Abstract
Objective
Homozygous loss-of-function mutations in the gene coding for the homeobox transcription factor (TF) PDX1 leads to pancreatic agenesis, whereas heterozygous mutations can cause Maturity-Onset Diabetes of the Young 4 (MODY4). Although the function of Pdx1 is well studied in pre-clinical models during insulin-producing β-cell development and homeostasis, it remains elusive how this TF controls human pancreas development by regulating a downstream transcriptional program. Also, comparative studies of PDX1 binding patterns in pancreatic progenitors and adult β-cells have not been conducted so far. Furthermore, many studies reported the association between single nucleotide polymorphisms (SNPs) and T2DM, and it has been shown that islet enhancers are enriched in T2DM-associated SNPs. Whether regions, harboring T2DM-associated SNPs are PDX1 bound and active at the pancreatic progenitor stage has not been reported so far.
Methods
In this study, we have generated a novel induced pluripotent stem cell (iPSC) line that efficiently differentiates into human pancreatic progenitors (PPs). Furthermore, PDX1 and H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) was used to identify PDX1 transcriptional targets and active enhancer and promoter regions. To address potential differences in the function of PDX1 during development and adulthood, we compared PDX1 binding profiles from PPs and adult islets. Moreover, combining ChIP-seq and GWAS meta-analysis data we identified T2DM-associated SNPs in PDX1 binding sites and active chromatin regions.
Results
ChIP-seq for PDX1 revealed a total of 8088 PDX1-bound regions that map to 5664 genes in iPSC-derived PPs. The PDX1 target regions include important pancreatic TFs, such as PDX1 itself, RFX6, HNF1B, and MEIS1, which were activated during the differentiation process as revealed by the active chromatin mark H3K27ac and mRNA expression profiling, suggesting that auto-regulatory feedback regulation maintains PDX1 expression and initiates a pancreatic TF program. Remarkably, we identified several PDX1 target genes that have not been reported in the literature in human so far, including RFX3, required for ciliogenesis and endocrine differentiation in mouse, and the ligand of the Notch receptor DLL1, which is important for endocrine induction and tip-trunk patterning. The comparison of PDX1 profiles from PPs and adult human islets identified sets of stage-specific target genes, associated with early pancreas development and adult β-cell function, respectively. Furthermore, we found an enrichment of T2DM-associated SNPs in active chromatin regions from iPSC-derived PPs. Two of these SNPs fall into PDX1 occupied sites that are located in the intronic regions of TCF7L2 and HNF1B. Both of these genes are key transcriptional regulators of endocrine induction and mutations in cis-regulatory regions predispose to diabetes.
Conclusions
Our data provide stage-specific target genes of PDX1 during in vitro differentiation of stem cells into pancreatic progenitors that could be useful to identify pathways and molecular targets that predispose for diabetes. In addition, we show that T2DM-associated SNPs are enriched in active chromatin regions at the pancreatic progenitor stage, suggesting that the susceptibility to T2DM might originate from imperfect execution of a β-cell developmental program.
Keywords: iPSC, T2DM, ChIP-seq, PDX1, SNPs, PP, GWAS
Highlights
-
•
PDX1 ChIP-seq analysis revealed 5664 target genes in human pancreatic progenitors, including unreported target genes.
-
•
Comparison of PDX1 profiles from PPs and adult human islets identified stage-specific PDX1 target gene sets.
-
•
T2DM-associated SNPs are enriched in active chromatin regions from iPSC-derived PPs.
-
•
Three SNPs fall into PDX1 occupied sites, located in intronic regions of the developmental regulatory TFs TCF7L2 and HNF1B.
1. Introduction
Diabetes mellitus is a group of heterogeneous disorders characterized by hyperglycemia and the progressive loss or dysfunction of the insulin-producing β-cells in the pancreas. Today, over 415 million people worldwide have been diagnosed with diabetes and the number is expected to rise to 642 million by 2040 [1]. There are four main types of diabetes mellitus: Type 1 Diabetes Mellitus (T1DM), Type 2 Diabetes Mellitus (T2DM), Gestational Diabetes and Maturity Onset Diabetes of the Young (MODY).
The most common form of diabetes is T2DM, a multifactorial disease that is caused by a complex interplay between genetic, epigenetic, and environmental factors. This form predominantly occurs in adulthood due to the failure of β-cells to produce sufficient amounts of insulin for maintenance of normoglycemia. For decades, analyses of natural genetic variation have been used to understand the pathogenesis of T2DM and to improve diagnosis and treatment for the people who suffer from this disease [1], [2].
MODY is inherited in an autosomal dominant fashion and presents early in life due to defects in developing enough β-cells or failure in function. There are at least 13 types of MODY that are diagnosed in 1–2% of diabetes patients, which are caused by mutations in several essential genes, such as GCK, HNF1A, HNF1B, and PDX1 [3].
Among these genes, PDX1 encodes one key TF, regulating β-cell development and function [4], [5]. In humans, the PDX1 gene is located on chromosome 13q12.1 and encodes for a protein of 283 amino acids. Typically for a TF it contains a transactivation domain and a homeodomain that binds to DNA. In mouse, the expression of Pdx1 is first evident at embryonic day (E) 8.5–9.0 and becomes restricted to β- and δ-cells in adult islets [6], [7], [8], [9]. Homozygous Pdx1 knockout mice form pancreatic buds but fail to develop a pancreas [10]. On the contrary, heterozygous Pdx1 knockout mice develop a pancreas but become diabetic in adulthood and β-cells increasingly undergo apoptosis [11], [12], [13]. In humans, PDX1 is expressed in the developing pancreas and heterozygous mutations in the PDX1 gene cause a strong form of monogenic diabetes, called MODY4 [14], [15]. Contrary to the numerous studies highlighting the importance of Pdx1 during mouse pancreas development, little is known about the role of this TF in human β-cell development, homeostasis and function. Specifically, it is important to unravel the PDX1 target gene program to understand its cell-type specific function during development and its contribution to MODY and T2DM in adulthood. Genome-wide association studies have identified multiple loci associated with the susceptibility to T2DM, including TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, and IGF2BP2 [16], [17]. It has been shown that islet enhancers are enriched in T2DM-associated SNPs [18]; however, it is unknown if these SNPs affect PDX1 bound cis-regulatory regions and play a role during pancreas development.
Here, we generated a novel iPSC line from a healthy female donor and confirmed its efficient in vitro pancreatic differentiation. We performed transcriptome analysis combined with ChIP-seq profiling of active H3K27ac histone modifications and PDX1 binding sites in PPs and compared these to adult islets to investigate stage-specific functions of PDX1 in progenitors and adult β-cells. Furthermore, through screening for T2DM-associated SNPs in active chromatin regions of PPs, we suggest that some SNPs might increase the diabetes risk by affecting pancreas and β-cell development.
2. Materials and methods
2.1. Ethics statement
The choice of appropriate human donors and the procedures for skin biopsy, isolation, and characterization of dermal fibroblasts were performed in accordance with study protocols approved by the Ethics Committee of the Medical Faculty of the Eberhard Karls University, Tübingen. The study design followed the principles of the Declaration of Helsinki. All study participants gave informed consent prior to entry into the study. All mice were housed in the facilities at the Helmholtz Zentrum München – German Research Center for Environmental Health (HMGU) and treated in accordance with the German animal welfare legislation and acknowledged guidelines of the Society of Laboratory Animals (GV-SOLAS) and of the Federation of Laboratory Animal Science Associations (FELASA). The teratoma generation procedure was approved by the Institutional Animal Care and Use Committee (IACUC) of HMGU and notified to the local regulatory supervisory authority.
2.2. Skin biopsy, isolation, and characterization of dermal fibroblasts
A full-thickness skin specimen was taken by punch biopsy from the upper arm in the deltoid muscle region. After removal of adipose tissue remnants and visible blood vessels, the sample was digested overnight at 4 °C with 10 U/mL dispase II (Roche Diagnostics, Mannheim, Germany) in 50 mM HEPES pH 7.4, 150 mM NaCl. Thereafter, the digest was incubated for 30 min at 37 °C under continuous shaking (1200 rpm). Using forceps, the dermis was separated from the epidermal layer, and fibroblasts were isolated from the dermis by digestion with 0.2% collagenase CLS I (Biochrom, Berlin, Germany) in DMEM, 10% BSA for 45 min at 37 °C under continuous shaking (1200 rpm). For purification of the fibroblasts, the digest was filtered through a 70 μm mesh and centrifuged. The pelleted cells were resuspended, grown for three days in DMEM, 10% FCS, and subsequently further expanded in Medium 106 supplemented with low serum growth supplement (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). Samples of dermal fibroblasts were tested for the presence of viruses, pathogenic to humans, i.e., HBV, HCV, and HIV, with Genesig PCR-based detection kits from Primerdesign Ltd. (Chandler's Ford, UK) and for the presence of mycoplasma with a PCR test kit from PanReac AppliChem (Darmstadt, Germany) and, in parallel, by DNA staining with DAPI. All cultures were found to be negative for the tested contaminants.
