Abstract
Objective
Regulation of fatty acid (FA) metabolism is central to adipocyte dysfunction during diet-induced obesity (DIO). Long-chain acyl-CoA synthetase-4 (ACSL4) has been hypothesized to modulate the metabolic fates of polyunsaturated FA (PUFA), including arachidonic acid (AA), but the in vivo actions of ACSL4 are unknown. The purpose of our studies was to determine the in vivo role of adipocyte ACSL4 in regulating obesity-associated adipocyte dysfunction.
Methods
We developed a novel mouse model with adipocyte-specific ablation of ACSL4 (Ad-KO) using loxP Cre recombinase technology. Metabolic phenotyping of Ad-KO mice relative to their floxed littermates (ACSL4floxed) was performed, including body weight and body composition over time; insulin and glucose tolerance tests; and energy expenditure, activity, and food intake in metabolic cages. Adipocytes were isolated for ex vivo adipocyte oxygen consumption by Clark electrode and lipidomics analysis. In vitro adipocyte analysis including oxygen consumption by Seahorse and real-time PCR analysis were performed to confirm our in vivo findings.
Results
Ad-KO mice were protected against DIO, adipocyte death, and metabolic dysfunction. Adipocytes from Ad-KO mice fed high-fat diet (HFD) had reduced incorporation of AA into phospholipids (PL), free AA, and levels of the AA lipid peroxidation product 4-hydroxynonenal (4-HNE). Additionally, adipocytes from Ad-KO mice fed HFD had reduced p53 activation and increased adipocyte oxygen consumption (OCR), which we demonstrated are direct effects of 4-HNE on adipocytes in vitro.
Conclusion
These studies are the first to elucidate ACSL4's in vivo actions to regulate the incorporation of AA into PL and downstream effects on DIO-associated adipocyte dysfunction. By reducing the incorporation of AA into PL and free fatty acid pools in adipocytes, Ad-KO mice were significantly protected against HFD-induced increases in adipose and liver fat accumulation, adipocyte death, gonadal white adipose tissue (gWAT) inflammation, and insulin resistance (IR). Additionally, deficiency of adipocyte ACSL4 expression in mice fed a HFD resulted in increased gWAT adipocyte OCR and whole body energy expenditure (EE).
Keywords: Adipocytes, Fatty acid metabolism, Obesity, Arachidonic acid, Polyunsaturated fatty acid
Highlights
-
•
ACSL4 expression is upregulated in murine white adipocytes during diet-induced obesity.
-
•
Mice with adipocyte-specific ablation of ACSL4 (Ad-KO) are protected against diet-induced obesity, adipocyte death and metabolic dysfunction.
-
•
Lipidomics profiling of isolated adipocytes from Ad-KO mice fed a high-fat diet (HFD) had reduced arachidonic acid (AA) in phospholipids.
-
•
Adipocytes from Ad-KO mice fed HFD had reduced free AA and levels of the AA lipid peroxidation product 4-hydroxynonenal (4-HNE).
-
•
Adipocytes from Ad-KO mice fed HFD had reduced p53 activation and increased adipocyte oxygen consumption (OCR).
-
•
P53 activation and inhibited adipocyte OCR are direct effects of 4-HNE on adipocytes in vitro.
1. Introduction
The prevalence of obesity, a condition which predisposes to the development of type 2 diabetes mellitus (T2DM), in the United States is approximately 35% in the adult population [1]. While the vast majority of obese individuals are afflicted with metabolic complications, not all such individuals develop these health problems [2]. These observations raise the fundamentally important question of understanding why weight gain causes metabolic dysregulation in some individuals but not others [3], [4]. One theory to explain the phenomenon of “healthy obesity” links metabolic resilience to optimal adipocyte function. To ameliorate the remarkable prevalence of obesity-associated metabolic complications, it is necessary to develop a greater understanding of the pathways that lead to obesity-associated adipocyte dysfunction. While fatty acid (FA) metabolism is central to the development of enlarged adipocytes in obesity, we have limited knowledge of the proteins that direct the incorporation of specific FA into phospholipids (PL) and neutral lipids, like triacylglycerol (TAG), as well as proteins that convert FA into a variety of bioactive lipid mediators. Polyunsaturated FA (PUFA) have multiple fates, including incorporation into PL, TAG, and cholesterol ester, and oxygenation by enzymatic (cyclooxygenase, lipoxygenase, cytochrome P450) and non-enzymatic mechanisms, specifically peroxidation of lipids by reactive oxygen species (ROS) to form aldehydes, such as 4-hydroxynonenal (4-HNE), that can detrimentally alter cellular metabolism [5], [6].
The long-chain acyl-CoA synthetase (ACSL) family of enzymes catalyze the addition of a coenzyme-A (CoA) group to a FA to form fatty acyl-CoAs, effectively “trapping” FAs within cells [7], [8]. Five ACSL isoforms can each activate and channel various FAs to different metabolic fates [7]. ACSL1 is the most abundant isoform expressed in white adipose tissue (WAT) and accounts for 80% of total ACSL activity in WAT [9], [10]. ACSL4 is expressed at a significantly lower level in adipocytes compared to ACSL1, but its role in adipocyte metabolism and obesity is unknown. Interestingly, ACSL4 is the only ACSL isoform whose enzymatic activity is directly inhibited by the class of anti-diabetic drugs thiazolidinediones (TZDs) through PPAR-γ-independent mechanisms, suggesting ACSL4's possible involvement in regulation of metabolism [11], [12]. Additionally, ACSL4 has a significantly greater in vitro preference for arachidonic acid (AA) and eicosapentanoic acid (EPA) as compared to other long-chain saturated and unsaturated FA, with a lower preference for stearic, palmitic, and linoleic acid (LA) as well [13]. Proposed functions of ACSL4 include intracellular lipid storage [14], cholesterol transport from the endoplasmic reticulum into the mitochondria [15], and regulation of AA and its metabolites [13], [16], [17], [18]. More recently, ACSL4 was demonstrated to be essential for the increased rates of lipid peroxidation necessary for ferroptosis, a specific mode of iron-dependent, non-apoptotic cell death [19], [20], [21].
The purpose of our studies was to determine the role of ACSL4 in regulating obesity-associated adipocyte dysfunction. To fulfill this objective, we used LoxP-Cre technology to generate a novel mouse model that specifically ablated ACSL4 in adipocytes in order to characterize in vivo mechanisms of ACSL4's effects in adipocytes during diet-induced obesity (DIO). This work presents the first in vivo elucidation of ACSL4's actions. In the context of DIO, adipocyte ACSL4 regulates adiposity, obesity-associated inflammation and metabolic complications, whole-body energy expenditure (EE), and isolated adipocyte oxygen consumption rate (OCR). Here we demonstrate that with DIO, gWAT adipocyte ACSL4 modulates AA in the free fatty acid (FFA) pool and incorporation into PL and promotes the production of the lipid peroxidation product 4-HNE.
2. Materials and methods
2.1. Generation of mice that lack ACSL4 specifically in adipocytes
Adipocyte-specific ACSL4 knockout mice (Ad-KO) were created using LoxP-Cre technology [22]. The gene-targeting vector was designed to produce a floxed ACSL4 gene on the X chromosome with loxP sites on either side of exons 3–4 at the University of North Carolina Animal Models Core using previously described methods [22]. The vector was constructed in a standard plasmid backbone containing neomycin phosphotransferase (neo) and thymidine kinase cassettes for positive and negative selection, respectively [22]. The targeting vector was electroporated into E14Tg2A (E14) embryonic stem cells, and the cells were grown in medium supplemented with G418 and ganciclovir [22]. Targeted cells were identified by PCR across both the 3′ and 5′ arms of homology using primers specific to neo and primers flanking the arms of homology. Targeted, neomycin-resistant cells were microinjected into blastocysts derived from mouse strain 129SJ to produce transmitting chimeras. Neo was then excised from the targeted allele by mating to mice expressing FLPo recombinase [23]. Mice homozygous for the targeted allele (Acsl4flox/Y) were identified by genotyping PCR and back-crossed six times to C57BL/6 mice. Heterozygous Acsl4flox/+ female and floxed male Acsl4flox/Y mice were sent to the Jean Mayer-U.S. Department of Agriculture Human Nutrition Research Center on Aging at Tufts University. DNA from these mice was sent to Charles River for C57BL/6J congenic analysis and the female and male with the greatest percentage of C57BL/6J microsatellite analysis (>99%) were mated together. From the progeny of this breeding pair, floxed male mice (Acsl4flox/Y) were then crossed with female C57BL/6 mice (Acsl4+/+) in which Cre expression is driven by the Adiponectin promoter purchased from Jackson Laboratories [B6; FVB-Tg (Adipoq-cre)1Evdr/J; Jackson Laboratories, Bar Harbor, ME] [24]. The next generation of progeny were produced by mating Acsl4flox/+/Cre + females to Acsl4flox/Y/Cre-males, and then subsequent matings to generate experimental mice were either between Acsl4flox/Y/Cre-males (ACSL4floxed) & Acsl4flox/flox/Cre + females (Ad-KO) or Acsl4flox/Y/Cre + males (Ad-KO) & Acsl4flox/flox/Cre-females (ACSL4floxed), which consistently yields equal numbers of males and females with (Ad-KO) and without Adipoq-Cre (ACSL4floxed).
2.2. Animal care and in vivo analysis
Experiments were conducted in a viral pathogen-free facility at the Jean Mayer U.S. Department of Agriculture Human Nutrition Research Center on Aging at Tufts University in accordance with Institutional Animal Care and Use Committees guidelines. All experimental Ad-KO and littermate ACSL4floxed mice were male. Mice were weaned at 3–4 weeks of age and fed ad libitum a purified standard chow diet (Teklad 2016S). Mice were individually caged at 6–7 weeks of age. At 8 weeks old, mouse body weight and body composition were measured by magnetic resonance (EchoMRI-700) and then mice were randomized to receive either a high-fat diet (HFD) containing 60% kcal from fat or a low-fat diet (LFD) containing 10% kcal from fat (Research Diets D12492 and D12450J, respectively) ad libitum for 12 weeks. Previous work has determined that 12 weeks on 60% HFD is the optimal amount of time for the greatest increase in adipocyte size, macrophage infiltration into WAT, and expression of pro-inflammatory cytokines without an increase in dead adipocytes that occurs with 16 weeks on 60% HFD [25], [26]. Body weight and body composition were measured every 2–4 weeks as indicated.
Mice were randomly selected for indirect calorimetry (TSE Systems) in metabolic cages to evaluate oxygen consumption, carbon dioxide production, food intake, and activity by infrared counts of all animal movement at 3 weeks of HFD feeding (11 weeks old) and at 7 weeks of HFD feeding (15 weeks old). Body composition by MRI was measured 3 days before mice were placed in metabolic cages for 96 h, and the first 24 h of measurement were excluded from data analysis while the mice acclimated. The resulting EE values and RER were calculated from these measurements. These experiments were conducted in two runs of mice in subsequent 5-day increments and data is adjusted for run (n = 7 per run; 3–4 mice per genotype per run).
