Abstract
Background
Papillary thyroid cancer (PTC) is currently the most commonly diagnosed endocrine malignancy. In addition, the sex- and age-adjusted incidence of PTC has exhibited a greater increase over the last 2 decades than in many other malignancies. Thus, discovering noninvasive specific serum biomarker to distinguish PTC from cancer-free controls in its early stages remains an important goal.
Material/Methods
Serum samples from 88 PTC patients and 80 cancer-free controls were randomly allocated into training or validation sets. Serum peptide profiling was performed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF-MS) after using weak cation exchange magnetic beads (WCX-MB), and the results were evaluated by use of ClinProTools™ Software. To distinguish PTC from cancer-free controls, quick classifier (QC), supervised neural network (SNN), and genetic algorithm (GA) models were established. The models were blindly validated to verify their diagnostic capabilities. The most discriminative peaks were subsequently identified with a nano-liquid chromatography-electrospray ionization-tandem mass spectrometry system.
Results
Six peptide ions were identified as the most discriminative peaks between the PTC and cancer-free control samples. The QC model exhibited satisfactory sensitivity and specificity among the 3 models that were validated. Two peaks, at m/z 2671.17 and m/z 1464.68, were identified as fragments of the alpha chain of fibrinogen, while a peak at m/z 1738.92 was a fragment of complement component 4A/B.
Conclusions
MS combined with ClinProTools™ software was able to detect peptide biomarkers in PTC patients. In addition, the constructed classification models provided a serum peptidome pattern for distinguishing PTC from cancer-free controls. Both fibrinogen α and complement C4A/B were identified as potential markers for diagnosis of PTC.
MeSH Keywords: Biological Markers, Carcinoma, Mass Spectrometry, Thyroid Nodule
Background
Thyroid cancer (TC) has a higher incidence than other common endocrine malignancies and it constitutes 3–4% of newly diagnosed cancers annually [1,2]. The incidence of TC has also increased substantially over the past 2 decades [3,4]. Papillary thyroid cancer (PTC) is the most common type of thyroid cancer diagnosed, and it accounts for up to 90% of all TC cases [5,6]. PTCs rarely exhibit clinically aggressive behavior and most remain indolent, with a disease-specific mortality rate of less than 5% [6]. Nonetheless, a small proportion of patients with PTC develop adverse symptoms, and a higher morbidity rate is associated with patients with advanced-stage PTC [6,7]. A risk of recurrence is also associated with PTC, with recurrence occurring in 10–30% of PTC cases according to the stage at diagnosis [8,9]. Thus, early and accurate diagnosis of PTC, as well as timely treatment, is critical for improving the long-term survival and recurrence incidence of PTC patients.
Physical examinations have had limited efficacy in the differentiation of benign thyroid nodules from malignant thyroid nodules unless there is an evident outcome. Fine-needle aspiration biopsy (FNAB) of the thyroid is commonly considered an effective screening test [10,11]. However, this is an invasive procedure and can have serious complications. Moreover, one of the major limitation of this method is a high occurrence of non-diagnostic results [12] and around 10–20% non-diagnostic rates [13–15]. At the same time, valuable biomarkers for papillary thyroid cancer have been sought by researchers, such as calprotectin, interleukin4, cytokeratin19, and galectin-3; unfortunately, these biomarkers either do not distinguish PTC from a benign nodule, or have poor predictive value [16–19]. Consequently, there is an increasing need to identify novel and clinically relevant biological markers to differentiate malignant thyroid nodules from benign lesions.
Proteomics is a powerful tool for identifying protein expression patterns that can differentiate among different disease states in individuals. Since blood is the main compartment of the body that flows through all tissues and carries components from all tissue types, a “proteome analysis” of blood serum samples should provide a pathological insight into tissue turnover. Moreover, access to serum samples is relatively straightforward compared with the collection of many other tissue types. Taken together, these factors support the use of proteomic analysis for serum samples, and they facilitate its use as a tool for the early diagnosis, treatment monitoring, and prognostic assessment of PTC cases.
Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF-MS) is a state-of-the-art technology in proteomic research that has been used for the identification and testing of disease-related proteins [20]. The use of weak cation exchange magnetic beads (WCX-MB) has developed as a complementary method by which small-molecule peptides can be captured by the large surface area of MBs [21]. The combination of MBs and MALDI-TOF-MS represents a unique approach for the detection of lower-molecular weight peptides and proteins in serum.