2.3. XM001 iPSCs generation
Primary fibroblasts from a healthy female donor were reprogrammed into pluripotent stem cells using a non-integrating Episomal iPSC Reprogramming Kit (Invitrogen, Cat. no. A14703). This kit contains a mixture of three vectors, which have the oriP/EBNA-1 (Epstein–Barr nuclear antigen-1) backbone that delivers six reprogramming factors: OCT4, SOX2, NANOG, LIN28, KLF4, and L-MYC [20]. Human fibroblasts, at 75–90% confluency, were transfected using the Amaxa 4D-Nucleofector transfection system and a nucleofector kit for human dermal fibroblasts (Lonza, Cat. no. VPD-1001), plated onto geltrex-coated culture dishes and incubated in supplemented fibroblast medium. The fibroblast medium contained knockout DMEM/F-12 (Life Technologies), 10% ESC-qualified FBS (Life Technologies), 1% MEM non-essential amino acids (Life Technologies), 10 μM HA-100 (Santa Cruz) and 4 ng/mL bFGF (Life Technologies). At 24 h after transfection, the medium was exchanged with N2B27 medium supplemented with 0.5 μM PD0325901 (Stemgent), 3 μM CHIR99021 (Stemgent), 0.5 μM A-83-01 (Stemgent), 10 μM HA-100 (Santa Cruz), 10 ng/mL hLIF (Life Technologies) and 100 ng/mL bFGF (Life Technologies). The basic N2B27 medium contained DMEM/F12 with HEPES (Life Technologies), 1 × N2 supplement (Life Technologies), 1 × B27 supplement (Life Technologies), 1% MEM non-essential amino acids, 1 × Glutamax (Life Technologies) and 1 × β-Mercaptoethanol (Life Technologies). On day 15 after transfection, the medium was changed to Essential 8 medium and monitored for the emergence of iPSC colonies. Around 3 weeks after transfection, undifferentiated iPSC colonies were picked and transferred onto fresh geltrex-coated culture dishes for expansion.
2.4. XM001 iPSCs characterization
DNA was extracted from iPSCs using standard procedure. Markers for the episomal backbone were amplified by semi-quantitative PCR to exclude transgene integration. Primers were as follows: oriP forward: TTCCACGAGGCTAGTGAACC. oriP reverse: TCGGGGGTGTTAGAGACAAC; EBNA-1 forward: ATCGTCAAAGCTGCACACAG. EBNA-1 reverse: CCCAGGAGTCCCAGTAGTCA. For karyotype analysis, we used the cells growing in logarithmic phase. These were fed with fresh medium the day before adding colcemid for 2 h. Cells were then trypsinized, treated with hypotonic solution (0.075 M KCl) for 20 min, and fixed with methanol:acetic acid (3:1). Metaphases were spread on microscope slides, and chromosomes were classified according to the International System for Human Cytogenic Nomenclature using the standard G banding technique. At least 20 metaphases were counted per cell line, and the final karyotype was stated if it was present in more than 85% of them. For teratomas, 2 × 106 iPSCs were injected into the right hind leg of immunocompromised NOD/SCID mice. Tumors were excised after 8 weeks, fixed, embedded in paraffin, sectioned and stained with hematoxylin/eosin [51].
2.5. XM001 iPSCs culture
XM001 iPSCs were cultured on 1:100 diluted Matrigel (BD Biosciences) in mTeSR™1 medium (Stem Cell Technologies). At ∼70–80% confluency, cultures were rinsed with 1 × DPBS without Mg2+ and Ca2+ (Invitrogen) followed by incubation with 1 × TrypLE Select Enzyme (Life Technology) for 3–5 min at 37 °C. Single cells were rinsed with mTeSR™1 medium, and spun at 1,000 rpm for 3 min. The resulting cell pellet was resuspended in mTeSR™1 medium supplemented with 10 μM Y-27632 (Sigma–Aldrich) and the single cell suspension was seeded at ∼1.5 × 105 cells/cm2 on Matrigel-coated surfaces. Cultures were fed every day with mTeSR™1 medium and differentiation was initiated 48 h following seeding, resulting in ∼90% starting confluency.
2.6. Pancreatic progenitor differentiation
2.6.1. S1: Definitive endoderm (3 d)
XM001 iPSCs plated on 1:100 diluted Matrigel were first rinsed with 1 × DPBS without Mg2+ and Ca2+ (Invitrogen) and then cultured in RPMI 1640 medium (Invitrogen) further supplemented with 1.2 g/L sodium bicarbonate (Sigma), 0.2% ESC-qualified FBS (Life Technologies), 100 ng/mL Activin-A (R&D Systems), and 20 ng/mL of Wnt3A (R&D Systems) for day 1 only. For the next 2 days, cells were cultured in RPMI with 0.5% FBS, 1.2 g/L sodium bicarbonate and 100 ng/mL Activin-A.
2.6.2. S2: Primitive gut tube (3 d)
Cells were rinsed with 1 × DPBS (without Mg2+ and Ca2+) once and then exposed to DMEM-F12 (Life Technologies) medium further supplemented with 2 g/L sodium bicarbonate, 2% FBS and 50 ng/mL of FGF7 (R&D Systems) for 3 days.
2.6.3. S3: Pancreatic progenitors (PP)/posterior foregut (4 d)
Cultures were maintained for 4 days in DMEM-HG medium (Life Technologies) supplemented with 0.25 μM SANT-1 (Sigma–Aldrich), 2 μM retinoic acid (RA; Sigma–Aldrich), 100 ng/mL of Noggin (R&D Systems), and 1% B27 (Invitrogen).
2.7. Immunofluorescence imaging
Cells were fixed with 4% paraformaldehyde for 30 min and then permeabilized in PBS containing 0.2% Triton X-100. Cells were blocked with PBS containing 3% BSA, and incubated with primary antibodies overnight at 4 °C. Then secondary antibodies were incubated for 1 h at room temperature after washing with PBS. Images were acquired on a TCS SP5 laser-scanning microscope (Leica). The following antibodies and dilutions were used: goat anti-OCT-3/4 (1:500, #sc-8628, Santa Cruz), goat anti-SOX2 (1:500, #sc-17320, Santa Cruz), mouse anti-TRA-1-60 (1:1000, #4746, Cell Signaling), mouse anti-TRA-1-81 (1:50, MAB4381, Millipore), mouse anti-SSEA4 (1:500, #4755, Cell Signaling), rabbit anti-FOXA2 (1:250, #8186, Cell Signaling), goat anti-SOX17 antibody (1:500, #GT15094, Acris/Novus), goat anti-PDX1 antibody (1:500, #AF2419, R&D Systems).
2.8. Flow cytometry
Cells were dissociated using 1 × TrypLE Select Enzyme and washed with cold FACS buffer (5% FBS in 1 × DPBS). Cells were fixed with 4% paraformaldehyde and permeabilized with donkey block solution (0.1% tween-20, 10% FBS, 0.1% BSA and 3% donkey serum) containing 0.5% saponin. The cells were incubated with goat anti-SOX17 antibody (1:500, #GT15094, Acris/Novus) and goat anti-PDX1 antibody (1:500, #AF2419, R&D Systems) for 30 min at room temperature and then stained with Alexa Fluor 555-conjugated donkey antibody directed against goat (1:500, #A21432, Invitrogen) for 30 min at room temperature. Flow cytometry was performed using FACS-Aria III (BD Bioscience). FACS data were analyzed using FlowJo.
2.9. RNA isolation and qPCR
Total RNA was extracted from cells with the miRNeasy mini kit (Qiagen). cDNA synthesis was performed with a high-capacity RNA-to-cDNA kit (Applied Biosystems). TaqMan qPCR was performed under standard conditions using ViiA7 (Applied Biosystems) and TaqMan Fast Advanced Master Mix (Applied Biosystems). Samples were normalized to housekeeping genes 18S ribosomal RNA (RN18S) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Taqman probes (Applied Biosystems): PDX1, Hs00236830_m1; SOX9, Hs01001343_g1; PTF1A, Hs00603586_g1; HNF1B, Hs01001602_m1; OCT4, Hs00999632_g1; NANOG, Hs04260366_g1; GAPDH, Hs02758991_g1; 18S, Hs99999901_s1; NKX6.1, Hs01055914_m1; NKX2.2, Hs00159616_m1.