At 9 weeks on diet (17 weeks old), a glucose tolerance test (GTT) was performed. 1 mg/kg body weight was injected intraperitoneally after a 6-hour morning fast, blood glucose was measured from the tail vein at 4 time points by glucometer, and a minimal amount of blood was collected at each time point for subsequent serum insulin analysis. At 11 weeks on diet (19 weeks old), an insulin tolerance test (ITT) was performed. 0.75 units of insulin/kg body weight was injected intraperitoneally after a 6-hour morning fast, and blood glucose was measured from the tail vein at six time points by glucometer.
Mice were sacrificed under Isoflurane by cardiac puncture at 20 weeks old (12 weeks on diet) and blood was collected by cardiac puncture, tissues were dissected, weighed, and snap frozen for analysis; or were fixed in zinc formalin, embedded in paraffin, and sectioned for histological analysis; or gWAT was used to isolate adipocytes as described below.
2.3. Serum biochemistry
At the time of sacrifice after a 6-hour morning fast, whole blood glucose measurements were made by glucometer (One Touch Ultra Blue, LifeScan, Inc). Serum from blood obtained at necropsy was removed after of low speed centrifugation. Insulin was determined by ELISA (Ultra-sensitive mouse insulin, Crystal Chem, Inc), and FFA was determined using a colormetric kit (Wako Diagnostics). Total TAG (glycerol phosphate oxidase, Beckman Coulter OSR6133) and total cholesterol (Aminoantipyrine/Phenol/Peroxidase, Beckman Coulter OSR6116) by enzymatic colorimetric endpoint assay (Beckman Coulter AU400).
2.4. Adipocyte isolation
Gonadal adipose fat pads were dissected from the mouse, weighed, and immediately submerged in Krebs-Ringer bicarbonate (KRB) buffer at pH 7.4 with 4% bovine serum albumin (BSA), 500 nM adenosine, and 5 mM glucose, and primary adipocytes were isolated by collagenase digestion (1 mg/mL KRB) as previously described [27], [28]. For mice fed LFD, fat pads from 2 to 3 mice per genotype were pooled to obtain enough cells for subsequent experiments. Cells were filtered through a 250-μm nylon mesh and washed. Adipocytes were washed an additional two times in KRB, then used for subsequent ex vivo experiments or added to a tube with silicon oil and centrifuged at 10,000g for 2 min, which separates KRB (bottom) from the adipocytes (top), and adipocytes were removed and frozen at −80 °C in Triazol for subsequent RNA analysis, in 10% SDS for western Blot, or in Med1 buffer for ACSL activity assay (described below). Fat cell number was determined by the method described by Goldrick where average cell diameter, determined using ImageJ software (NIH), is used to calculate fat cell volume, which can then be multiplied by triglyceride density to calculate the average fat cell weight [29].Then, total neutral lipid was measured as previously described [30]. Finally, fat cell number was calculated by the total lipid content divided by the average fat cell weight [29].
2.5. Ex vivo isolated adipocyte oxygen consumption
Isolated adipocyte OCR was measured using a Clark-type electrode digital model 10 controller with a constant stir bar (Rank Brothers) and was water jacketed to maintain adipocytes at 37 °C, and data was recorded by a PowerLab 4/30 data acquisition system (AD Instruments) as previously described with modifications [31], [32], [33]. The electrode was pre-calibrated with oxygenated water at 37 °C and then with 500 μL oxygenated KRB without cells to detect electrode drift. After the third wash with KRB as described above, isolated adipocytes were diluted 1:3 in KRB, and a 500 μL aliquot of adipocytes were transferred to the electrode chamber and air sealed. Basal respiration was recorded for 15 min and then NaCN was injected for a final concentration of 1 mM and measured for 10 min to inhibit oxygen consumption. Results were calculated based on the assumption that oxygenated water contains 225 μmol/l O2, and data were corrected for electrode drift and normalized for fat cell number calculated as described above. Isolated adipocytes from one ACSL4floxed and one Ad-KO littermate were isolated after 12 weeks on diet and oxygen consumption measured per day to minimize variability for a total of 4 subsequent days (n = 4 mice/genotype).
2.6. Adipose tissue Acyl-CoAs extraction and quantification
Acyl-CoAs were extracted and analyzed using a modified version of a previously described method [34]. Briefly, gWAT was ground to a homogenous powder using a liquid nitrogen-chilled mortar and pestle. Approximately 30–40 mg (wet mass) of each pulverized tissue sample was rapidly transferred to a pre-weighed chilled Eppendorf tube before the addition of extraction solvent and the internal standard. For acyl-CoAs, 1 ml of extraction solvent was used; solvent was composed of 1:1 of 100 mM KH2PO4 and organic mixture (3:1:1 ACN, IPA: Methanol). The internal standard was 20 ng/ml of C17-CoA. The samples were sonicated on ice, followed by centrifugation at 4 °C. Clean extract was recovered and dried under a gentle stream of nitrogen. Dried sample was reconstituted with cold 1:1 (methanol: water) and 10 μl was injected on the LC-MS. Chromatographic separations were performed with an Agilent Technologies (Santa Clara, CA) 1200 HPLC system equipped with a Waters (MA, USA) Acquity UPLC® BEH C18 column (2.1 × 100 mm, 1.7 μm) and a 2.1 × 5 mm VanGuard™ pre-column using the following conditions: mobile phase A was 10% acetonitrile, 90% MS water, mobile phase B was acetonitrile. Both mobile phases contained 15 mM ammonium hydroxide. The gradient program for LC-CoA started at 0% B which was held for 3 min while LC eluent was directed to the waste during this period. %B was ramped linearly to 30% B over 3 min, then ramped to 40% B over 6 more minutes, then held at 100% B for 4 min, after which the column was reconditioned using 100% A for 6 min. The total run time was 22 min and the flow rate was 0.25 ml/min. Metabolites were quantified by measuring the metabolite peak area as a ratio to its 13C internal standard analog and was normalized to the tissue weight.
2.7. Isolated adipocyte lipidomics
Adipocytes were isolated as described above with two washes with KRB containing BSA and the final wash with KRB without BSA. Adipocytes with buffer were added to a tube with silicon oil and centrifuged at 10,000g for 2 min, then adipocytes were removed minimalizing amount of silicon oil in the sample, and frozen in liquid nitrogen and stored at −80 °C. Shotgun lipidomics analysis of isolated adipocytes was performed at the Center for Metabolic Origins of Disease at Sanford Burnham Presbyterian Medical Discovery Institute. The adipocyte samples were added to 200 μL of PBS in an Eppendorf tube and were homogenized for 1 min by using a disposable soft tissue homogenizer. An aliquot of 6 μL was pipetted to determine the protein content (BCA protein assay kit, Thermo Scientific, Rockford, IL). The rest of homogenate was accurately transferred into a disposable glass culture test tube, and a mixture of lipid internal standards was added prior to lipid extraction for quantification of all reported lipid species. Lipid extraction was performed by using a modified Bligh and Dyer procedure as described previously [35]. The aqueous phase solution was loaded onto a HybridSPE cartridge to recover and enrich lyso phospholipids [36], and the chloroform phase was evaporated under nitrogen. Both of the extract from aqueous and organic phases were re-suspended into a volume of 500 μL of chloroform/methanol (1:1, v/v) per mg of protein and flushed with nitrogen, capped, and stored at −20 °C for lipid analysis. Part of the extract solution from organic phase was separately derivatized to determine 4-hydroxyalkenal species [37], cholesterols [38], diacylglycerol species [39], and non-esterified fatty acids [40]. For ESI direct infusion analysis, lipid extract or derivatives were further diluted to a final concentration of ∼500 fmol/μL, and the mass spectrometric analysis was performed on a QqQ mass spectrometer (Thermo TSQ QUANTIVA, San Jose, CA) equipped with an automated nanospray device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, NY). Identification and quantification of lipid molecular species were performed using an automated software program [35], [41]. Data were normalized per mg of protein.
2.8. ACSL enzyme activity assay
ACSL enzyme specific activity was measured as previously described [10] in isolated adipocytes homogenized in Medium 1 (Med1) buffer (10 mM Tris pH 7.4, 1 mM EDTA, 0.25 M sucrose, 1 mM DTT), and centrifuged at 40,000rpm for 1 h at 4 °C to pellet membranes, which were re-suspended in 250 μL Med1 buffer. BCA assay performed to determine protein content. ACSL enzyme activity assay performed with 50 μM [1–14C] palmitic acid, 10 mM ATP, and 0.25 mM CoA with total membrane fractions (1–4 μg protein) [10]. Data were normalized per micrograms of protein determined by BCA assay.
2.9. Liver triacylglycerol and cholesterol
Liver TAG and cholesterol were determined by modified Folch procedure [42], [43], [44]. Snap frozen, wet liver was homogenized in 2:1 chloroform:methanol with overnight incubation and subsequent extraction with magnesium chloride; the organic layer was evaporated under nitrogen at 37 °C to complete dryness and saponified by incubating with ethanolic potassium hydroxide solution at 60 °C for 1 h and adding magnesium sulfate. For TAG, the resultant supernatant was evaluated for liberated glycerol (Sigma). For cholesterol, the supernatant was evaluated by enzymatic, colormetric kit (Inifinity Cholesterol Liquid Stable Reagent, Thermo Scientific). Measurements were made in triplicate per sample and data was normalized by protein content determined by BCA assay.
2.10. Real-time PCR
Adipose tissue and liver RNA was extracted using Qiagen RNEasy Lipid Mini kits, and isolated adipocyte RNA was extracted using Qiagen RNEasy Plus Micro kit according to manufacturer's instructions. RNA was quantified and checked for purity using the Nanodrop spectrophotometer (Nanodrop 1000, Wilmington, DE). cDNA was generated from equal amounts of RNA, and real-time quantitative PCR was performed using SYBR Green (Applied Biosystems 7300, Carlsbad, CA). Fold-changes were calculated as 2–ΔΔCT, with Cyclophilin A used as the endogenous control. Primer sequences are listed in Supplementary Table 1.
2.11. Western blot
Isolated adipocyte protein was prepared as previously described where previously frozen isolated adipocytes in 10% SDS were homogenized in equal amount of lysis buffer without SDS 0.1 mM Na3VO4, 50 mM HEPES, 2 mM EDTA, 50 mM NaF, 1 mM benzamidine, 5 μL/mL Protease Inhibitor (Sigma), ph 7.4, plus red blood cell lysis buffer (Sigma), then incubated for 1 h at 37 °C with vortexing, centrifuged at 14,000g for 10 min at room temperature, and the internatent containing protein between the fat cake and the pellet was removed for subsequent analysis [45]. Adipose tissue and liver were homogenized in RIPA buffer with protease (Halt) and phosphatase (PhoStop) inhibitors at 100 mg/100 μL and 10 mg/100 μL, respectively. Homogenates were centrifuged at 12,400g for 10 min at room temperature, fat free homogenate removed, and centrifugation repeated to ensure all fat is removed from sample.