Therefore, in the present study, serum samples were collected from PTC patients and cancer-free controls (e.g., healthy individuals and benign thyroid node (BTN) patients) and these were profiled by MALDI-TOF-MS following WCX-MB fractionation and then were analyzed by ClinProTools™ 3.0 (CPT) software (Bruker Daltonics). Three classification models were also established and evaluated based on differentially expressed peptides. These models were further verified with an independent validation set.
Material and Methods
Patient selection and preparation of serum samples
This study was approved by the local institutional review board and ethics committee and written informed consent was obtained from each participant. We enrolled a total of 168 participants, including 88 PTC patients (63 females, 25 males), 31 BTN patients (23 females, 8 males), and 49 healthy individuals (35 females, 14 males). The median age of the PTC group was 47 years (range: 21–65). The cancer-free control group was matched with the PTC group according to age and sex. The patients that were pathologically diagnosed with PTC or BTN were confirmed independently by 2 expert pathologists. Patients with other tumors types, as well as inflammatory diseases, were excluded from this study. An overview of the study is provided in Figure 1.
Figure 1.
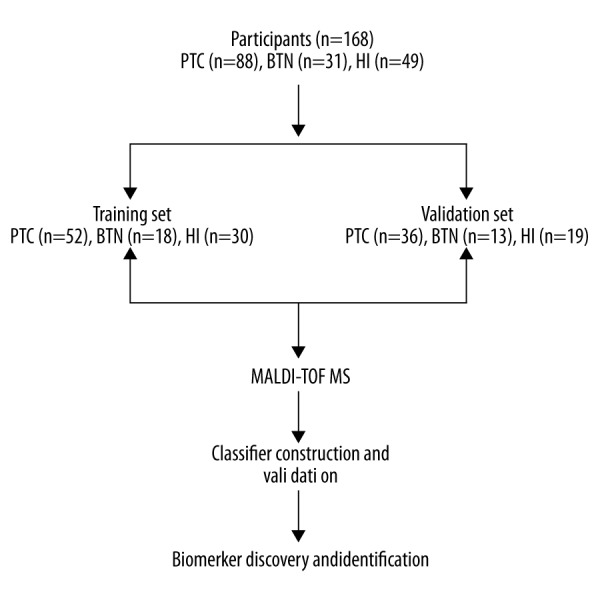
A flowchart of this study. PTC – papillary thyroid cancer; BTN – benign thyroid node; HI – healthy individuals.
Blood samples were collected prior to any therapeutic intervention at the General Hospital of Jinan Military Command Region (China). Briefly, the blood samples were collected in 5-mL vacutainer tubes and were incubated for 30 min at room temperature for clotting. The samples were subsequently centrifuged at 2000 rpm for 15 min. Isolated serum samples were stored in Eppendorf tubes at −80°C for further analysis.
Peptide isolation by magnetic beads
Serum samples were fractionated by MB-WCX (Bruker Daltonics, Bremen, Germany) according to a standard protocol (manufacturer’s instruction). Briefly, each 5-μL serum sample was mixed with 10 μL of MBs and 10 μL of buffer in a 200-μL tube. After mixing, the sample tubes were placed into a magnetic separator, and 1 min later the supernatant in each tube was carefully removed. After the samples were bound to the MBs and washed 3 times with 100 μL of MB cleaning buffer, 5 μL of MB elution buffer was added to the samples bound to the magnetic beads. The suspended fluid samples were then transferred into new 10-μL tubes after complete separation of the MBs from the supernatants was achieved. Five μL of stable buffer was added to each tube before they were stored at −20°C for MS analyses.
MALDI-TOF-MS analysis
One μL of each prepared sample was pipetted onto a polished steel target (Bruker Daltonics, Bremen, Germany). After air-drying the samples, 1 μL of a matrix solution of a-cyano-4-hydroxycinnamic acid (3 mg/ml in 2% trifluoroacetic acid/50% acetonitrile) was applied to each spot. Droplets were dried at room temperature and then immediately analyzed with an AutoflexIII MALDI-TOF mass spectrometer (Bruker Daltonics) with FlexControl software (version 3.4; Bruker). Mass spectra were obtained in a positive ion linear mode at a mass range of m/z 1–10 kDa. The MS analysis ion source settings for ion source 1 and ion source 2 were 19.64 kV and 18.34 kV, respectively. A profile was obtained by adding 20 spectra of 60 laser shots, each with a laser power of 70%. A standard calibration mixture of peptides and proteins was used for mass calibration of the instrument prior to MS analysis. All of the tests were performed in a blinded manner. To evaluate the stability and repeatability of the MALDI-TOF mass spectrometer, a quality control sample (pooled from 10 PTC patient serum samples) was used to run 6 within-run assays during the experiment.