SYBR qPCR was performed under standard conditions using ViiA7 (Applied Biosystems) and SYBR™ Green PCR Master Mix (Applied Biosystems). Samples were normalized to the housekeeping gene ACTB. Primers were as follows:
HHEX forward: ACGGTGAACGACTACACGC.
HHEX reverse: CTTCTCCAGCTCGATGGTCT;
MEIS1 forward: GGGCATGGATGGAGTAGGC.
MEIS1 reverse: GGGTACTGATGCGAGTGCAG;
ONECUT1 forward: GAACATGGGAAGGATAGAGGCA.
ONECUT1 reverse: GTAGAGTTCGACGCTGGACAT;
RFX6 forward: AAGCAGCGGATCAATACCTGT.
RFX6 reverse: ACCGTGGTAAGCAAACTCCTT;
ACTB forward: CCCAGAGCAAGAGAGG.
ACTB reverse: GTCCAGACGCAGGATG.
2.10. ChIP-seq
XM001 PP cells (2 × 106 cells) were cross-linked in 1% formaldehyde in culture medium for 10 min at room temperature. The cross-linking reaction was stopped by the addition of glycine to a final concentration of 125 mM. For chromatin fragmentation, cells were resuspended in lysis buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5% SDS) and sonicated in a Covaris S220 sonicator with a duty cycle of 2%, a peak incident power of 105 W and 200 cycles per burst for 20 min. The fragmented chromatin was diluted 1:5 in IP-Buffer (10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.1% Na-Desoxycholate, 140 mM NaCl, H2O, Protease Inhibitors) and directly used for immunoprecipitation. For PDX1 ChIP, 60% of the chromatin (equivalent of 1.2 × 106 cells) was processed in two parallel ChIPs using magnetic beads, preloaded with 3 μl goat anti PDX1 antibody (kindly provided by C. Wright), for each ChIP. For H3K27ac ChIP, 40% of the chromatin (equivalent 0.4 × 106 cells) was processed in three parallel IPs using magnetic beads, preloaded with 3 μg anti H3K27ac antibody (Diagenode, C15410174). For all ChIPs, antibody incubation was performed at 4 °C for 5 h. Beads were then washed 5 times using (1.) IP-Buffer, (2.) Washing-Buffer 1 (500 mM NaCl, 50 mM Tris-HCl (pH 8.0), 0.1% SDS, 1% NP-40), (3.) Washing-Buffer 2 (250 mM LiCl, 50 mM Tris-HCl (pH 8.0), 0.5% Na-Deoxycholate, 1% NP-40) and (4. & 5.) Washing-Buffer 3 (10 mM Tris-HCl (pH 8.0), 10 mM EDTA). Subsequently, protein-DNA complexes were eluted from the beads in Elution-Buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% SDS) at 65 °C for 20 min. Cross-links were reversed at 65 °C overnight, and DNA was purified for library construction with the MicroPlex kit (Diagenode, C05010010). Libraries were sequenced on an Illumina HiSeq 1500 (50 bp, single end).
2.11. ChIP-seq data analysis
Raw reads from PDX1 and H3K27ac ChIP-seq were processed with Trimmomatic (0.35) to remove low quality bases and potential adapter contamination. Next, reads were aligned to hg19 genome using bowtie2 (2.2.6) with very-sensitive option and duplicate reads were removed using samtools (1.3). Binding sites for PDX1 were then called using GEM [52] and filtered, after visual inspection, using a Q-value cut-off of 10−4 and overlapping regions were merged. Regions of H3K27ac enrichment were called using BCP-HM [53] with default parameters. PDX1 binding sites and H3K27ac enriched regions were further filtered by removing blacklist regions [54]. Regions and binding sites within 20 kb of a TSS or within a gene body were annotated with the respective gene using bedtools (2.26.0). Motif analysis, annotations with genomic features, and pathway enrichment analysis were performed using HOMER [55]. For publicly available data sets, raw reads were downloaded from online repositories and processed as described above (GSE54471 [22], GSE58686 [24], E-MTAB-1143 [28], E-MTAB-1919 [18]). For clustering of H3K27ac profiles, first the reads that fall in the intersection of enriched regions from all samples to be compared were counted and normalization factors were calculated using DESeq2. Next, the union of all identified regions from all samples to be compared was merged and reads falling into these regions were counted. Reads were then processed with DESeq2 using normalization factors calculated before and normalized read counts were used to calculate Pearson correlation coefficients that were subsequently subjected to hierarchical clustering. To calculate enrichment of H3K27ac at PDX1-bound sites, three sets of random sites were generated by shuffling all 8088 PDX1 binding sites using bedtools (2.26.0) shuffle. Then, H3K27ac reads were counted and the ratio of the mean read count at PDX1 sites over the mean read count at shuffled sites was calculated. For visualization, BIGWIG files, normalized to 1 × sequencing depth (Reads Per Genomic Content) using deepTools bamCoverage, were used. H3K27ac and PDX1 tracks for adult islets were generated from merged BAM files containing reads from all available experiments. To generate high confidence PDX1 binding sites, the overlap of PDX1 sites from progenitor profiles (XM001, [22], [24]) or islet profiles [18], [28] was calculated, and sites found in two out of three progenitor data sets or three out of four islet data sets were considered as high confidence binding sites.
2.12. Enrichment of T2DM SNPs
To obtain positions and disease association P-values of T2DM-associated SNPs, DIAGRAM 1000G GWAS meta-analysis Stage 1 Summary statistics [17] was downloaded and filtered to remove all SNPs with an association P-value below 5 × 108. For islet H3K27ac, the union of enriched regions from all 3 available experiments [18], [22] was generated and subsequently, regions closer than 1000 bp were merged. Enrichment P-values were calculated using bedtools (2.26.0) fisher and the SNPs overlapping regions of interest were identified using bedtools (2.26.0) intersect. SNPs within 20 kb of a TSS or within a gene body were annotated with the respective gene using bedtools (2.26.0).
2.13. Affymetrix microarray
For gene profiling, total RNA was extracted using miRNeasy Mini kit (Qiagen, #217004), RNA integrity was checked using Agilent 2100 Bioanalyzer (Agilent RNA 6000 Pico Kit), and cDNA was amplified with the Ovation PicoSL WTA System V2 (Nugen, 3312) in combination with the Encore Biotin Module (Nugen, USA). Amplified cDNA was hybridized on GeneChip™ Human Gene 2.0 ST arrays (Affymetrix, 902113). Expression console (v.1.3.0.187, Affymetrix) was used for quality control. All subsequent computational analysis was performed in R using Bioconductor packages. Expression data were RMA normalized using the oligo package (version 1.38.0) and probe sets were annotated using the package hugene20sttranscriptcluster.db (version 8.5.0). Differential expression analyses were performed using the limma package (version 3.30.7) and P-values were adjusted for multiple testing by Benjamini-Hochberg correction. A gene was considered as differentially expressed if the adjusted p-value (FDR) was below a threshold of 0.05 and the fold-change was greater than or equal to 2. Functional enrichments were conducted using HOMER [55]. Gene set enrichment analysis was performed using GSEA 3.0 [56], [57] with genes ranked by log2 ratio between XM001 PPs and XM001 iPSCs.
2.14. Accession number
Microarray and ChIP-seq data have been submitted to the GEO database at NCBI under the accession number GSE106950.
3. Results
3.1. Efficient differentiation of XM001 iPSCs into pancreatic progenitors
To generate the XM001 induced pluripotent stem cell (iPSC) line, primary adult skin fibroblasts from a healthy female donor were reprogrammed into iPSCs by transfection with three episomal plasmids encoding human OCT4, SOX2, NANOG, LIN28, KLF4, and L-MYC. (Suppl. Figure 1A) [19], [20]. We established the reprogramming protocol including three stages: transfection, reprogramming and expansion (Suppl. Figure 1B). Colonies with stem cell-like characteristics appeared around day 15 post-infection and were handpicked by day 21 and subjected to quality controls. XM001 iPSCs were episomal vector integration-free and of normal karyotype (Suppl. Figure 1C–D). Next, we confirmed the expression of several pluripotency markers (TRA-1-60 and TRA-1-81, SSEA-3, and SSEA-4) and pluripotency TFs (OCT4 and SOX2) by immunostainings in XM001 iPSCs (Suppl. Figure 2). To rigorously demonstrate multi-lineage potency of the generated iPSC line, we injected 2 × 106 iPSCs subcutaneously into None-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice to generate teratomas. The histological analysis of the induced teratomas revealed that tissues from all three germ layers were differentiated, confirming pluripotency of our XM001 iPSCs in vivo (Suppl. Figure 3). These data show successful reprogramming of fibroblasts into pluripotent iPSCs that stably display human embryonic stem cell (hESC)-like characteristics and multi-lineage potency.