Equal amounts of protein (determined by BCA assay) of the protein lysates were solubilized and denatured by boiling for 5 min in 1X SDS solution with 2-mercaptoethanol, which were resolved by loading into wells of 7.5% polyacrylamide gel (Mini Protean TGX, BioRad) and run with 1X tris-glycine-SDS buffer, transferred to nitrocellulose membrane, and blocked with 5% bovine serum albumin in tris-buffered saline with 0.2% tween overnight at 4 °C. For demonstration of the presence or lack of ACSL4 protein content, an affinity purified antibody generously given to us by Dr. Diana Stafforni (University of Utah) previously described [46] (1:500 dilution for adipocytes; 1:2,000 for adipose tissue and liver) for 1.5 h at room temperature. The secondary antibody, donkey anti-rabbit horseradish peroxidase (GE Healthcare UK) was diluted 1:10,000 prior to 1 h membrane incubation at room temperature, detected by chemiluminescence (Supersignal West Pico, Fisher), and images captured with a sensitive camera (Fluorchem Q, Alpha Innotech). Blots were then stripped (Restore Western Blot Stripping Buffer, Thermo Scientific), blocked overnight, and reprobed for normalization with 1:1,000 dilution of GAPDH or Actin antibody raised in rabbit for 1 h at room temperature (Cell Signaling). P53 (Cell Signaling) and UCP1 (AbCam) were detected with a purchased antibody per manufacturer's instructions.
2.12. Immunohistochemistry
gWAT was fixed and stained for Mac-2 as previously described [26]. Liver was fixed and stained for hematoxylin and eosin (H&E) as previously described [47].
2.13. In vitro experiments
Cell culture: 3T3-L1 pre-adipocytes were seeded onto tissue culture plates in 10% fetal bovine serum (FBS) in DMEM high glucose and no phenol red (basal medium) and allowed to become confluent. Following confluence, cells were fed basal medium for an addition 48 h prior to starting differentiation. Differentiation medium consisted of dexamethasone (1 uM), rosiglitazone (1 uM), 3-isobutyl-1-methylxanthine (IBMX) (500 uM), and insulin (10 ug/mL) in the basal medium. Cells were fed differentiation medium for 2 days, then given basal media supplemented with 10 ug/mL insulin for an addition 2 days. Cells were fed new basal medium every two days for a total of 8 days post differentiation.
Seahorse metabolic measurements: 3T3-L1 cells were grown and differentiated in the 96-well culture plate provided with the Seahorse XFe96 flux pack. After 8 days differentiation, the cells were incubated with a modified Seahorse assay medium (MgSO4 0.8 mM, CaCl2 1.8 mM, NaCl 143 mM, KCl 5.4 mM, NaH2PO4, 0.91 mM, l-Glutamine 2 mM, glucose 25 mM, pyruvate 1 mM, without any other amino acids or supplements) for 1 h at 37 °C in the absence of CO2 prior to the Seahorse assay. 4-HNE was diluted to the various concentrations in phosphate buffered saline (PBS) and loaded into the Seahorse cartridge along with oligomycin (1 uM), FCCP (1 uM), Antimycin A and rotenone (15 uM) (Sigma Aldrich). Measurement of oxygen consumption rate (OCR) were recorded with a XF96 Seahorse (Agilent).
2.14. Statistical analysis
Data were analyzed using SAS 9.3 (Cary, NC) by two-way ANOVA with Tukey's HSD for multiple comparisons (where appropriate) or repeated measures ANOVA for time series data, unless otherwise indicated. Gene expression data were analyzed by Student's t-test with a Bonferroni adjustment for the appropriate number of comparisons. Data were expressed as mean ± standard error of the mean. Differences at values of p ≤ 0.05 were considered significant.
3. Results
3.1. Generation of mice that lack ACSL4 specifically in adipocytes
Mice with loxP sequences inserted in introns flanking exons 3 and 4 in the ACSL4 gene were mated to mice expressing a Cre transgene driven by the Adiponectin promoter [B6; FVB-Tg (Adipoq-cre)1Evdr/J; Jackson Laboratories, Bar Harbor, ME] [24] to generate adipocyte-specific Acsl4 knockout mice (Ad-KO) [Supplementary Fig. 1]. Littermates without the Cre transgene (ACSL4floxed) were used as controls. All experiments were conducted using male mice.
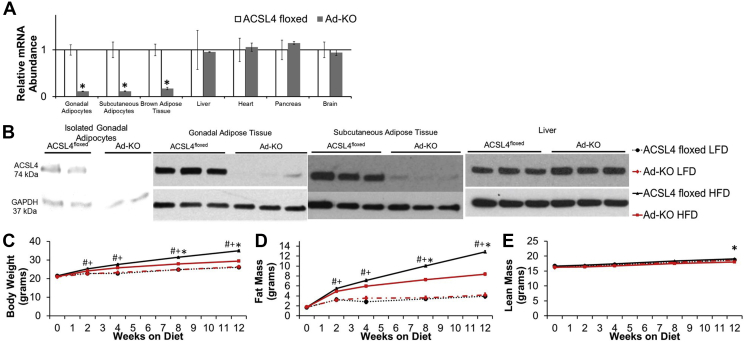
Adipocyte deficiency of ACSL4 was confirmed by dramatically reduced ACSL4 mRNA and protein expression in adipocytes isolated from gWAT, whole gWAT subcutaneous adipose tissue (scWAT), and BAT of Ad-KO mice as compared to ACSL4floxed littermates. [Figure 1A,B; Supplementary Figs. 2b and c, Supplementary Fig. 5b]. Consistent with the tissue specificity of the Adipoq-Cre transgene, no differences in ACSL4 mRNA expression were noted in several non-adipose tissues nor protein expression in the liver of the two lines of mice [Figure 1A,B].
Figure 1.
Mice that lack ACSL4 specifically in adipocytes are protected against high fat diet-induced obesity. (A) ACSL4 RNA expression quantified by real-time PCR relative to Cyclophilin A in isolated adipocytes and metabolically relevant tissues in 20 week old males fed LFD. n = 3–7 mice per group; *p ≤ 0.05 ACSL4floxed vs. Ad-KO. Student's t-test. (B) Representative immunoblot against ACSL4 in isolated gonadal adipocytes, whole gonadal adipose tissue, whole subcutaneous adipose tissue, and liver of 20-week-old males. (C) Body weight, (D) fat mass, and (E) lean mass from 8 to 20 weeks of age in male mice fed LFD or HFD; n = 11–12 mice/group LFD, 28–31 mice/group HFD; #p ≤ 0.05 ACSL4floxed LFD vs. HFD, +p ≤ 0.05 Ad-KO LFD vs. Ad-KO HFD, and *p ≤ 0.05 ACSL4floxed HFD vs. Ad-KO HFD; Repeated measures ANOVA with Tukey's HSD for multiple comparisons. Data represent ± SEM.
3.2. Ad-KO mice were protected against diet-induced obesity
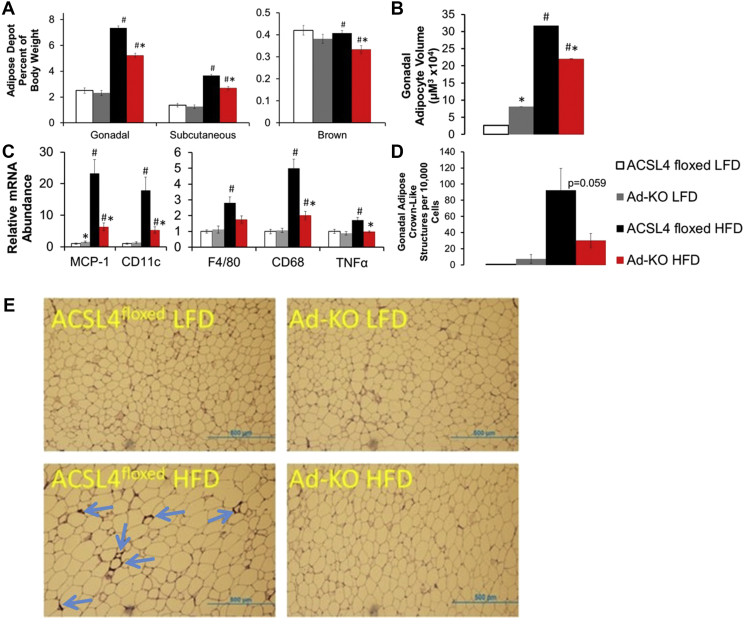
To determine the role of adipocyte ACSL4 expression in DIO, both lines of mice were fed a LFD or a HFD for 12 weeks. On LFD, no differences in body composition, fat pad weights, serum insulin, TAG, free FA (FFA), or hepatic TAG levels were observed between the two lines of mice [Figure 1, Figure 2A, and Table 1]. However, when fed a HFD, Ad-KO mice gained 16% less body weight compared to controls [Figure 1C]. MRI measurements of body fat demonstrated that Ad-KO mice fed a HFD, compared to ACSL4floxed littermates fed a HFD, had a significant 35% reduction in body fat with only a minimal change in lean mass after 12 weeks of HFD feeding [Figure 1D,E]. Consistent with the body composition observations on a HFD, at sacrifice, the weights of scWAT and gWAT of Ad-KO mice fed HFD were reduced by ∼45%, and gWAT adipocyte size was decreased [Figure 2A,B]. BAT weight was also decreased 35% compared to controls fed HFD [Figure 2A]. In mice fed HFD, Ad-KO mice had reduced liver weight and accumulated significantly less liver TAG, but not cholesterol [Table 1]. Serum cholesterol, but not TAG or FFA, was significantly reduced in Ad-KO mice compared to controls fed a HFD [Table 1].
Figure 2.
Ad-KO mice fed HFD are protected against markers of adipocyte inflammation and cell death. (A) Adipose tissue depot weight determined at time of sacrifice after 12 weeks on diet. n = 11–12 mice/group LFD, 28–31 mice/group. *p ≤ 0.05 ACSL4floxed HFD vs. Ad-KO within diet; #p ≤ 0.05 LFD vs. HFD within genotype; Two-way ANOVA with Tukey's HSD for multiple comparisons. (B) Isolated average gonadal adipocyte volume at time of sacrifice after 12 weeks on diet. n = 6–12 mice/group; *p ≤ 0.0001 ACSL4floxed HFD vs. Ad-KO within diet; #p ≤ 0.0001 LFD vs. HFD within genotype; Student's t-test after log transformation. (C) Gonadal adipose tissue inflammatory genes of interest mRNA expression quantified by real-time PCR relative to Cyclophilin A. n = 7–8 mice per group. *p < 0.0125 FL vs. KO within diet; #p < 0.0125 LFD vs. HFD within genotype. Student's t-test with Bonferroni correction for 4 comparisons. (D) Crown-like structure (arrows) abundance determined by (E) MAC-2 staining in gonadal adipose tissue of 20 week old male mice; n = 3–4 mice/group LFD 12–15 mice/group HFD. #p ≤ 0.05 LFD vs. HFD within genotype. Student's t-test. Data represent ± SEM.
Table 1.