Peptide identification
Sequences of the differential peptides were separated and identified with nano-liquid chromatography-electrospray ionization-tandem mass spectrometry (nano-LC/ESI-MS/MS) by using an Aquity UPLC system (Waters) and a LTQ orbitrap XL mass spectrometer (Thermo Fisher) equipped with a nano-ESI source. Briefly, the peptide solutions were loaded onto a C18 trap column (nanoACQUITY) (180 μm×20 mm×5 μm) at a flow rate of 15 μL/min. The desalted peptides were then enriched by a C18 analytical column (nanoACQUITY) (75 μm×150 mm×3.5 μm symmetry) at a flow rate of 400 nL/min. Mobile phases A (5% acetonitrile, 0.1% formic acid) and B (95% acetonitrile, 0.1% formic acid) were applied to the analytical columns. Gradient elution profiles were: 5%B / 50%B / 80%B / 80%B / 5%B / 5%B over 100 min. The MS instrument was operated in a data-dependent mode. A full scan ranged from 400 m/z to 2000 m/z with a resolution of 100 000. MS/MS spectra were limited to 2 consecutive scans per precursor ion, followed by 60 s of dynamic exclusion. The obtained chromatograms were analyzed with BioworksBrower3.3.1 SPI, and mass lists were determined based on a database search with Sequest™ [IPI Human (3.45)]. The parent ion and fragment mass relative accuracy values were set at 50 ppm and 1 Da, respectively.
Statistical analysis
CPT software was used to analyze spectra of the serum samples. Data analysis was performed with raw-data pretreatment, including baseline subtraction on spectra, total ion current (TIC) normalization, recalibration, and smoothing. The signal-to-noise ratio was set greater than 5, and a mass shift of no more than 0.1% was determined to align the spectra. Peaks were screened by using the t test or the Wilcoxon rank sum test (according to the normality of data distribution). P-values less than 0.05 indicated statistically significant differences. Receiver operating characteristic (ROC) curve and area under the ROC curve (AUC) values were calculated to assess the discriminative capacities of the serum peptide ions identified (Figure 2). Three different algorithms – quick classifier (QC), supervised neural network (SNN), and genetic algorithm (GA) – were used for model analyses and for the selection of peptide peaks. Recognition capacity and 20% leave-one-out cross-validation were calculated for all 3 algorithms as indicators of their performance. A total of 100 samples (52 from PTC patients and 48 from cancer-free controls) were used as training samples for the classification models. To determine the clinical applicability of the constructed models, external validation was performed for a set of samples obtained from 36 PTC patients and 32 cancer-free controls. Sensitivity and specificity values were calculated for the QC, SNN, and GA models.
Figure 2.
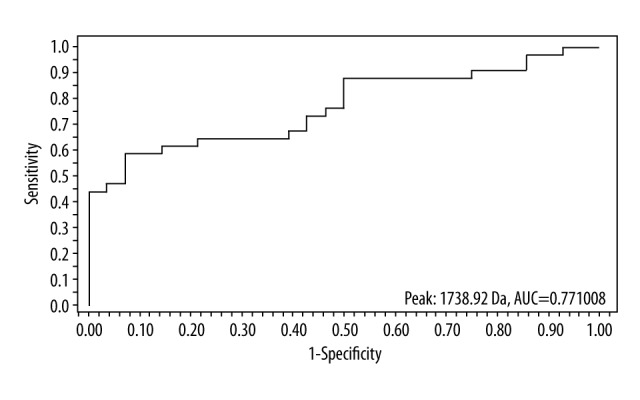
ROC curve of peak m/z 1738.92.
Results
Serum peptide profiling
Serum samples from 10 PTC patients were pooled and 6 within-run assays were conducted with MALDI-TOF-MS. The coefficient of variation (CV) values of selected peaks ranged from 14.30% to 24.99% (Supplementary Table 1), and this range is considered acceptable for the analysis of complex biological samples [22].