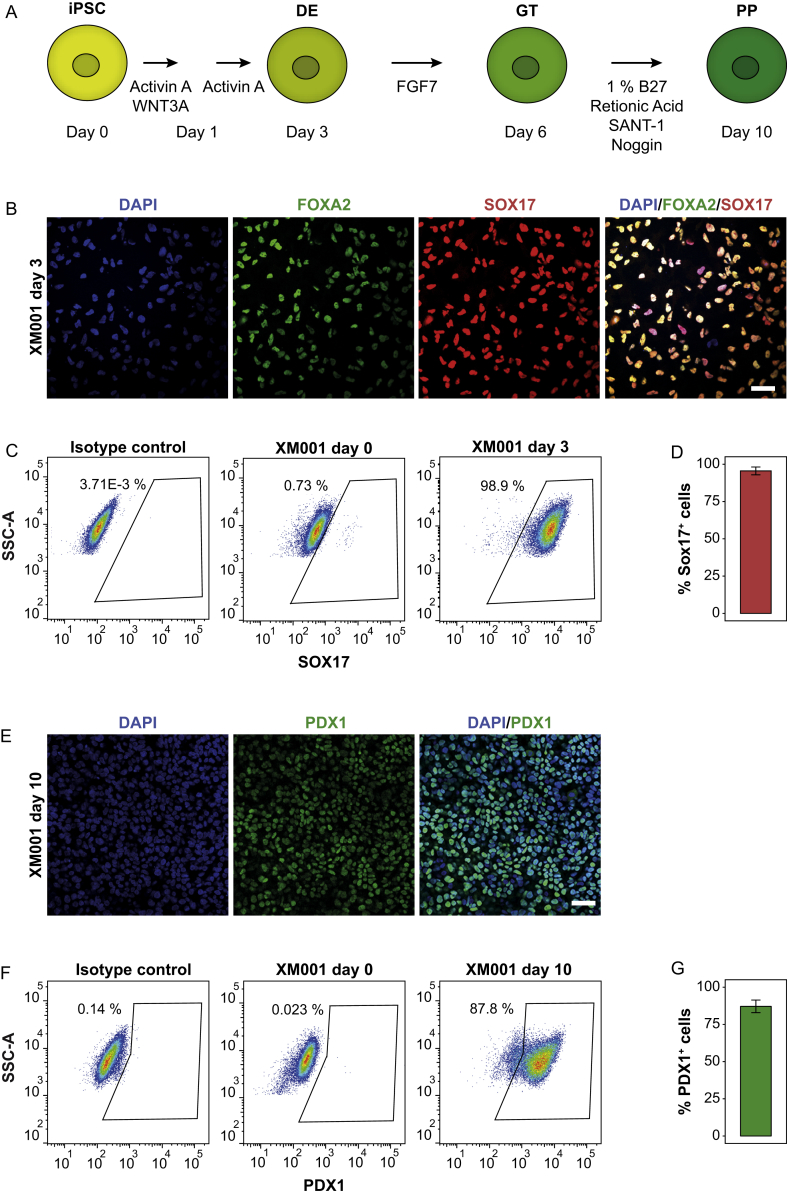
Next, we differentiated XM001 iPSCs into pancreatic progenitors (PP) using a previously established protocol recapitulating pancreas development through three main stages: definitive endoderm (DE), primitive gut tube (GT), and PPs of the posterior foregut (Figure 1A) [21]. Specific markers, corresponding to the DE (FOXA2 and SOX17) and PP (PDX1) stages were verified by immunostaining and flow cytometry at day 3 and 10, respectively. At the DE stage, cells co-expressed FOXA2 and SOX17 (Figure 1B) and flow-cytometry analysis showed that approximately 95% of the cells were positive for the DE marker SOX17 (Figure 1C–D). At the PP stage, the number of PDX1-positive cells was around 85% as measured by flow cytometry and shown by immunostaining (Figure 1E–G).
Figure 1.
Efficient differentiation of XM001 iPSCs into pancreatic progenitors. (A) Schematic of iPSC-derived pancreatic progenitor differentiation protocol. (B) Immunostaining for FOXA2 and SOX17 on day 3. Scale bar indicates 50 μm. (C) Representative FACS plot of SOX17+ cells at DE stage. A differentiated sample stained with only the secondary antibody and XM001 iPSCs were used as negative controls. (D) FACS quantification of the percentage of SOX17+ cells at DE stage (n = 3). (E) Immunostaining for PDX1 on day 10. Scale bar indicates 50 μm. (F) Representative FACS plot of PDX1+ cells at the PP stage. A differentiated sample stained with only the secondary antibody and XM001 iPSCs were used as negative controls. (G) FACS quantification of the percentage of PDX1+ cells at PP stage (n = 3).
3.2. mRNA profiles of XM001 PPs show expression of pancreas specific genes
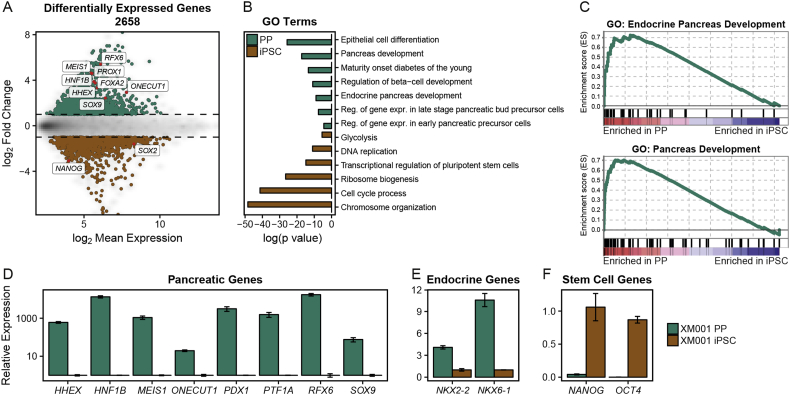
To characterize global gene expression changes during PP differentiation, we performed Affymetrix microarray analysis on XM001 cells at the pluripotency and PP stages. In total, we identified 2658 differentially expressed genes between iPSCs and PPs (FC ≥ 2, FDR ≤ 5%), including important pancreatic genes such as HNF1B, SOX9, ONECUT1, HHEX, FOXA2 and RFX6 that are upregulated in PPs, as well as pluripotency-associated genes including NANOG and SOX2 that are specifically expressed in iPSCs and downregulated in PPs (Figure 2A). Moreover, gene set enrichment analysis as well as unbiased gene ontology (GO) term and pathway analysis showed significant enrichment of terms associated with pancreas development in PP cells, while terms associated with embryonic stem cells, cell cycle and proliferation were downregulated during the differentiation process (Figure 2B–C). To verify the microarray results, we validated expression changes of important genes involved in pancreas development (HHEX, HNF1B, MEIS1, ONECUT1, PDX1, PTF1A, RFX6, and SOX9), endocrine formation (NKX2-2, NKX6-1) and stem cell maintenance (NANOG, OCT4) by quantitative PCR (qPCR) (Figure 2D–F). Taken together, the XM001 iPSCs were efficiently differentiated into PPs after 10 days of in vitro differentiation characterized by the expression of lineage-specific genes and the downregulation of pluripotency genes.
Figure 2.
mRNA profiles of XM001 PPs show expression of pancreas specific genes. (A) MA plot showing the mean log2 expression against the log2-fold change of the microarray data obtained from XM001 iPSCs and PPs. Genes with significantly different expression (log2 fold change ≥ 1 and adjusted p-value ≤ 0.05) are drawn in color. Green depicts increased expression in PPs, whereas brown depicts increased expression in iPSCs. (B) Bar graph of p-values from selected GO terms and KEGG and Reactome pathways from differentially expressed genes, shown in (A). (C) Gene set enrichment analysis of pancreas related GO terms. (D–F) qPCR validation of the expression of pancreas (D), endocrine (E) and stem cell (F) related genes in XM001 iPSCs and PPs (n = 3).
Correct specification of tissue-specific enhancers is crucial for the differentiation of individual cell types [22]. In this context, the histone modification H3K27ac is strongly associated with active enhancers and promoters [23]. Therefore, we analyzed this histone modification in XM001 cells after 10 days of differentiation using chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) and found that key pancreatic genes are marked by an active chromatin configuration. Wang et al. [22] thoroughly characterized the active H3K27ac chromatin mark at several stages of pancreatic endoderm differentiation (embryonic stem cells (ESCs), definitive endoderm (DE), gut tube (GT), posterior foregut (FG; corresponds to PP stage of this study) and late pancreatic progenitors/pancreatic endoderm (PE)) and showed that the chromatin state at lineage-specific enhancers is vital for developmental competence [22]. We used H3K27ac profiles from hESC in vitro differentiations into pancreatic progenitors [22] to compare with our H3K27ac profile from differentiated XM001 iPSCs. Unsupervised hierarchical clustering showed that after 10 days of differentiation, the active chromatin landscape in XM001 iPSCs was reminiscent of that in FG and PE stage derived from hESCs, indicating that our iPSC-derived PPs represent an early stage of pancreas development, shortly after PDX1 induction (Suppl. Figure 4).