Ad-KO mice fed HFD are protected against markers of metabolic syndrome and nonalcoholic steatohepatitis
| Low-fat diet |
High-fat diet |
|||
|---|---|---|---|---|
| ACSL4floxed | Ad-KO | ACSL4floxed | Ad-KO | |
| Serum TAG (mg/dL) | 73.1 ± 3.5 | 72.4 ± 3.7 | 67.7 ± 2.9 | 69.3 ± 3.2 |
| Serum cholesterol (mg/dL) | 114.4 ± 5.6 | 111.0 ± 5.6 | 184.5 ± 4.3b | 151.3 ± 4.9b,a |
| Serum FFA (μmol/L) | 360.3 ± 18.0 | 337.3 ± 19.0 | 376.1 ± 14.7 | 337.2 ± 16.5 |
| Serum glucose (mg/dL) | 169.7 ± 12.3 | 196.0 ± 11.1 | 206.20 ± 17.4b | 194.6 ± 18.6 |
| Serum insulin (ng/mL) | 0.41 ± 0.052 | 0.39 ± 0.049 | 1.84 ± 0.15b | 0.93 ± 0.13b,a |
| Liver weight (grams) | 0.934 ± 0.034 | 0.920 ± 0.034 | 1.066 ± 0.021b | 0.870 ± 0.026b,a |
| Liver TAG (mg/mg protein) | 0.194 ± 0.0469 | 0.134 ± 0.0469 | 0.592 ± 0.0524b | 0.350 ± 0.0469b,a |
| Liver cholesterol (μg/mg protein) | 35.17 ± 5.56 | 37.06 ± 5.56 | 34.02 ± 6.21 | 45.62 ± 5.56 |
TAG, cholesterol, FFA, and glucose n = 9–15 mice/group; Insulin 5-10 mice/group; Liver weight n = 11–12 mice/group LFD, 26–31 mice/group HFD; Liver TAG and cholesterol n = 8 mice/group.
p ≤ 0.05 ACSL4floxed HFD vs. Ad-KO within diet.
p ≤ 0.05 LFD vs. HFD within genotype. Two-way ANOVA with Tukey's HSD for multiple comparisons.
3.3. Ad-KO mice fed HFD were protected against markers of gWAT inflammation, adipocyte death, and insulin resistance
We next determined whether ablation of adipocyte ACSL4 protected against the development of obesity-associated WAT inflammation. Real-time PCR analysis of gWAT from ACSL4floxed and Ad-KO mice fed HFD demonstrated that gWAT from Ad-KO mice had significantly decreased mRNA abundance of macrophage chemotactic factor (MCP-1) and macrophage markers F4/80 and CD68, consistent with reduced macrophage content, and CD11c, a marker for dendritic cells which has been linked to insulin resistance (IR) [48] [Figure 2C]. Additionally, we observed protection against HFD-induced expression of the cytokine tumor necrosis factor-α (TNF-α) in gWAT from Ad-KO mice fed HFD [Figure 2C]. Our laboratory has previously documented that increased rates of adipocyte death and the formation of crown-like structures (CLS) correlate with gWAT inflammation, systemic IR, and liver fat content [25], [26]. Ad-KO gWAT from mice fed HFD had 67% fewer CLS compared to gWAT from ACSL4floxed mice fed HFD (p = 0.059) [Figure 2D,E].
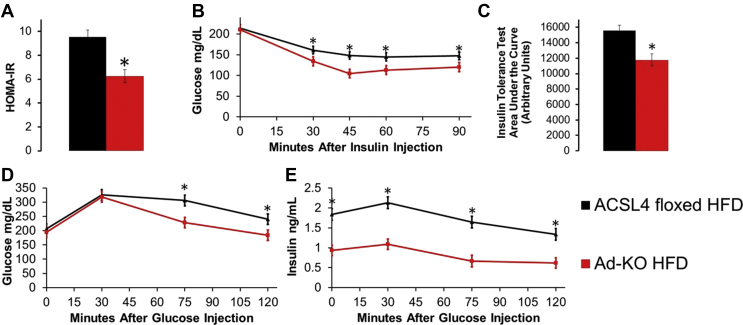
We next investigated measures of glucose and insulin homeostasis. Fasting glucose did not differ between the two lines of mice fed a HFD, but Ad-KO mice fed HFD compared to ACSL4floxed littermates fed HFD had a significant 49% reduction in fasting serum insulin levels [Table 1] and 34% lower homeostatic model assessment to determine insulin resistance (HOMA-IR) [Figure 3A]. Additionally, Ad-KO mice had improved insulin sensitivity as determined by reduced area under the curve for insulin tolerance tests (ITT) compared to ACSL4floxed littermates fed HFD [Figure 3B,C]. During a glucose tolerance test (GTT), Ad-KO mice fed HFD also exhibited significantly reduced blood glucose levels at 75 and 120 min and dramatically lower serum insulin at every time point compared to ACSL4floxed littermates fed HFD [Figure 3D,E]. In summary, consistent with reduced adiposity, gWAT inflammation, and adipocyte death, Ad-KO mice fed a HFD, as compared to ACSL4floxed littermates, were significantly protected against IR [Figure 3].
Figure 3.
Ad-KO mice fed HFD are protected against insulin resistance. (A) Homeostatic model assessment to quantify insulin resistance (HOMA-IR) from serum at time of sacrifice after 12 weeks of HFD feeding; n = 14–15 mice/group. (B) Insulin tolerance test (ITT) and (C) ITT area under the curve after 11 weeks of HFD feeding; n = 16–19 mice/group. Glucose tolerance test (GTT) (D) blood glucose over time and (E) serum insulin over time after 9 weeks of HFD feeding; n = 7–8 mice/group glucose; 5–6 mice/group insulin. *p < 0.05 ACSL4floxed HFD vs. Ad-KO HFD; Repeated measures ANOVA ITT and GTT, Student's t-test HOMA-IR and ITT AUC. Data represent ± SEM.
3.4. Ad-KO mice fed HFD had higher whole-body EE and isolated adipocyte oxygen consumption
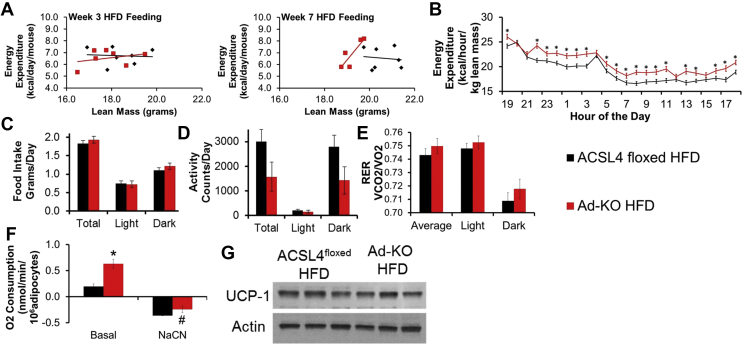
As a first step to determine the mechanisms by which Ad-KO mice were protected against DIO, EE was determined before changes in lean mass occurred, which does not change until week 9–12 of HFD feeding [Figure 1E]. Notably, after 3 weeks of HFD feeding, differences were not observed in metabolic rates between ACSL4floxed and Ad-KO littermates [Figure 4A]. However, after 7 weeks of HFD feeding, rates of EE were 10% higher in Ad-KO mice as compared to ACSL4floxed littermates [Figure 4A,B]. Additionally, using multiple linear regression methods previously described [49], [50], we found the most significantly predictive model after including genotype, body weight, fat mass, lean body mass, and experimental run order (described in methods) was energy expenditure (kcal/day/mouse) = −0.076–1.76(Run) + 0.69(Genotype) + 0.42 (Lean Mass) [model p < 0.0001; Run p < 0.0001, Genotype p = 0.06, lean mass p = 0.048]. These alterations in EE occurred in the absence of differences in food intake, activity, respiratory exchange ratio (RER), or lean mass at 7 weeks of HFD feeding [Figure 4C–E, Figure 1E].
Figure 4.
Ad-KO mice on HFD have higher whole-body energy expenditure and isolated adipocyte oxygen consumption without increased UCP1 expression. (A) Energy expenditure per mouse per day vs. lean mass measured at 3 weeks of HFD feeding (left) and 7 weeks of HFD feeding (right); n = 5–7 mice/group. (B) Energy expenditure per kg lean mass per at 7 weeks on HFD; n = 5–7 mice/group; *p ≤ 0.05 ACSL4floxed HFD vs. Ad-KO; Repeated measures ANOVA. (C) food intake, (D) activity, and (E) respiratory exchange ratio measured by TSE Metabolic Chambers for 72 h mass in 15 week-old male mice on HFD for 7 weeks; n = 5–7 mice/group. (F) Oxygen consumption in isolated gonadal adipocytes measured by Clark Electrode in 20 week-old male mice fed HFD for 12 weeks. Rate of basal respiration with 5 mM glucose was measured for 15 min and then 1 mM NaCN added to shut off respiration for 10 min; n = 4 mice/group; **p ≤ 0.01 ACSL4floxed HFD vs. Ad-KO HFD; Two-way ANOVA. (G) Representative immunoblot against UCP1 in gWAT from 20-week-old male mice fed HFD for 12 weeks. Data represent ± SEM.
We hypothesized that alterations in gonadal adipocyte oxygen consumption (OCR) could contribute to the differences in EE. To address this, gWAT adipocytes were isolated from mice, and we determined OCR using a Clark electrode. In isolated adipocytes from both ACSL4floxed and Ad-KO mice fed LFD, adipocyte OCR was very low (data not shown), but OCR was easily measured in adipocytes from HFD-fed mice. This is consistent with previous studies in mice and humans demonstrating that obesity is associated with increased white adipocyte OCR [51], [52]. Remarkably, isolated adipocytes from Ad-KO mice fed HFD had 3.2-fold higher rates of basal OCR in the presence of glucose compared to adipocytes from ACSL4floxed mice fed HFD [Figure 4F].
Previous studies have suggested that FA from adipocyte lipolysis can increase adipocyte OCR [52], [53]. Therefore, we measured rates of lipolysis and total adipocyte fatty acid content in ACSL4floxed and Ad-KO adipocytes, which did not statistically differ; thus, alterations in lipolysis or FFA did not contribute to differences in adipocyte OCR [Supplementary Figs. 4a and b]. Another possible contributor to increased adipocyte OCR is the conversion of white adipocytes to brite or “brown-like” adipocytes and associated increased uncoupling protein 1 (UCP1) expression [54]. However, western blot analysis of gWAT lysates probed for UCP1 revealed no differences in protein expression [Figure 4G], and analysis of gene expression demonstrated no significant differences in Ucp1 and Pgc1α RNA abundance in scWAT [Supplementary Fig. 2c] or BAT [Supplementary Fig. 5a]. Therefore, “browning” of white adipocytes was not a contributor to increased rates of basal OCR in adipocytes from Ad-KO mice fed HFD.