CPT software identified a total of 97 unique protein peaks between the PTC patient samples and the cancer-free control samples. In a univariate statistical analysis, 6 peptide peaks exhibited significant differences between the 2 sets of samples (Table 1). ROC curves were used to further analyze the diagnostic capacity of these 6 peaks. In particular, the peptides with m/z 5905.22, 2671.17, 1464.68, 1738.92, 6630.37, and 1978.22 had the highest discriminative power between the PTC patients and the cancer-free controls and their AUC values ranged from 0.75–0.83 (Table 1).
Table 1.
Relative intensities (mean ±SD) of the most discriminative peaks (m/z) in the serum samples obtained from PTC patients versus cancer-free controls.
| Mass (m/z) | PTC patients | Cancer-free controls | P* | AUC** |
|---|---|---|---|---|
| 5905.22 | 16±9.17 | 28.45±11.79 | <0.000001 | 0.83 |
| 2671.17 | 15.46±6.64 | 21.59±9.07 | 0.0000106 | 0.78 |
| 1464.68 | 6.43±3.06 | 10.99±4.32 | 0.000792 | 0.76 |
| 1738.92 | 6.91±2.52 | 4.25±2.14 | 0.000113 | 0.77 |
| 6630.37 | 2.46±2.14 | 1.2±0.61 | 0.000350 | 0.76 |
| 1978.22 | 2.21±0.66 | 3.0±0.67 | 0.000244 | 0.75 |
Calculated based on a t-test or the Wilcoxon test;
area under the ROC curve.
Establishment of classification models and blind validation
It has been observed that the use of multiple disease markers provides more reliable differentiation compared with use of a single marker. Therefore, 3 mathematical algorithms – QC, SNN, and GA – were established to generate prediction models based on a training set containing randomly selected samples. The combinations of peaks that were used by each of the models are listed in Table 2. Both recognition capacity and cross-validation for all 3 models were evaluated (Table 3). To verify the accuracy of these 3 models, additional samples obtained from 36 PTC patients and 32 cancer-free controls were analyzed. The sensitivity and specificity of the classification of these 2 groups of samples by the QC, SNN, and GA models were 84.38%, 87.50%, and 90.32%, and 78.13%, 72.41%, and 65.52%, respectively (Table 3). The QC model exhibited the best efficiency in distinguishing PTC from the cancer-free controls.
Table 2.
The m/z peak values that were included in each of the classification models.
| Quick Classifier | Supervised Neural Network | Genetic algorithm |
|---|---|---|
| 1059.91 | 3098.22 | 5905.22 |
| 1135.04 | 5353.75 | 3264.81 |
| 1464.68 | 2955.75 | 2936.13 |
| 1738.92 | 7463.16 | |
| 1978.13 | 5753.7 | |
| 2028.76 | 1059.91 | |
| 2381.62 | 1449.6 | |
| 2671.17 | 2381.62 | |
| 2955.75 | 1464.68 | |
| 2098.22 | 4057.3 | |
| 3264.81 | 2772.7 | |
| 3904.96 | 3244.66 | |
| 3922.06 | 2663.66 | |
| 4250.2 | 4213.07 | |
| 4697.85 | 5905.22 | |
| 5905.22 | 6663.57 | |
| 6630.37 | 7008.3 | |
| 1978.22 |
Table 3.
Diagnostic performances of the QC, SNN, and GA models.