3.3. Characterization of PDX1 binding in XM001 PP cells
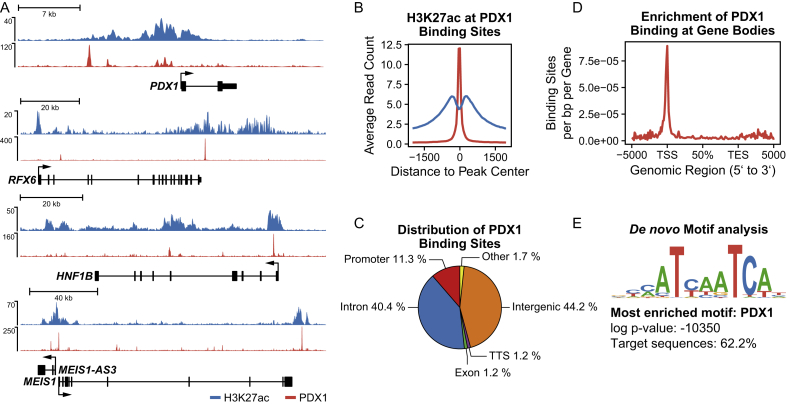
Due to the crucial role of PDX1 in the specification and differentiation of the pancreatic lineages, characterization of its transcriptional targets is important to understand human pancreas development. To do so, we employed ChIP-seq analysis of PDX1 to profile genome-wide PDX1 binding sites in XM001 iPSCs after 10 days of differentiation, a time point at which PDX1 is robustly expressed in approximately 85% of differentiated cells (Figure 1G). We identified a total of 8088 PDX1-bound regions that are associated with 5664 genes which have PDX1 binding sites within 20 kb of their TSS or within their gene body. These include PDX1 itself and other pancreatic genes, such as RFX6, MEIS1, and the MODY gene HNF1B (Figure 3A). Moreover, the active histone modification H3K27ac was enriched at the PDX1-bound sites (Figure 3B and Suppl. Figure 5), which were predominantly found in intergenic (44.2%) and intronic (40.4%) regions. However, with 11.3% of the binding sites occurring in promoter regions, PDX1 was enriched at the transcription start sites (TSS) of its target genes (Figure 3C–D). De novo motif analysis was used to discover enriched motifs within peak regions. The most significantly enriched motif was found in around 62% of all peak sequences and matched the PDX1 consensus motif, confirming the efficiency and specificity of the PDX1 ChIP-seq approach (Figure 3E). We also compared our data to previous published datasets of PDX1 ChIP-seq from hESC in vitro-derived PPs [22], [24] and found that the profiles of PDX1 binding show obvious differences that might reflect hESC and iPSC differences, genetic background variations and differences in the in vitro differentiation stage or protocol (Suppl. Figure 6).
Figure 3.
Characterization of PDX1 binding in XM001 PP cells. (A) ChIP-seq data tracks showing the enrichment of H3K27ac (blue) and PDX1 (red) at the loci of important pancreatic genes. (B) Average ChIP-seq Signal of H3K27ac (blue) and PDX1 (red) at PDX1 binding sites shows enrichment of H3K27ac at PDX1-bound sites. (C) Distribution of PDX1 binding sites among genomic features. PDX1 binds predominantly to intergenic, intronic, and promoter regions. (D) Meta-genomic plot of the enrichment of PDX1 at the transcriptional start sites (TSS) of its target genes displayed as binding sites per base pair (bp) per gene over the genomic regions of all RefSeq genes. (E) Most enriched motif discovered by de novo motif analysis resembles the known PDX1 consensus sequence and is identified in 62.2% of all PDX1-bound sequences.
3.4. Functional characterization of PDX1-bound genes
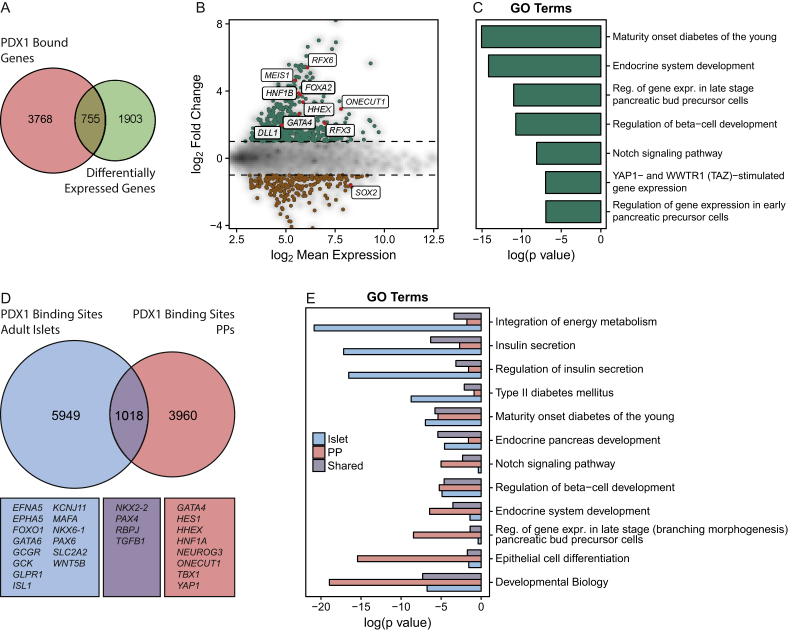
Next, we examined the expression levels and function of PDX1-bound genes in XM001 PP cells. From the 5664 PDX1-bound genes, 4523 were covered by the microarray. In XM001 PP cells, 755 PDX1-bound genes were differentially expressed between the iPSC and PP stage, which accounts for 28.4% of the total differentially expressed genes (Figure 4A–B). Notably, we found up-regulation of RFX3 and DLL1 among PDX1-bound genes during the differentiation process. The mouse orthologues of these genes are known to be involved in endocrine differentiation [25], [26], [27], suggesting a potential role of these genes also during human pancreas development. GO term analysis revealed an association of PDX1-bound genes enriched in XM001 PP cells with pancreas, endocrine and β-cell development as well as Notch and Hippo signaling, suggesting that PDX1 induces a pancreatic program and primes for endocrine differentiation (Figure 4C).
Figure 4.
Functional characterization of PDX1-bound genes. (A) Venn diagram depicting the overlap of PDX1-bound genes and the differentially expressed genes identified by microarray analysis. (B) MA plot showing the log2 fold change over the mean log2 expression of PDX1-bound genes. Differentially expressed genes are displayed in color. Green indicates enrichment in PPs, whereas brown indicates enrichment in iPSCs. (C) Bar chart of log10 p-values from enriched GO terms and KEGG and Reactome pathways of PDX1-bound genes upregulated in PPs. (D) Venn diagram showing the overlap of high confidence PDX1 binding sites from adult human islets and PPs and some annotated genes. (E) Bar chart of log10 p-values from enriched gene ontology and KEGG/Reactome pathways from genes bound in adult islets and/or PPs.
Based on the tempo-spatial pattern of Pdx1 expression in mouse that shows ubiquitous expression in all pancreatic progenitors and restricted expression in mature β- and δ-cells, we hypothesized that the molecular programs controlled by Pdx1 might be stage specific. To understand if PDX1 carries out different functions during development and in adult β-cells, we compared PDX1 binding sites from PPs and adult human islets. To that end, we derived a set of high confidence PDX1 binding sites for PPs and adult islets, comprised of PDX1 sites, consistently bound in multiple datasets from this study and previous studies from hESC-derived PPs and islets cells [18], [22], [24], [28]. Of the 4978 high confidence binding sites in PP cells, 1018 (20.4%) were identified in the islet binding data (Figure 4D). This indicates that in PP cells and adult islets only a minority of PDX1 binding sites are conserved, while the majority of sites change during the differentiation and maturation processes. Correspondingly, among the genes near PP-specific high confidence binding sites, we found important genes for pancreas development, such as GATA4, HES1, HHEX, HNF1A, NEUROG3, ONEUCUT1, TBX1, and YAP1, while genes near islet-specific binding sites, including GLPR1, KCNJ11, SLC2A2, NKX6-1, and MAFA, are associated with mature β-cell functions, e.g. insulin secretion, glucose sensing and β-cell maturation and identity (Figure 4D–E). Genes in the vicinity of shared binding sites are implicated in endocrine development and include NKX2-2, PAX4, RBPJ, and TGFB1. Of note, we also observed genes that show stage-specific binding of PDX1 that can occur in combination with common binding sites. These include HNF1B, RFX3, and RFX6, MNX1, FOXA2, the MEG3 locus, and other important pancreatic genes (Figure 5C and Suppl. Figure 7). Together, these results suggest that PDX1 binding sites change during the process of pancreas and endocrine cell development to meet the functional requirements of the different stages during development and homeostasis.