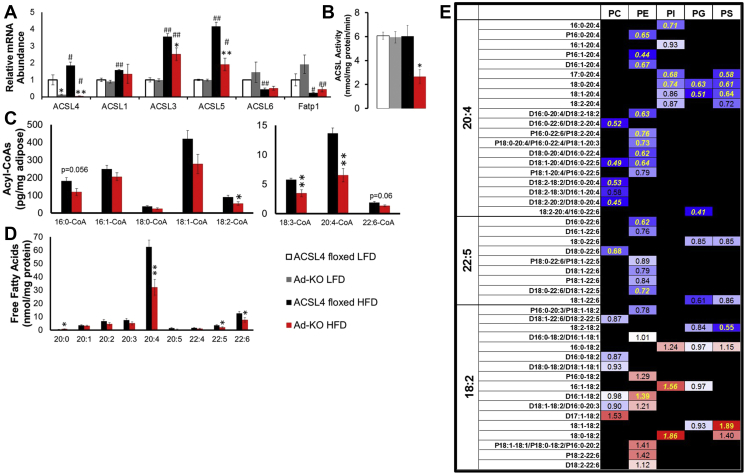
3.5. Ad-KO mice fed HFD have alterations in adipocyte ACSL activity, n-6 PUFA fatty acyl-CoA formation, and FFA and phospholipid composition
Next, we performed studies to elucidate the molecular mechanisms that protect Ad-KO mice against DIO in isolated adipocytes. First, we determined that there was no compensation by other ACSL isoforms due to ACSL4 knockout in adipocytes from mice fed LFD [Figure 5A]. We found that gWAT ACSL4 mRNA expression increased with ingestion of a HFD compared to adipocytes from LFD-fed mice [Figure 5A]. Notably, Ad-KO adipocytes were protected against HFD-induced increases in ACSL1, -3, and -5 gene expression [Figure 5A], which may explain, at least in part, our observation that in the context of HFD-feeding, isolated adipocytes from Ad-KO mice fed HFD had 56% lower total ACSL activity compared to isolated adipocytes from ACSL4floxed mice fed HFD [Figure 5B]. We also demonstrated that differences in adipocyte metabolism were not due to differences in lipogenic or adipogenic gene expression [Supplementary Fig. 4c].
Figure 5.
Ad-KO mice fed HFD have alterations in adipocyte ACSL activity, n-6 PUFA fatty acyl-CoA formation, and FFA and phospholipid composition. (A) ACSL isoform gene expression in isolated gonadal adipocytes of 20-week old male ACSL4floxed or Ad-KO mice fed LFD or HFD for 12 weeks RNA expression quantified by real-time PCR relative to Cyclophilin A. n = 5–8 mice per group. *p ≤ 0.05 ACSL4floxed vs. Ad-KO within diet; **p ≤ 0.0125 ACSL4floxed vs. Ad-KO within diet; #p ≤ 0.05 LFD vs. HFD within genotype; ##p ≤ 0.0125 LFD vs. HFD within genotype; Student's t-test. (B) Total ACSL enzyme activity in isolated gonadal adipocytes of 20 week old males fed LFD or HFD for 12 weeks. n = 4–6 mice/group *p ≤ 0.05 ACSL4floxed vs. Ad-KO within diet. Student's t-test. (C) Long-chain acyl-CoA species determined in gonadal adipose tissue of male ACSL4floxed or Ad-KO mice on HFD for 12 weeks as determined by two-way ANOVA. *p ≤ 0.05 ACSL4floxed vs. Ad-KO within diet; **p ≤ 0.0125 ACSL4floxed vs. Ad-KO within diet. n = 7–8 mice/group. (D) Isolated adipocyte FFA species (no species less than 20 carbons have significant differences) as determined by two-way ANOVA. *p ≤ 0.05 ACSL4floxed vs. Ad-KO within diet; **p ≤ 0.0125 ACSL4floxed vs. Ad-KO within diet. n = 6–9 mice/group. (E) Fold-change of isolated adipocyte arachidonic acid, DHA, and linoleic acid phospholipid species. Significant differences between groups represented in yellow (p ≤ 0.05) and italicized (p ≤ 0.01) as determined by two-way ANOVA (Black boxes = Species not detected). n = 6–9 mice/group. Data represent mean ± SEM.
To specifically investigate ACSL4's direct actions on the regulation of acyl-CoA formation in vivo, we examined ACSL4's effects on long-chain fatty acyl-CoA levels. Interestingly, 18:2-, 18:3- and 20:4-CoA concentrations were decreased 41%, 40%, and 52%, respectively, in gWAT from Ad-KO mice fed HFD compared to ACSL4floxed fed HFD [Figure 5C]. No differences in gWAT acyl-CoAs were observed between ACSL4floxed and Ad-KO mice fed LFD [Supplementary Fig. 6]. These data indicated an interaction between dietary fat and the relative consequences of ACSL4 activity in vivo for 18:2-, 18:3-, and 20:4-CoAs in gWAT in the context of DIO.
Lipidomics analysis of isolated adipocytes from the two lines of mice fed HFD was performed to determine ACSL4's role in regulating the fate of specific FA within different lipid classes specifically within adipocytes. Remarkably, this analysis revealed that adipocytes from Ad-KO mice fed HFD had a reduced ability to incorporate AA into all species of PL [Figure 5E]. Several species of PL also had reduced incorporation of 22:6 into Ad-KO adipocytes as well [Figure 5E]. Interestingly, some species of PL that had reduced AA also had concomitant increased incorporation of LA, including PE D16:1–18:2 (0.59 nmol/mg protein ACSL4floxed vs. 0.82 nmol/mg protein Ad-KO) compared to PC D16:1–20:4 (0.48 nmol/mg protein ACSL4floxed vs. 0.32 nmol/mg protein Ad-KO), PI 18:0–18:2 (0.13 nmol/mg protein ACSL4floxed vs. 0.24 nmol/mg protein Ad-KO) compared to PI 18:0–20:4 (5.80 nmol/mg protein ACSL4floxed vs. 4.32 nmol/mg protein Ad-KO), and PS 18:1–18:2 (0.13 nmol/mg protein ACSL4floxed vs. 0.24 nmol/mg protein Ad-KO) compared to PS 18:1–20:4 (0.69 nmol/mg protein ACSL4floxed vs. 0.44 nmol/mg protein Ad-KO) [Figure 5E]. Consistent with these data and the actions of ACSL4 to “trap” AA within cells [55], [56], the FFA pool in isolated adipocytes had 48% and 44% decreases in 20:4 (62.4 nmol/mg protein ACSL4floxed vs. 32.2 nmol/mg protein Ad-KO) and the less abundant 22:6 (12.5 nmol/mg protein ACSL4floxed vs. 7.7 nmol/mg protein Ad-KO), respectively [Figure 5D]. The FA composition of other lipid classes including TAG, diacylglycerol, and cardiolipin was not altered between adipocytes from the two lines of mice fed HFD [Supplementary Table 2].
Notably, other logical pathways of PUFA metabolism, such as the pattern of oxylipin metabolites, which are defined as oxygenated metabolites derived from arachidonic and linoleic acid that includes eicosanoids [57], [58], were not altered in gWAT from the two lines of mice fed HFD [Supplementary Fig. 7]. Analysis of gWAT did not reveal any significant differences in free metabolites between ACSL4floxed and Ad-KO mice fed HFD, including metabolites of cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome p450 enzymes [Supplementary Fig. 7]. The absolute levels of total oxylipins, which includes both esterified and free oxylipins, are significantly increased as compared to levels of free oxylipins (Supplementary Fig. 7, Supplementary Table 3). Analysis of oxylipin levels revealed that four out of fifty-five total oxylipins, were identified as significantly different in gWAT from ACSL4floxed and Ad-KO mice fed HFD; however, these differences were not consistent with alterations in a specific metabolic pathway of oxylipin production or pro/anti-inflammatory balance [Supplementary Fig. 7]. These data strongly suggest that alterations in oxylipin metabolism in gWAT did not contribute to differences between ACSL4floxed and Ad-KO mice fed HFD. In summary, lipidomics analysis of isolated adipocytes demonstrated that ACSL4 had a specific role in generating AA-CoA, which ultimately resulted in incorporation of AA into PL. Adipocytes from Ad-KO mice fed HFD had reductions in AA in both PL and FFA pools.
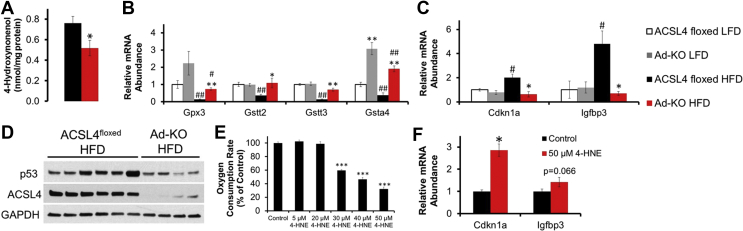
3.6. Adipocytes from Ad-KO mice fed HFD are protected against increased 4-HNE production, reductions in glutathione-mediated detoxification gene expression, and 4-HNE mediated p53 activation
One detrimental metabolic fate of PUFA is lipid peroxidation by ROS and degradation into toxic aldehydes, including 4-HNE [6], [59]. Previous studies have demonstrated that HFD feeding significantly increased 4-HNE concentrations, particularly in gWAT [60]. Free-radical ROS can abstract the bisallylic protons on all polyunsaturated lipids, which then leads to peroxidation. However, increased numbers of unsaturated double bonds make lipid species more susceptible to peroxidation. Thus, AA has a ∼4-fold higher rate of lipid peroxidation than does LA due to the extra two double bonds [61], [62]. In conjunction with reduced incorporation of AA into PL and FFA pools, lipidomics profiling demonstrated a significant 32% decrease in 4-HNE concentration in adipocytes from Ad-KO mice fed HFD compared to controls [Figure 6A].
Figure 6.
Adipocytes from Ad-KO mice fed HFD are protected against increased 4-HNE production, reductions in glutathione-mediated detoxification gene expression, and 4-HNE mediated p53 activation. (A) Isolated adipocyte 4-HNE content as determined by two-way ANOVA. *p ≤ 0.05 ACSL4floxed vs. Ad-KO within diet. (B) Glutathione-mediated detoxification genes of interest in isolated gonadal adipocytes of 20-week old male ACSL4floxed or Ad-KO mice fed LFD or HFD for 12 weeks RNA expression quantified by real-time PCR relative to Cyclophilin A. n = 5–8 mice per group. *p ≤ 0.05 ACSL4floxed vs. Ad-KO within diet; **p ≤ 0.0125 ACSL4floxed vs. Ad-KO within diet; #p ≤ 0.05 LFD vs. HFD within genotype; ##p ≤ 0.0125 LFD vs. HFD within genotype; Student's t-test with Bonferroni correction for 4 comparisons. (C) p53 downstream target genes in isolated gonadal adipocytes of 20-week old male ACSL4floxed or Ad-KO mice on LFD or HFD for 12 weeks RNA expression quantified by real-time PCR relative to Cyclophilin A. n = 5–7 mice per group. *p < 0.0125 ACSL4floxed vs. Ad-KO within diet; #p < 0.0125 LFD vs. HFD within genotype; Student's t-test with Bonferroni correction for 4 comparisons. (D) Representative immunoblot against p53 and ACSL4 in gonadal adipose tissue from 20-week-old male mice fed HFD for 12 weeks. (E) Oxygen consumption rate (OCR) determined by XFe96 Seahorse in 3T3-L1 adipocytes with increasing concentrations of 4-HNE treatment represented as a percentage of basal OCR normalized to the control OCR values. n = 8 wells for control and 6–8 wells per treatment. (F) Real-time PCR of p53 downstream target genes in 3T3-L1 adipocytes treated with 50 μM 4-HNE for 6 h. N = 5–6 wells per treatment; *p ≤ 0.05 treatment vs. control; Student's t-test. Data represent mean ± SEM.
