halaban R, Cheng E, Zhang Y, Moellmann G, Hanlon D, Michalak M, Setaluri V, Hebert D N. Proc Natl Acad Sci USA. 1997;94:6210–6215. doi: 10.1073/pnas.94.12.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes E A, Hammond C, Cresswell P. Proc Natl Acad Sci USA. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huppa J B, Ploegh H L. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, Kaung G, Kobayashi S, Kopito R R. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 12.Bebok Z, Mazzochi C, King S A, Hong J S, Sorscher E J. J Biol Chem. 1998;273:29873–29878. doi: 10.1074/jbc.273.45.29873. [DOI] [PubMed] [Google Scholar]
- 13.de Virgilio M, Weninger H, Ivessa N E. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- 14.Johnston J A, Ward C L, Kopito R R. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosse C A, Meadows L, Luckey C J, Kittlesen D J, Huczko E L, Slingluff C L, Shabanowitz J, Hunt D F, Engelhard V H. J Exp Med. 1998;187:37–48. doi: 10.1084/jem.187.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang M, Omura S, Bonifacino J S, Weissman A M. J Exp Med. 1998;187:835–846. doi: 10.1084/jem.187.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petájá-Repo U E, Hogue M, Laperriere A, Walker P, Bouvier M. J Biol Chem. 2000;275:13727–13736. doi: 10.1074/jbc.275.18.13727. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T, Seko A, Kitajima K, Inoue Y, Inoue S. Biochem Biophys Res Commun. 1993;194:1124–1130. doi: 10.1006/bbrc.1993.1938. [DOI] [PubMed] [Google Scholar]
- 19.Kitajima K, Suzuki T, Kouchi Z, Inoue S, Inoue Y. Arch Biochem Biophys. 1995;319:393–401. doi: 10.1006/abbi.1995.1309. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, Kitajima K, Emori Y, Inoue Y, Inoue S. Proc Natl Acad Sci USA. 1997;94:6244–6249. doi: 10.1073/pnas.94.12.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki T, Park H, Kitajima K, Lennarz W J. J Biol Chem. 1998;273:21526–21530. doi: 10.1074/jbc.273.34.21526. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki T, Park H, Hollingsworth N M, Sternglanz R, Lennarz W J. J Cell Biol. 2000;149:1039–1052. doi: 10.1083/jcb.149.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki T, Park H, Kwofie M A, Lennarz W J. J Biol Chem. 2001;276:21601–21607. doi: 10.1074/jbc.M100826200. [DOI] [PubMed] [Google Scholar]
- 24.Park H, Sternglanz R. Chromosoma. 1998;107:211–215. doi: 10.1007/s004120050299. [DOI] [PubMed] [Google Scholar]
- 25.Park H, Sternglanz R. Yeast. 1999;15:35–41. doi: 10.1002/(SICI)1097-0061(19990115)15:1<35::AID-YEA340>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 26.Makarova K S, Aravind L, Koonin E V. Protein Sci. 1999;8:1714–1719. doi: 10.1110/ps.8.8.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers J H, Hanaoka F. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 28.Kohler A, Cascio P, Leggett D S, Woo K M, Goldberg A L, Finley D. Mol Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann K, Bucher P. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 30.Bertolaet B L, Clarke D J, Wolff M, Watson M H, Henze M, Divita G, Reed S I. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 31.Buchberger A, Howard M J, Proctor M, Bycroft M. J Mol Biol. 2001;307:17–24. doi: 10.1006/jmbi.2000.4462. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu K, Tani M, Watanabe H, Nagamachi Y, Niinaka Y, Shiroishi T, Ohwada S, Raz A, Yokota J. FEBS Lett. 1999;456:295–300. doi: 10.1016/s0014-5793(99)00966-7. [DOI] [PubMed] [Google Scholar]
- 33.Ponting C P. Biochem J. 2000;351:527–535. [PMC free article] [PubMed] [Google Scholar]
- 34.Bordallo J, Plemper R K, Finger A, Wolf D H. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bays N W, Gardner R G, Seelig L P, Joazeiro C A, Hampton R Y. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 36.Deak P M, Wolf D H. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 37.Tabb M M, Tongaonkar P, Vu L, Nomura M. Mol Cell Biol. 2000;20:6062–6073. doi: 10.1128/mcb.20.16.6062-6073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]