Figure 5.
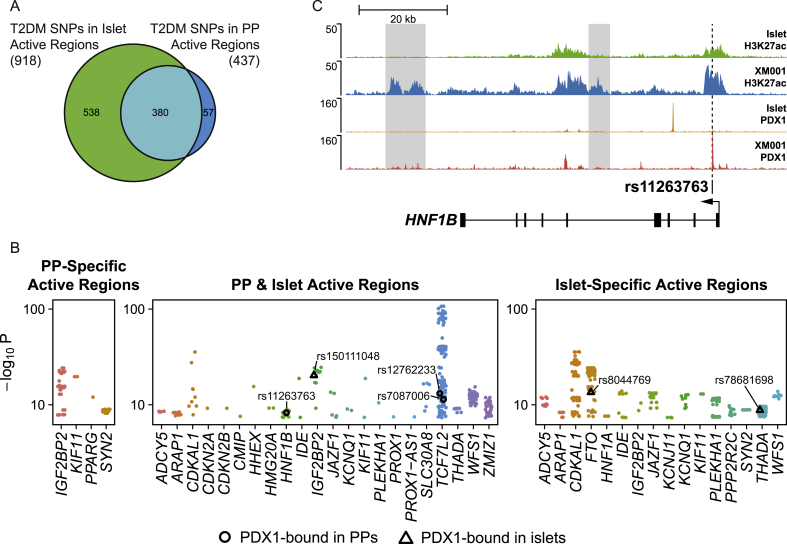
Analysis of T2DM SNPs in XM001 PPs and adult islets. (A) Venn diagram depicting the overlap of T2DM SNPs found in active regions of adult islets and XM001 PPs. (B) P-values of T2DM SNPs near known T2DM-associated genes found in active regions of adult islets [18], [22] and XM001 PPs. P-values of SNPs bound by PDX1 in PPs and islets are shown as black circles and triangles, respectively. (C) ChIP-seq data tracks showing H3K27ac and PDX1 from PP and islet [18], [22], [28] cells at the HNF1B locus. The SNP rs11263763 is located in a PP specific PDX1 binding site in the first intron of HNF1B. The PP specific enhancer regions are shaded in gray.
3.5. Analysis of T2DM SNPs in XM001 PPs and adult islets
In a worldwide effort, genome-wide association studies (GWAS) identified more than 300 genes that are linked to T2DM [29]. Of these genes, 104 (32%) were bound by PDX1 in XM001 PPs. While harsh mutations in MODY genes lead to monogenic forms of diabetes, an increased susceptibility to T2DM is linked to SNPs occurring mostly in non-coding regions. Moreover, it has been shown that islet enhancers are enriched in T2DM-associated SNPs [18], [30]. Therefore, we analyzed whether XM001 PP active chromatin regions, as determined by enrichment of H3K27ac, are also enriched in T2DM-associated SNPs and if there are different variants in XM001 PPs and adult islets. To this end, we used the DIAGRAM 1000G GWAS meta-analysis data [17] and identified SNPs with a high association significance (1788 SNPs, P ≤ 5 × 108) that overlap with active chromatin marks in XM001 PPs and adult islets [18], [22]. We found a significant enrichment of T2DM-associated SNPs in active chromatin of both PPs (437 SNPs, P = 1.0156 × 10−106) and islets (918 SNPs, P = 8.2266 × 10−263). Closer examination of the enriched SNPs showed that of the 918 SNPs found in islet active chromatin, 380 fall in regions that are also enriched in H3K27ac in the XM001 PP cells. However, we could also identify 57 SNPs in regions, active specifically in PPs (Figure 5A). When we mapped the SNPs to genes, we identified 857 SNPs in the vicinity of known genes (20 kb up-/downstream of or within a gene body), and these included mostly known T2DM-associated genes, e.g. TCF7L2, IGF2BP2, HNF1B, and FTO (Figure 5B). While most of these genes were associated with SNPs in regulatory regions active in PP and islets, some genes were associated with SNPs in cell type-specific active regions. SNPs in islet-specific regions were found near the genes FTO, HNF1A, KCNJ11, PPP2R2C, and SYN2 and SNPs in PP-specific regions near SYN2, IGF2BP2, KIF11, and PPARG. Of note, the PPARG locus is the only T2DM locus were a T2DM-associated SNP falls exclusively into a PP-specific active region and that does not have any T2DM-associated SNP in a commonly active or islet-specific active site. The enrichment of T2DM SNPs in active regulatory regions of PP cells raises the possibility that the susceptibility to T2DM is, at least in parts, already established during pancreatic development, possibly through miss-regulation of key genes and developmental programs.
Interestingly, when overlapping the disease-associated SNPs with PDX1 binding sites in PP and islet cells, we found only six SNPs that lie in the PDX1-bound sites (3 SNPs in PP PDX1 sites and 3 SNPs in islet PDX1 sites) (Figure 5B). The SNP rs8044769, SNP rs78681698 and SNP rs150111048 are in PDX1-bound regions in islets and are near FTO, THADA, and IGF2BP2. Another 3 SNPs near HNF1B and TCF7L2 are found in PDX1-bound regions of PP cells. At the HNF1B locus, the SNP rs11263763 is located in a PP-specific PDX1 binding site in the first intron of HNF1B (Figure 5C). In addition, there are PP-specific active enhancer regions, in the 4th intron and downstream of HNF1B, suggesting that HNF1B might be regulated in different ways during development and adulthood. At the TCF7L2 locus, the SNPs rs12762233 and rs7087006 are located in PP-specific PDX1 binding sites that may regulate TCF7L2 expression during development. SNPs near the TCF7L2 locus have the highest association to T2DM and mutations of HNF1B can cause MODY5. Thus, mutations in cis-regulatory regions of these genes may predispose to diabetes. This further supports the notion that the risk for T2DM might be linked to early pancreas development.
4. Discussion
Even though, a recent study confirmed that the key events of pancreatic endocrine cell formation in vivo are mimicked during in vitro differentiation [31], the efficient generation of functional and mature β-cells for cell-replacement therapy of diabetes still remains a challenge [32], [33], [34]. To produce such functional cells fulfilling the physiological properties of mature islets, it is important to understand the molecular mechanisms and gene regulatory networks governing the specification of early human pancreatic lineages that later differentiate into functional β-cells. In this context, understanding the temporal progression of key pancreatic TF target genes during development is valuable information that helps to better recapitulate the developmental steps required for the differentiation of functional β-cells in vitro. However, the access to primary human material of early pancreas development is very limited and thus, in vitro approaches have to be employed to study these early steps in a human system.
Although it is well known that Pdx1 plays a crucial role during mouse pancreas development, its stage-specific role in human pancreatic progenitors and adult β-cells has not been thoroughly studied. To address this question, we combined integrated analysis of genome-wide PDX1 binding sites and H3K27ac together with mRNA profiling in PPs derived from a novel human iPSC line.
Global characterization of PDX1 binding sites in iPSC-derived PPs identified numerous target sites and associated genes, including developmental regulatory factors, signaling molecules and factors important for mature β-cell function as well as many of the genes, differentially expressed between iPSCs and PPs. To our knowledge, this is the first genome-wide characterization of PDX1 binding in iPSC-derived PPs and comparison with previous PDX1 datasets from hESC-derived PPs of similar stages shows significant differences in PDX1 binding [22], [24]. There are many possible explanations for this discrepancy in PDX1 binding, including differences in the efficiency of differentiation, factors used in the in vitro differentiation protocol, and the exact developmental stage at the time of the experiments. This could explain many of the differences between the XM001 data from this study and the data from Wang et al. [22], as these cells are slightly further in development and already express NKX6.1. Moreover, the developmental competence of a given cell line has implications on the differentiation of the cells and might favor a set of target sites over another. Despite the differences, however, sites bound by PDX1 in multiple datasets are useful to identify high confidence stage-specific target genes.