In addition to reducing AA, which can be metabolized to form 4-HNE, we also investigated whether adipocyte ACSL4 regulated expression of genes involved in glutathione-mediated detoxification of lipid peroxidation products [6]. Of note, isolated adipocytes from Ad-KO mice fed a HFD had improved glutathione-mediated detoxification gene expression [Figure 6B]. These genes (GPX3, GSTT2/3, and GSTA4) are critical for the detoxification of lipid peroxides and α- and β-aldehydes, such as the lipid peroxidation product 4-HNE [6]. 4-HNE is detoxified in cells by several pathways, including the formation of adducts with glutathione catalyzed by glutathione-S-transferases (GST). In particular, glutathione-S-transferase A-4 (GSTA4) detoxification of 4-HNE is a major determinant of intracellular 4-HNE concentrations [6], [63], [64]. Increased expression of GSTA4 is particularly notable because its expression was upregulated independent of diet in adipocytes from Ad-KO mice in mice fed both LFD and HFD [Figure 6B].
4-HNE has also been demonstrated to increase activation of p53 [63], [65], a master regulator of the DNA damage response, cell cycle arrest, and to promote obesity-associated WAT inflammation and IR [72], [73]. Therefore, we then investigated the association of 4-HNE levels and p53 in adipocytes from Ad-KO mice. RT-PCR analysis of isolated adipocytes from Ad-KO mice fed HFD, as compared to ACSL4floxed, demonstrated significantly reduced expression of genes regulated by p53, including cyclin-dependent kinase inhibitor 1 (CDKN1A or p21) and insulin growth factor binding protein 3 (IGFBP3) [Figure 6C]. Activation of p53 may be secondary to an increase in its protein expression, and western blot analysis revealed that gWAT from Ad-KO mice fed HFD also had decreased p53 expression compared to controls fed HFD [Figure 6D]. We also noted that ACSL4 adipocyte deficiency also reduced HFD-induced increases in expression of Cdkn1a in subcutaneous adipose tissue [Supplementary Fig. 2e]. These data suggest that ACSL4 ablation, in the context of DIO, is associated with protection against p53 activation and its downstream effects on inflammation.
To confirm that 4-HNE can activate p53 in adipocytes, we found that incubation of 3T3-L1 adipocytes for 6-hours with 4-HNE increased Cdkn1a mRNA levels 2.9-fold (p = 0.003) and Igfbp3 mRNA expression approached significance (p = 0.066), consistent with downstream p53 activation [Figure 6F]. Previously, we noted that after HFD feeding for 7 weeks, Ad-KO mice were protected against a reduction in systemic EE that was observed in ACSL4floxed littermates [Figure 4A] and subsequently we demonstrated that isolated adipocytes from Ad-KO mice had 3.2-fold higher basal OCR compared to adipocytes from ACSL4floxed mice [Figure 5A], suggesting that alterations in adipocyte OCR may contribute to the observed differences in systemic EE. Since previous studies have demonstrated that incubation of mitochondria or other non-adipocyte cells with 4-HNE reduced OCR [66], [67] and knockdown of GSTA4 in 3T3-L1 adipocytes reduced adipocyte OCR as well [68], we were led to hypothesize that the increased levels of 4-HNE in ACSL4floxed adipocytes may depress OCR. To directly investigate this hypothesis, we incubated 3T3-L1 adipocytes with increasing concentrations of 4-HNE for 6 h and observed a dose-dependent decrease in adipocyte OCR where adipocyte OCR was reduced up to 70% compared to control with 50 μM 4-HNE treatment (in the absence of cell death as determined by XTT assay data not shown) [Figure 6E]. In summary, in vitro incubation of adipocytes with increasing concentrations of 4-HNE reduced adipocyte OCR and increased p53 activation. These studies are consistent with a role for 4-HNE in activating p53 and directly reducing gWAT adipocyte OCR in ACSL4floxed adipocytes that have ACSL4 expression compared to adipocytes from Ad-KO mice that do not have ACSL4 expression.
4. Discussion
While many studies have focused on the detrimental role of saturated fatty acids [69], most HFDs also include high dietary PUFA as well, but we have a more limited understanding of the role of n-6 PUFA, such as LA, which can be converted within the cells of the body into AA. Presently, we have a limited understanding of the role of cellular AA, in the modulation of obesity-associated adipocyte dysfunction. Previous in vitro studies have suggested that ACSL4 has a preference for activating AA to form AA-CoA, effectively “trapping” AA within cells, which can then be directed to incorporation into PL and TAG [17], [56], [70]. Additionally, oxylipin metabolites of AA, including prostaglandins, leukotrienes, and epoxyeicosatrienoic acids (EET), have been implicated in adipose tissue inflammation [5]. In addition to serving as a precursor for eicosanoid synthesis, AA incorporated into PL can undergo lipid peroxidation, ultimately resulting in toxic lipid peroxidation products, including 4-HNE [6], [62]. Interestingly, recent studies have demonstrated that lipid peroxidation of AA incorporated into PL specifically by ACSL4 mediates ferroptosis, an iron-dependent non-apoptotic mode of cell death [19], [20], [21].
Here, we have demonstrated that, in the context of HFD feeding, expression of ACSL4 in adipocytes of mice was significantly increased [Figure 5A] and determined to be a critical regulator of AA incorporation into PL [Figure 5E], which is consistent with prior in vitro studies [20], [21], [56], [70]. Notably, we found that adipocyte ACSL4 expression regulated adipocyte OCR and systemic EE in mice on a HFD. Also, we made the novel observation that ACSL4 expression also regulates levels of the lipid peroxidation product 4-HNE [Figure 6A], and ultimately, the development of DIO-associated adipocyte dysfunction. Cellular levels of 4-HNE are regulated by both the rate of formation of 4-HNE from PUFA, including LA and AA, and detoxification of 4-HNE by GSTA4, a major determinant of intracellular 4-HNE concentrations [65], [71], [72]. Since adipocytes from Ad-KO mice fed HFD had reduced cellular levels of AA [Figure 5D,E] as well as increased expression of GSTA4 [Figure 6B], adipocytes from Ad-KO mice fed HFD possess the two aforementioned complementary pathways to reduce cellular 4-HNE concentrations. These observations are important because previous studies in cultured adipocytes demonstrated that knockdown of GSTA4 resulted in increased lipid peroxidation, mitochondrial oxidative stress, and reduced adipocyte OCR [68], [71], which significantly parallels our in vivo observations of adipocytes from ACSL4floxed mice fed HFD but not in adipocytes from Ad-KO mice fed HFD. Also, of related interest, genetic disruption of GSTA4 in mice resulted in increased adipose tissue levels of 4-HNE, obesity, and WAT inflammation [72]. Interestingly, adipocytes of Ad-KO mice fed a HFD were at least partially protected against the increased mRNA expression of two additional ACSL isoforms, ACSL3 and ACSL5, whose expression could also contribute to obesity-associated adipocyte dysfunction.
In the obese state, there is increased production of ROS in gWAT of mice [73], which likely contributes to the documented increased concentrations of toxic aldehydes in gWAT, including 4-HNE [60]. Both 20:4 (AA) and 22:6 (DHA) PUFA contain bis-allylic groups where a methylene group is located in between two double bonds, and are oxidized and undergo lipid peroxidation at 2–3 times the rate per bis-allylic bond compared to 18:2 (LA) PUFA [61], [62]. Thus, even though 18:2 content was not reduced in the PL of adipocytes from Ad-KO mice fed HFD, the reduction of 20:4 content, and corresponding reduction of reactive bis-allylic protons, in adipocyte PL, from Ad-KO mice fed a HFD may explain, in part, the reduction in 4-HNE. An alternative but related potential mechanism for the role of ACSL4 in adipocyte dysfunction, under investigation in our laboratory, comes from studies which have found that the lipid peroxidation of PL-AA in ferroptosis occurs secondary to the actions of a lipooxygenase [21].
Adipocyte OCR is regulated in many complex ways as was demonstrated in previous studies [31], [52]. Remarkably, our data demonstrated that adipocytes from Ad-KO mice chronically fed HFD for 12 weeks had significantly reduced levels of 4-HNE and increased rates of adipocyte OCR compared to adipocytes from ACSL4floxed mice [Figure 4F]. We hypothesized that the alterations in adipocyte OCR is a significant contributor to our observation that with HFD feeding, Ad-KO mice were protected against a reduction in systemic EE that was noted in ACSL4floxed mice [Figure 4A].
We also observed that adipocyte expression of ACSL4 was associated with increased activation of p53, which we hypothesized was due to the increased 4-HNE levels [Figure 6C,D]. Consistent with these previously published studies, we incubated 3T3-L1 adipocytes with 4-HNE that resulted in a 2.86-fold increase in Cdkn1a mRNA expression (p = 0.009) [Figure 6F]. Importantly, increased activation of p53 within adipocytes of mice has been demonstrated to increase both WAT inflammation and IR [74], [75], which is consistent with our observations of reduced p53 expression, WAT inflammation, and IR.
A necessary question that stems from our observations is how specifically does adipocyte PL remodeling result in alterations in 4-HNE levels and adipocyte dysfunction. We hypothesize that one possible scenario to explain the phenotype of Ad-KO mice fed a HFD is that ablation of ACSL4 expression in adipocytes reduced the availability of cellular AA that can be converted into 4-HNE secondary to associated increases in oxidative stress. In the adipocytes from ACSL4floxed mice (control mice), increased concentrations of 4-HNE contributed to adipocyte dysfunction through several potential pathways, including activation of p53, redox potential, and inflammation, including the production of cytokines such as TNF-α. It is also known that incubation of cultured adipocytes with TNF-α reduces GSTA4 mRNA expression [71]. Thus, in addition to reduced substrate for lipid peroxidation, adipocytes from Ad-KO mice fed HFD also had reduced TNF-α [Figure 2C] and higher levels of GSTA4 expression [Figure 6B], which suggests an improved ability to detoxify 4-HNE from adipocytes. Additionally, we hypothesized that increased levels of 4-HNE resulted in altered mitochondrial function and reduced adipocyte OCR. Protection against increased levels of adipocyte 4-HNE may then protect against activation of p53, WAT inflammation, and reductions in adipocyte OCR.
The phenotypic improvements of ACSL4 ablation on DIO in our mice are highlighted by the observation that reductions in body weight of obese humans with metabolic complications, as modest as 5%, progressively improve systemic IR, intra-abdominal adipose tissue mass, and hepatic steatosis [76]. Since TZDs have been demonstrated to potently block the catalytic activity of ACSL4 by a non-PPARγ mechanism, our results document a potentially novel mechanism by which this class of drugs may possibly act to ameliorate adipocyte dysfunction [11], [12]. Importantly, and of translational interest, in various micro-array studies in humans, ACSL4 expression in adipose tissue and/or adipocytes has been demonstrated to be increased with obesity and IR [77], [78].