Among the PDX1-bound genes were RFX3 and DLL1 that are important for mouse pancreas development [25], [26], [27]. RFX3 is a member of the regulatory factor × family of transcription factors that contain a highly conserved winged helix DNA-binding domain [35]. It has been shown that Rfx3 is required for the differentiation and function of mature β-cells and regulates the expression of the glucokinase gene in mice [25]. DLL1 is a member of the delta/serrate/jagged family of Notch ligands. Mice deficient for the Delta-like 1 gene (Dll1) show increased expression of Ngn3, which, in turn, causes an accelerated and increased commitment to the endocrine lineage and depletes the progenitor pool, ultimately leading to pancreatic hypoplasia [26], [27]. Whether these genes play important roles in human pancreas development has not been characterized. Therefore, our analysis suggests a possible evolutionary conserved function of RFX3 and DLL1 during human pancreas development. In contrast, other PDX1-bound genes such as TIMP2, MAFB, and SIX2, are known to be specifically express in human, but not in mouse β-cells [36], [37]. To address the function of newly identified PDX1 target genes during development, the XM001 iPSCs has been modified to express CRISPR/Cas9 under the control of an inducible promoter, which will allow gene functional studies in the future [38].
PDX1 is a master regulator of early pancreatic development and is expressed in all PPs in vivo but becomes restricted to β-and δ-cells during later stages of endocrine differentiation. Therefore, it is likely, that PDX1 has different functions and targets at different stages of development. When we compared genome-wide PDX1 binding sites from iPSC-derived PPs and adult human islets, only 20.4% of the identified PDX1 targets were common in both cell populations. Interestingly, the function of genes near common and specific binding sites differed markedly and reflects cellular function. PDX1-bound genes in islets were related to β-cell-specific functions. For example, the K+ channel KCNJ11 as well as the glucose transporter SLC2A2 are essential for adult β-cell function, whereas NKX6-1 and MAFA are important for β-cell identity and maturation. On the other hand, in PP cells PDX1-bound genes, such as GATA4 and ONECUT1, are associated with processes and pathways in early pancreas development. This shows that, depending on the developmental stage, PDX1 target sites change to activate molecular programs that are required for stage-specific functions.
The presence of H3K27ac in PP cells at regions carrying T2DM-associated SNPs indicates that these regions participate in the regulation of gene activity. Variation in these regions potentially interferes with the regulation of T2DM-relevant genes at the PP stage and might therefore impair developmental programs and lead to reduce β-cell mass or function at birth that contributes to an increased susceptibility to T2DM. It has been shown that β-cell mass in children is highly variable between individuals [39] and diabetic patients show decreased β-cell mass [40]. One of the SNPs that we detected in a PP-specific PDX1 binding site, rs11263763, lies in the first intron of HNF1B and has been shown to reduce HNF1B expression [41]. This gene has several SNPs in the first and second intron that are linked to T2DM. During mouse pancreatic development, Hnf1β is involved in controlling proliferation and survival of multipotent pancreatic progenitors and deletion of the gene causes pancreatic hypoplasia. Hnf1β is also important for the specification of endocrine progenitors through regulation of Ngn3 expression [42]. In humans, some mutations in HNF1B cause MODY5 [43]. We also identified the SNPs rs12762233 and rs7087006 within PP-specific PDX1 binding sites at the TCF7L2 locus. This gene, also known as Tcf4, is a member of the HMG-box containing T-cell factor (Tiff)/Lymphoid enhancer factor (Lef) transcription factor family of DNA-binding proteins and acts downstream of the canonical Wnt pathway [44]. GWAS studies have identified TCF7L2 as the locus conveying the highest risk for developing T2DM in European [45]. In mouse, Tcf7l2 is an important regulator of β-cell development [46], whereas in human it regulates β-cell survival and function [47]. Genetic and metabolic studies show that people carrying TCF7L2 polymorphisms have insulin secretion defects in the presence of normal incretin plasma levels [48]. HNF1B and TCF7L2 are therefore important developmental regulatory factors and SNPs in PDX1-bound regions in these loci might impair early developmental processes that result in a higher risk to develop T2DM later in adulthood. The three SNPs we found in islet-specific binding sites are near the FTO, THADA, and IGF2BP2 genes, suggesting that these SNPs might modulate adult β-cell function.
In summary, our study provides a PDX1 regulated human pancreas developmental program that can be used to investigate the effects of PDX1 mutations, e.g. in point mutations in the transactivation domain found in T2DM patients, on pancreas developmental and islet PDX1 program [49], [50]. Also, we find more than 430 T2DM-associated SNPs in active regulatory regions and 32% of T2DM genes to be bound by PDX1 in XM001 PPs, demonstrating that PDX1 occupancy is an efficient way to identify important pancreatic developmental and disease genes. This provides the base for further disease modeling in vitro using stem cell and gene editing technologies to understand the causal pathomechanisms underlying diabetes.
Author contribution
X.W. performed and analyzed iPSC experiments. M.S. performed ChIP-seq experiments and computational analyses. X.W. and S.C. generated XM001 iPSCs. A.H., F.M., H.S., and H-U.H. provided cells for iPSC generation. G.L. and T.M. performed karyotyping. F.M.C. and G.S. provided infrastructure for ChIP-seq experiments and contributed to ChIP-seq analysis. M.I., J.B., and M.H.A. performed microarray experiments and initial analysis of microarray data. M.R. and C.V.E.W. provided the goat anti PDX1 antibody, used for ChIP experiments. X.W. and M.S. wrote the manuscript. I.B. and M.B. reviewed and edited the manuscript. H.L. wrote the manuscript and conceived the work. H.L. is the guarantor of this work.
Acknowledgements
We thank A. Raducanu and A. Böttcher for comments and discussions and A. Theis, B. Vogel, J. Beckenbauer, and K. Diemer for their technical support. We also thank Stefan Krebs and the sequencing unit of the Laboratory of Functional Genome Analysis (LAFUGA) at the Gene Center of the LMU. This work was supported from the Helmholtz Portfolio Theme 'Metabolic Dysfunction and Common Disease’ (J.B.) and the Helmholtz Alliance ‘Aging and Metabolic Programming, AMPro’ (J.B.). This work was funded in part by the German Center for Diabetes Research (DZD e.V.) and by the European Union's Seventh Framework Programme for Research, Technological Development and Demonstration under grant agreement No. 602587 (http://www.hum-en.eu/).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.01.011.
Conflict of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplement Figure 1: Reprogramming of skin fibroblasts into XM001 iPSCs. (A) Scheme showing the reprogramming factors for the generation of the XM001 iPSC line. (B) Reprogramming protocol of skin fibroblasts into integration-free iPSCs using episomal vectors. (C) Normal karyotype (46, XX) of the XM001 iPSC clone. (D) Semi-quantitative PCR confirms that episomal vectors did not integrate into the genomic DNA of the selected XM001 iPSC clone. H9 hESCs were used as a negative control, whereas transfected fibroblasts at day 6 were used as positive controls.
Supplement Figure 2: XM001 iPSCs are pluripotent in vitro. XM001 iPSCs display typical morphological hESC-like characteristics and uniformly express several pluripotency markers. Scale bar indicates 50 μm.
Supplement Figure 3: XM001 iPSCs are pluripotent in vivo. Hematoxylin and eosin staining of tissue sections of teratomas generated from the XM001 iPSCs. All germ layer-derivatives are clearly detectable in vivo. Scale bar indicates 100 μm.
Supplement Figure 4: Hierarchical clustering of H3K27ac at different stages. Hierarchical clustering of Pearson correlation coefficients from H3K27ac data from XM001 and published datasets [22] from embryonic stem cells (ES), definitive endoderm (DE), gut tube (GT), posterior foregut (FG), and pancreatic endoderm (PE) cells.
Supplement Figure 5: H3K27ac is enriched at PDX1 binding sites. Read count of H3K27ac at the 8088 PDX1 binding sites in XM001 and at three sets of 8088 random sites of the same length distribution. H3K27ac is enriched >5.6 × at PDX1-bound sites compared to the random sites. P-value for all comparisons <2.2 × 10−16.
Supplement Figure 6: Comparison of PDX1 binding in XM001 PPs with previous datasets. Venn diagram showing the overlap of PDX1 binding sites between XM001 PP cells, hES-derived PP cells from Teo et al. [24] and hES-derived PE cells from Wang et al. [22].
Supplement Figure 7: Differential binding of PDX1 in XM001 PPs and adult islets. ChIP-seq data tracks showing H3K27ac and PDX1 from XM001 PP and adult islet cells [18], [22], [28] at several loci with stage specific PDX1 binding.