5. Conclusions
In conclusion, this study documents the first in vivo elucidation of ACSL4's actions. It is critical to note that ACSL4 represents a small fraction of total ACSL expression in adipocytes, but eliminating its contribution, in the context of DIO, resulted in major whole-body physiological effects. Adipocyte ACSL4 regulates AA incorporation into PL, which is highly susceptible to peroxidation, and forms detrimental products, such as 4-HNE. By reducing the incorporation of AA into PL and FFA pools in adipocytes, Ad-KO mice were significantly protected against HFD-induced increases in adipose and liver fat accumulation, adipocyte death, gWAT inflammation, and IR. Additionally, deficiency of adipocyte ACSL4 expression in mice fed a HFD increased adipocyte OCR, which we demonstrated are direct effects of 4-HNE on adipocytes in vitro, and whole body EE.
Author contributions
Conceptualization, E.A.K., R.A.C., and A.S.G; Methodology, E.A.K., E.L.K., D.K., M.J.L., and A.S.G; Investigation, E.A.K., A.R.R., Q.W.Y., D.S., M.A.E., J.D.G., T.A.B., C.W., and N.R.M.; Writing – Original Draft, E.A.K. and A.S.G.; Writing – Review & Editing, E.A.K., A.R.R., N.R.M., R.A.C., and A.S.G.; Funding Acquisition, A.S.G.; Resources, J.W.N., X.H.; Visualization, E.A.K.; Supervision, E.A.K. and A.S.G.
Acknowledgements
The authors would like to thank Dr. Don Smith and the Comparative Biology Unit at the HNRCA for excellent animal care, Ira Gray (University of California) for critical analysis of oxylipin data, Dr. Stefania Lamon-Fava, Dr. Deanna M. Salter, and Alex Coston (HNRCA) for critical reading of this manuscript and discussion.
A.S.G. receives support from NIDDK R01 DK098606-02, NIDDK RO1 DK108722 01A1, NIDDK Boston Nutrition Obesity Research Center (P30-DK-46200), NIH T32 DK062032-24, and the U.S. Department of Agriculture, Agricultural Research Service, under agreement no. 58-1950-7-70. Dr. Greenberg is also the recipient of the Robert C and Veronica Atkins endowed Professorship in Nutrition and Metabolism at Tufts Medical School. A.S.G. is the guarantor of this submitted manuscript.
D.K. receives support from NIDDK K01 DK094943, R01 DK108797, and a Charles Hood Foundation Grant. J.W.N. support was provided by the NIH West Coast Metabolomics Center Grant U24 DK097154 and USDA Intramural project 2032-51530-022-00D. USDA is an equal opportunity provider and employer. R.A.C. receives support from the NIH (DK59935) and E.L.K. receives support from the NIH (DK107481).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.01.012.
Conflict of interest
Authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Ogden C.L., Carroll M.D., Kit B.K., Flegal K.M. Prevalence of childhood and adult obesity in the United States, 2011–2012. Journal of the American Medical Association. 2014;311(8):806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Primeau V., Coderre L., Karelis A.D., Brochu M., Lavoie M.E., Messier V. Characterizing the profile of obese patients who are metabolically healthy. International Journal of Obesity (London) 2011;35(7):971–981. doi: 10.1038/ijo.2010.216. [DOI] [PubMed] [Google Scholar]
- 3.Samocha-Bonet D., Dixit V.D., Kahn C.R., Leibel R.L., Lin X., Nieuwdorp M. Metabolically healthy and unhealthy obese–the 2013 stock conference report. Obesity Reviews. 2014;15(9):697–708. doi: 10.1111/obr.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefan N., Haring H.U., Hu F.B., Schulze M.B. Metabolically healthy obesity: epidemiology, mechanisms, and clinical implications. Lancet Diabetes Endocrinology. 2013;1(2):152–162. doi: 10.1016/S2213-8587(13)70062-7. [DOI] [PubMed] [Google Scholar]
- 5.Masoodi M., Kuda O., Rossmeisl M., Flachs P., Kopecky J. Lipid signaling in adipose tissue: connecting inflammation & metabolism. Biochimica et Biophysica Acta. 2015;1851(4):503–518. doi: 10.1016/j.bbalip.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Hauck A.K., Bernlohr D.A. Oxidative stress and lipotoxicity. The Journal of Lipid Research. 2016 doi: 10.1194/jlr.R066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mashek D.G., Li L.O., Coleman R.A. Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidology. 2007;2(4):465–476. doi: 10.2217/17460875.2.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L.O., Mashek D.G., An J., Doughman S.D., Newgard C.B., Coleman R.A. Overexpression of rat long chain acyl-coa synthetase 1 alters fatty acid metabolism in rat primary hepatocytes. The Journal of Biological Chemistry. 2006;281(48):37246–37255. doi: 10.1074/jbc.M604427200. [DOI] [PubMed] [Google Scholar]
- 9.Mashek D.G., Li L.O., Coleman R.A. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. The Journal of Lipid Research. 2006;47(9):2004–2010. doi: 10.1194/jlr.M600150-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Ellis J.M., Li L.O., Wu P.C., Koves T.R., Ilkayeva O., Stevens R.D. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metabolism. 2010;12(1):53–64. doi: 10.1016/j.cmet.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.H., Lewin T.M., Coleman R.A. Expression and characterization of recombinant rat Acyl-CoA synthetases 1, 4, and 5. Selective inhibition by triacsin C and thiazolidinediones. The Journal of Biological Chemistry. 2001;276(27):24667–24673. doi: 10.1074/jbc.M010793200. [DOI] [PubMed] [Google Scholar]
- 12.Askari B., Kanter J.E., Sherrid A.M., Golej D.L., Bender A.T., Liu J. Rosiglitazone inhibits acyl-CoA synthetase activity and fatty acid partitioning to diacylglycerol and triacylglycerol via a peroxisome proliferator-activated receptor-gamma-independent mechanism in human arterial smooth muscle cells and macrophages. Diabetes. 2007;56(4):1143–1152. doi: 10.2337/db06-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang M.J., Fujino T., Sasano H., Minekura H., Yabuki N., Nagura H. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(7):2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X., Gopalacharyulu P., Seppanen-Laakso T., Ruskeepaa A.L., Aye C.C., Carson B.P. Insulin signaling regulates fatty acid catabolism at the level of CoA activation. PLoS Genetics. 2012;8(1) doi: 10.1371/journal.pgen.1002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duarte A., Poderoso C., Cooke M., Soria G., Cornejo Maciel F., Gottifredi V. Mitochondrial fusion is essential for steroid biosynthesis. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0045829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maloberti P.M., Duarte A.B., Orlando U.D., Pasqualini M.E., Solano A.R., Lopez-Otin C. Functional interaction between acyl-CoA synthetase 4, lipooxygenases and cyclooxygenase-2 in the aggressive phenotype of breast cancer cells. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0015540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwata H., Hara S. Inhibition of long-chain acyl-CoA synthetase 4 facilitates production of 5, 11-dihydroxyeicosatetraenoic acid via the cyclooxygenase-2 pathway. Biochemical and Biophysical Research Communications. 2015;465(3):528–533. doi: 10.1016/j.bbrc.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 18.Klett E.L., Chen S., Edin M.L., Li L.O., Ilkayeva O., Zeldin D.C. Diminished acyl-CoA synthetase isoform 4 activity in INS 832/13 cells reduces cellular epoxyeicosatrienoic acid levels and results in impaired glucose-stimulated insulin secretion. Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.M113.481077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon S.J., Winter G.E., Musavi L.S., Lee E.D., Snijder B., Rebsamen M. Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chemical Biology. 2015;10(7):1604–1609. doi: 10.1021/acschembio.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nature Chemical Biology. 2017;13(1):91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kagan V.E., Mao G., Qu F., Angeli J.P., Doll S., Croix C.S. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nature Chemical Biology. 2017;13(1):81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L.O., Ellis J.M., Paich H.A., Wang S., Gong N., Altshuller G. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. The Journal of Biological Chemistry. 2009;284(41):27816–27826. doi: 10.1074/jbc.M109.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y., Wang C., Sun H., LeRoith D., Yakar S. High-efficient FLPo deleter mice in C57BL/6J background. PLoS One. 2009;4(11):e8054. doi: 10.1371/journal.pone.0008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eguchi J., Wang X., Yu S., Kershaw E.E., Chiu P.C., Dushay J. Transcriptional control of adipose lipid handling by IRF4. Cell Metabolism. 2011;13(3):249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cinti S., Mitchell G., Barbatelli G., Murano I., Ceresi E., Faloia E. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. The Journal of Lipid Research. 2005;46(11):2347–2355. doi: 10.1194/jlr.M500294-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Strissel K.J., Stancheva Z., Miyoshi H., Perfield J.W., 2nd, DeFuria J., Jick Z. Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes. 2007;56(12):2910–2918. doi: 10.2337/db07-0767. [DOI] [PubMed] [Google Scholar]
- 27.Viswanadha S., Londos C. Optimized conditions for measuring lipolysis in murine primary adipocytes. The Journal of Lipid Research. 2006;47(8):1859–1864. doi: 10.1194/jlr.D600005-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Shaughnessy S., Smith E.R., Kodukula S., Storch J., Fried S.K. Adipocyte metabolism in adipocyte fatty acid binding protein knockout mice (aP2-/-) after short-term high-fat feeding: functional compensation by the keratinocyte [correction of keritinocyte] fatty acid binding protein. Diabetes. 2000;49(6):904–911. doi: 10.2337/diabetes.49.6.904. [DOI] [PubMed] [Google Scholar]
- 29.Goldrick R.B. Effects of insulin on glucose metabolism in isolated human fat cells. The Journal of Lipid Research. 1967;8(6):581–588. [PubMed] [Google Scholar]
- 30.Dole V.P., Meinertz H. Microdetermination of long-chain fatty acids in plasma and tissues. The Journal of Biological Chemistry. 1960;235:2595–2599. [PubMed] [Google Scholar]
- 31.Wang T., Si Y., Shirihai O.S., Si H., Schultz V., Corkey R.F. Respiration in adipocytes is inhibited by reactive oxygen species. Obesity (Silver Spring) 2010;18(8):1493–1502. doi: 10.1038/oby.2009.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraus D., Yang Q., Kong D., Banks A.S., Zhang L., Rodgers J.T. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508(7495):258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulinilkunnil T., He H., Kong D., Asakura K., Peroni O.D., Lee A. Adrenergic regulation of AMP-activated protein kinase in brown adipose tissue in vivo. The Journal of Biological Chemistry. 2011;286(11):8798–8809. doi: 10.1074/jbc.M111.218719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blachnio-Zabielska A.U., Koutsari C., Jensen M.D. Measuring long-chain acyl-coenzyme A concentrations and enrichment using liquid chromatography/tandem mass spectrometry with selected reaction monitoring. Rapid Communications in Mass Spectrometry: rapid Communications in Mass Spectrometry. 2011;25(15):2223–2230. doi: 10.1002/rcm.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M., Han X. Multidimensional mass spectrometry-based shotgun lipidomics. Methods in Molecular Biology. 2014;1198:203–220. doi: 10.1007/978-1-4939-1258-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C., Wang M., Han X. Comprehensive and quantitative analysis of lysophospholipid molecular species present in obese mouse liver by shotgun lipidomics. Analytical Chemistry. 2015;87(9):4879–4887. doi: 10.1021/acs.analchem.5b00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M., Fang H., Han X. Shotgun lipidomics analysis of 4-hydroxyalkenal species directly from lipid extracts after one-step in situ derivatization. Analytical Chemistry. 2012;84(10):4580–4586. doi: 10.1021/ac300695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng H., Jiang X., Han X. Alterations in lipid homeostasis of mouse dorsal root ganglia induced by apolipoprotein E deficiency: a shotgun lipidomics study. Journal of Neurochemistry. 2007;101(1):57–76. doi: 10.1111/j.1471-4159.2006.04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M., Hayakawa J., Yang K., Han X. Characterization and quantification of diacylglycerol species in biological extracts after one-step derivatization: a shotgun lipidomics approach. Analytical Chemistry. 2014;86(4):2146–2155. doi: 10.1021/ac403798q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M., Han R.H., Han X. Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach. Analytical Chemistry. 2013;85(19):9312–9320. doi: 10.1021/ac402078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang K., Cheng H., Gross R.W., Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Analytical Chemistry. 2009;81(11):4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frayn K.N., Little R.A., Threlfall C.J. Protein metabolism after unilateral femoral fracture in the rat, and comparison with sham operation. British Journal of Experimental Pathology. 1980;61(5):474–478. [PMC free article] [PubMed] [Google Scholar]
- 43.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. The Journal of Biological Chemistry. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 44.Spriet L.L., Heigenhauser G.J., Jones N.L. Endogenous triacylglycerol utilization by rat skeletal muscle during tetanic stimulation. Journal of Applied Physiology. 1986;60(2):410–415. doi: 10.1152/jappl.1986.60.2.410. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Sullivan S., Trujillo M., Lee M.J., Schneider S.H., Brolin R.E. Perilipin expression in human adipose tissues: effects of severe obesity, gender, and depot. Obesity Research. 2003;11(8):930–936. doi: 10.1038/oby.2003.128. [DOI] [PubMed] [Google Scholar]
- 46.Cao Y., Dave K.B., Doan T.P., Prescott S.M. Fatty acid CoA ligase 4 is up-regulated in colon adenocarcinoma. Cancer Research. 2001;61(23):8429–8434. [PubMed] [Google Scholar]
- 47.Perfield J.W., 2nd, Lee Y., Shulman G.I., Samuel V.T., Jurczak M.J., Chang E. Tumor progression locus 2 (TPL2) regulates obesity-associated inflammation and insulin resistance. Diabetes. 2011;60(4):1168–1176. doi: 10.2337/db10-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patsouris D., Li P.P., Thapar D., Chapman J., Olefsky J.M., Neels J.G. Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metabolism. 2008;8(4):301–309. doi: 10.1016/j.cmet.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaiyala K.J., Schwartz M.W. Toward a more complete (and less controversial) understanding of energy expenditure and its role in obesity pathogenesis. Diabetes. 2011;60(1):17–23. doi: 10.2337/db10-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tschop M.H., Speakman J.R., Arch J.R., Auwerx J., Bruning J.C., Chan L. A guide to analysis of mouse energy metabolism. Nature Methods. 2011;9(1):57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hallgren P., Sjostrom L., Hedlund H., Lundell L., Olbe L. Influence of age, fat cell weight, and obesity on O2 consumption of human adipose tissue. American Journal of Physiology. 1989;256(4 Pt 1):E467–E474. doi: 10.1152/ajpendo.1989.256.4.E467. [DOI] [PubMed] [Google Scholar]
- 52.Lee Y.S., Kim J.W., Osborne O., Oh da Y., Sasik R., Schenk S. Increased adipocyte O2 consumption triggers HIF-1alpha, causing inflammation and insulin resistance in obesity. Cell. 2014;157(6):1339–1352. doi: 10.1016/j.cell.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yehuda-Shnaidman E., Buehrer B., Pi J., Kumar N., Collins S. Acute stimulation of white adipocyte respiration by PKA-induced lipolysis. Diabetes. 2010;59(10):2474–2483. doi: 10.2337/db10-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kajimura S., Spiegelman B.M., Seale P. Brown and beige fat: physiological roles beyond heat generation. Cell Metabolism. 2015;22(4):546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mashek D.G., Coleman R.A. Cellular fatty acid uptake: the contribution of metabolism. Current Opinion in Lipidology. 2006;17(3):274–278. doi: 10.1097/01.mol.0000226119.20307.2b. [DOI] [PubMed] [Google Scholar]
- 56.Kuch E.-M., Vellaramkalayil R., Zhang I., Lehnen D., Brugger B., Stremmel W. Differentially localized acyl-CoA synthetase 4 isoenzymes mediate the metabolic channeling of fatty acids towards phosphatidylinositol. Biochimica et Biophysica Acta. 2013;1841:227–239. doi: 10.1016/j.bbalip.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Gabbs M., Leng S., Devassy J.G., Monirujjaman M., Aukema H.M. Advances in our understanding of oxylipins derived from dietary PUFAs. Advances in Neurology. 2015;6(5):513–540. doi: 10.3945/an.114.007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strassburg K., Huijbrechts A.M., Kortekaas K.A., Lindeman J.H., Pedersen T.L., Dane A. Quantitative profiling of oxylipins through comprehensive LC-MS/MS analysis: application in cardiac surgery. Analytical and Bioanalytical Chemistry. 2012;404(5):1413–1426. doi: 10.1007/s00216-012-6226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Csala M., Kardon T., Legeza B., Lizak B., Mandl J., Margittai E. On the role of 4-hydroxynonenal in health and disease. Biochimica et Biophysica Acta. 2015;1852(5):826–838. doi: 10.1016/j.bbadis.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 60.Long E.K., Olson D.M., Bernlohr D.A. High-fat diet induces changes in adipose tissue trans-4-oxo-2-nonenal and trans-4-hydroxy-2-nonenal levels in a depot-specific manner. Free Radical Biology and Medicine. 2013;63:390–398. doi: 10.1016/j.freeradbiomed.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.RTaOCE Holman. The rates of oxidation of unsaturated fatty acids and esters. Journal of the American Oil Chemists Society. 1947;24(4):127–129. [Google Scholar]
- 62.Else P.L., Kraffe E. Docosahexaenoic and arachidonic acid peroxidation: it's a within molecule cascade. Biochimica et Biophysica Acta. 2015;1848(2):417–421. doi: 10.1016/j.bbamem.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 63.Awasthi Y.C., Yang Y., Tiwari N.K., Patrick B., Sharma A., Li J. Regulation of 4-hydroxynonenal-mediated signaling by glutathione S-transferases. Free Radical Biology and Medicine. 2004;37(5):607–619. doi: 10.1016/j.freeradbiomed.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 64.Singhal S.S., Singh S.P., Singhal P., Horne D., Singhal J., Awasthi S. Antioxidant role of glutathione S-transferases: 4-Hydroxynonenal, a key molecule in stress-mediated signaling. Toxicology and Applied Pharmacology. 2015;289(3):361–370. doi: 10.1016/j.taap.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaudhary P., Sharma R., Sharma A., Vatsyayan R., Yadav S., Singhal S.S. Mechanisms of 4-hydroxy-2-nonenal induced pro- and anti-apoptotic signaling. Biochemistry. 2010;49(29):6263–6275. doi: 10.1021/bi100517x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Picklo M.J., Amarnath V., McIntyre J.O., Graham D.G., Montine T.J. 4-Hydroxy-2(E)-nonenal inhibits CNS mitochondrial respiration at multiple sites. Journal of Neurochemistry. 1999;72(4):1617–1624. doi: 10.1046/j.1471-4159.1999.721617.x. [DOI] [PubMed] [Google Scholar]
- 67.Roede J.R., Jones D.P. Reactive species and mitochondrial dysfunction: mechanistic significance of 4-hydroxynonenal. Environmental and Molecular Mutagenesis. 2010;51(5):380–390. doi: 10.1002/em.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Curtis J.M., Hahn W.S., Stone M.D., Inda J.J., Droullard D.J., Kuzmicic J.P. Protein carbonylation and adipocyte mitochondrial function. Journal of Biological Chemistry. 2012;287(39):32967–32980. doi: 10.1074/jbc.M112.400663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang E.Y., Leone V.A., Devkota S., Wang Y., Brady M.J., Chang E.B. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. JPEN - Journal of Parenteral and Enteral Nutrition. 2013;37(6):746–754. doi: 10.1177/0148607113486931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Golej D.L., Askari B., Kramer F., Barnhart S., Vivekanandan-Giri A., Pennathur S. Long-chain acyl-CoA synthetase 4 modulates prostaglandin E(2) release from human arterial smooth muscle cells. The Journal of Lipid Research. 2011;52(4):782–793. doi: 10.1194/jlr.M013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Curtis J.M., Grimsrud P.A., Wright W.S., Xu X., Foncea R.E., Graham D.W. Downregulation of adipose glutathione S-transferase A4 leads to increased protein carbonylation, oxidative stress, and mitochondrial dysfunction. Diabetes. 2010;59(5):1132–1142. doi: 10.2337/db09-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh S.P., Niemczyk M., Saini D., Awasthi Y.C., Zimniak L., Zimniak P. Role of the electrophilic lipid peroxidation product 4-hydroxynonenal in the development and maintenance of obesity in mice. Biochemistry. 2008;47(12):3900–3911. doi: 10.1021/bi702124u. [DOI] [PubMed] [Google Scholar]
- 73.Furukawa S., Fujita T., Shimabukuro M., Iwaki M., Yamada Y., Nakajima Y. Increased oxidative stress in obesity and its impact on metabolic syndrome. Journal of Clinical Investigation. 2004;114(12):1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minamino T., Orimo M., Shimizu I., Kunieda T., Yokoyama M., Ito T. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nature Medicine. 2009;15(9):1082–1087. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 75.Vergoni B., Cornejo P.J., Gilleron J., Djedaini M., Ceppo F., Jacquel A. DNA Damage and the activation of the p53 pathway mediate alterations in metabolic and secretory functions of adipocytes. Diabetes. 2016;65(10):3062–3074. doi: 10.2337/db16-0014. [DOI] [PubMed] [Google Scholar]
- 76.Magkos F., Fraterrigo G., Yoshino J., Luecking C., Kirbach K., Kelly S.C. Effects of moderate and subsequent progressive weight loss on metabolic function and adipose tissue biology in humans with obesity. Cell Metabolism. 2016;23(4):591–601. doi: 10.1016/j.cmet.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hardy O.T., Perugini R.A., Nicoloro S.M., Gallagher-Dorval K., Puri V., Straubhaar J. Body mass index-independent inflammation in omental adipose tissue associated with insulin resistance in morbid obesity. Surgery for Obesity and Related Diseases: official Journal of the American Society for Bariatric Surgery. 2011;7(1):60–67. doi: 10.1016/j.soard.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodriguez-Acebes S., Palacios N., Botella-Carretero J.I., Olea N., Crespo L., Peromingo R. Gene expression profiling of subcutaneous adipose tissue in morbid obesity using a focused microarray: distinct expression of cell-cycle- and differentiation-related genes. BMC Medical Genomics. 2010;3(61) doi: 10.1186/1755-8794-3-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.