References
- 1.International Diabetes Federation . 2015. IDF diabetes atlas. [Google Scholar]
- 2.Prasad R.B., Groop L. Genetics of type 2 diabetes—pitfalls and possibilities. Genes. 2015;6(1):87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heuvel-Borsboom H., de Valk H.W., Losekoot M., Westerink J. Maturity onset diabetes of the young: seek and you will find. Netherlands Journal of Medicine. 2016:193–200. [PubMed] [Google Scholar]
- 4.Jennings R.E., Berry A.A., Strutt J.P., Gerrard D.T., Hanley N.A. Human pancreas development. Development. 2015;142(18):3126–3137. doi: 10.1242/dev.120063. [DOI] [PubMed] [Google Scholar]
- 5.Bastidas-Ponce A., Scheibner K., Lickert H., Bakhti M. Cellular and molecular mechanisms coordinating pancreas development. Development. 2017;144(16):2873–2888. doi: 10.1242/dev.140756. [DOI] [PubMed] [Google Scholar]
- 6.Ahlgren U., Jonsson J., Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development (Cambridge, England) 1996;122(5):1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 7.Guz Y., Montminy M.R., Stein R., Leonard J., Gamer L.W., Wright C.V. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development (Cambridge, England) 1995;121(1):11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 8.Offield M.F., Jetton T.L., Labosky P.A., Ray M., Stein R.W., Magnuson M.A. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development (Cambridge, England) 1996;122(3):983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 9.Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes & Development. 1998;12(12):1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 11.Brissova M., Shiota M., Nicholson W.E., Gannon M., Knobel S.M., Piston D.W. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. Journal of Biological Chemistry. 2002;277(13):11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- 12.Johnson J.D., Ahmed N.T., Luciani D.S., Han Z., Tran H., Fujita J. Increased islet apoptosis in Pdx1+/- mice. Journal of Clinical Investigation. 2003;111(8):1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holland A.M., Góñez L.J., Naselli G., MacDonald R.J., Harrison L.C. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of β-cells in the adult pancreas. Diabetes. 2005;54(9):2586–2595. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- 14.Staffers D.A., Ferrer J., Clarke W.L., Habener J.F. Early-onset type-ll diabetes mellitus (MODY4) linked to IPF1. Nature Genetics. 1997;17(2):138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 15.Stoffers D.A., Stanojevic V., Habener J.F. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant negative isoprotein. Journal of Clinical Investigation. 1998;102(1):232–241. doi: 10.1172/JCI2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Li H., Loos R.J.F., Yu Z., Ye X., Chen L. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57(10):2834–2842. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott R.A., Scott L.J., Mägi R., Marullo L., Gaulton K.J., Kaakinen M. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017:db161253. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasquali L., Gaulton K.J., Rodríguez-Seguí S.A., Mularoni L., Miguel-Escalada I., Akerman İ. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nature Genetics. 2014;46(2):136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;107(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Yu J., Chau K.F., Vodyanik M.A., Jiang J., Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezania A., Bruin J.E., Riedel M.J., Mojibian M., Asadi A., Xu J. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61(8):2016–2029. doi: 10.2337/db11-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang A., Yue F., Li Y., Xie R., Harper T., Patel N.A. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16(4):386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teo A.K.K., Tsuneyoshi N., Hoon S., Tan E.K., Stanton L.W., Wright C.V.E. PDX1 binds and represses hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Reports. 2015;4(4):578–590. doi: 10.1016/j.stemcr.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ait-Lounis A., Bonal C., Seguin-Estevez Q., Schmid C.D., Bucher P., Herrera P.L. The transcription factor Rfx3 regulates -cell differentiation, function, and glucokinase expression. Diabetes. 2010;59(7):1674–1685. doi: 10.2337/db09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim W., Shin Y.-K., Kim B.-J., Egan J.M. Notch signaling in pancreatic endocrine cell and diabetes. Biochemical and Biophysical Research Communications. 2010;392(3):247–251. doi: 10.1016/j.bbrc.2009.12.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apelqvist A., Li H., Sommer L., Beatus P., Anderson D.J., Honjo T. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400(6747):877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 28.Khoo C., Yang J., Weinrott S.A., Kaestner K.H., Naji A., Schug J. Research resource: the pdx1 cistrome of pancreatic islets. Molecular Endocrinology (Baltimore, Md.) 2012;26(3):521–533. doi: 10.1210/me.2011-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Research. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corradin O., Scacheri P.C. Enhancer variants: evaluating functions in common disease. Genome Medicine. 2014;6(10):85. doi: 10.1186/s13073-014-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramond C., Glaser N., Berthault C., Ameri J., Kirkegaard J.S., Hansson M. Reconstructing human pancreatic differentiation by mapping specific cell populations during development. eLife. 2017;6 doi: 10.7554/eLife.27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159(2):428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature Biotechnology. 2014;32(11):1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 34.Russ H.a, Parent A.V., Ringler J.J., Hennings T.G., Nair G.G., Shveygert M. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. The EMBO Journal. 2015;34(13):e201591058. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reith W., Ucla C., Barras E., Gaud a., Durand B., Herrero-Sanchez C. RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Molecular and Cellular Biology. 1994;14(2):1230–1244. doi: 10.1128/mcb.14.2.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.-M., Andréasson A.-C., Sun X. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism. 2016;24(4):593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xin Y., Kim J., Okamoto H., Yancopoulos G.D., Lin C., Correspondence J.G. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metabolism. 2016;24:608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 38.Yumlu S., Stumm J., Bashir S., Dreyer A.-K., Lisowski P., Danner E. Gene editing and clonal isolation of human induced pluripotent stem cells using CRISPR/Cas9. Methods. 2017;121–122:29–44. doi: 10.1016/j.ymeth.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Meier J.J., Butler A.E., Saisho Y., Monchamp T., Galasso R., Bhushan A. β-cell replication is the primary mechanism subserving the postnatal expansion of β-cell mass in humans. Diabetes. 2008;57(6):1584–1594. doi: 10.2337/db07-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. (January) [DOI] [PubMed] [Google Scholar]
- 41.Painter J.N., O'Mara T.A., Batra J., Cheng T., Lose F.A., Dennis J. Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Human Molecular Genetics. 2015;24(5):1478–1492. doi: 10.1093/hmg/ddu552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Vas M.G., Kopp J.L., Heliot C., Sander M., Cereghini S., Haumaitre C. Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development. 2015;142(5):871–882. doi: 10.1242/dev.110759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horikawa Y., Iwasaki N., Hara M., Furuta H., Hinokio Y., Cockburn B.N. Mutation in hepatocyte nuclear factor–1β gene (TCF2) associated with MODY. Nature Genetics. 1997;17(4):384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 44.Lickert H., Domon C., Huls G., Wehrle C., Duluc I., Clevers H. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development (Cambridge, England) 2000;127(17):3805–3813. doi: 10.1242/dev.127.17.3805. [DOI] [PubMed] [Google Scholar]
- 45.McCarthy M.I., Zeggini E. Genome-wide association studies in type 2 diabetes. Current Diabetes Reports. 2009:164–171. doi: 10.1007/s11892-009-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.da Silva Xavier G., Mondragon A., Sun G., Chen L., McGinty J.A., French P.M. Abnormal glucose tolerance and insulin secretion in pancreas-specific Tcf7l2-null mice. Diabetologia. 2012;55(10):2667–2676. doi: 10.1007/s00125-012-2600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu L., Sauter N.S., Schulthess F.T., Matveyenko A.V., Oberholzer J., Maedler K. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008;57(3):645–653. doi: 10.2337/db07-0847. [DOI] [PubMed] [Google Scholar]
- 48.Schäfer S.A., Tschritter O., Machicao F., Thamer C., Stefan N., Gallwitz B. Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia. 2007;50(12):2443–2450. doi: 10.1007/s00125-007-0753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Chen S., Burtscher I., Sterr M., Hieronimus A., Machicao F. Generation of a human induced pluripotent stem cell (iPSC) line from a patient carrying a P33T mutation in the PDX1 gene. Stem Cell Research. 2016;17(2):273–276. doi: 10.1016/j.scr.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Wang X., Chen S., Burtscher I., Sterr M., Hieronimus A., Machicao F. Generation of a human induced pluripotent stem cell (iPSC) line from a patient with family history of diabetes carrying a C18R mutation in the PDX1 gene. Stem Cell Research. 2016;17(2):292–295. doi: 10.1016/j.scr.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Li W., Wang X., Fan W., Zhao P., Chan Y.C., Chen S. Modeling abnormal early development with induced pluripotent stem cells from aneuploid syndromes. Human Molecular Genetics. 2012;21(1):32–45. doi: 10.1093/hmg/ddr435. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y., Mahony S., Gifford D.K. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Computational Biology. 2012;8(8) doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xing H., Mo Y., Liao W., Zhang M.Q. Genome-wide localization of protein-DNA binding and histone modification by a bayesian change-point method with ChIP-seq data. PLoS Computational Biology. 2012;8(7) doi: 10.1371/journal.pcbi.1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for Macrophage and B Cell identities. Molecular Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.