| Diagnostic parameter | Quick Classifier | Supervised Neural Network | Genetic algorithm |
|---|---|---|---|
| Recognition capacity (%) | 90.95 | 89.44 | 93.98 |
| Cross validation (%) | 82.90 | 81.50 | 70.28 |
| Sensitivity (%) | 84.38 | 87.50 | 90.32 |
| Specificity (%) | 78.13 | 72.41 | 65.52 |
Identification of peptide peaks
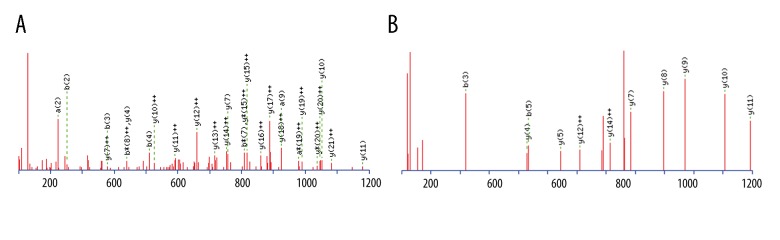
The peptide peaks with the greatest discriminatory power were identified next. The corresponding peptide ions were m/z 5905.22, m/z 2671.17, m/z 1464.68, and m/z 1738.92. Peptide peaks m/z 2671.17 and m/z 1464.68 had amino acid sequences of K. SYKMADEAGSEADHEGTHSTKRGHA.K and DSGEGDFLAEGGGVR.G, respectively, which corresponded to fragments of fibrinogen alpha chain protein (Figure 3A). The Mascot score was 196. Precursor ion m/z 1738.92 corresponded to a fragment of complement component 4A/B (Figure 3B) with the sequence R.NGFKSHALQLNNRQI.R. The Mascot score was 64. In contrast, the amino acid sequence of peptide peak m/z 5905.22 could not be identified. An unknown modification on this peptide may have contributed to this result [23].
Figure 3.
Identification of serum peptides present at higher levels in PTC patients. Sequencing of select serum peptides was achieved with LTQ-Orbitrap-MS/MS. (A) Peak m/z 2671.17 (sequence: K.SYKMADEAGSEADHEGTHSTKRGHA.K) was identified as a fragment of fibrinogen alpha chain with an ion score of 196. (B) Peak m/z 1738.92 (sequence: R.NGFKSHALQLNNRQI.R) was identified as a fragment of Complement C4B with an ion score of 64.
Discussion
Currently, PTC is the most common type of endocrine cancer diagnosed, and it usually presents as a nodule in the thyroid gland. A diagnosis of PTC typically depends on diagnostic imaging, although it can be difficult to distinguish between PTC and benign nodules. Hence, a need exists for the discovery and identification of biomarkers of PTC. More recently, characterization of low-molecular weight serum peptides has been an area of active research. MALDI-TOF MS is a user-friendly method that is able to perform peptide profiling with high sensitivity, resolution, and accuracy, albeit in a low-throughput manner. The aim of the present study was to identify serum peptides present in PTC patients and not in cancer-free controls in order to assess the utility of using peptide profiling for the detection of PTC.
To the best of our knowledge, this is the first study to analyze differences in serum peptide profiles among PTC patients and cancer-free controls by using WCX-MB fractionation followed by MALDI-TOF-MS, with the analysis of data performed by CPT software. Six peptides were identified that exhibited higher discriminative capacities. Moreover, 3 of the peptides were identified based on both univariate and multivariate (QC) statistical analyses (2671.17 Da, 1464.68 Da, and 1738.92 Da peaks) (Table 1). Based on the observation that combinations of biomarkers can increase sensitivity and specificity of detection [24,25], we employed different algorithms to generate classification models. As a result, different combinations of peptide peaks were used to generate 3 classification models. Differences in the algorithms that were used to establish the models account for the differences in the combination of peptides they evaluated [26]. It has been demonstrated that classification models tend to achieve better results with data that are similar to those which they were originally constructed from compared with data that derive from a new set of samples [27]. Thus, cross-validation did not adequately assess the diagnostic capacity of the models examined. For this reason, external validation has been identified as an important step in defining the diagnostic capacity of generated models [28]. Therefore, independent test sets were used to evaluate the accuracy of the classification models generated in this study. The best differentiating capacity and satisfactory values of sensitivity (84.38%) and specificity (78.13%) were associated with the QC model. However, it is also possible that the use of all 3 models could provide a more reliable approach for PTC screening.
Application of nano-LC/ESI-MS/MS resulted in the identification of 2 potential PTC serum biomarkers: fibrinogen a and complement C4A/B. Fibrinogen that is synthesized by hepatocytes plays a significant role in blood coagulation and mediates platelet aggregation [29]. Fibrinogen has also been shown to enhance the proliferation and migration of malignant tumor cells in patients [30,31], and higher levels of fibrinogen α chain fragments have been detected in biliary tract cancer [32], liver cancer [33], and gastric cancer [34]. However, by contrast, we found fibrinogen α at significantly lower concentrations in PTC sera relative to the controls. It is noted fibrinogen has low levels in patients after operation [35] and patients with a higher concentration of plasma fibrinogen levels had a worse prognosis [36]. Then, the lower levels of fibrinogen may be related to indolent character and good prognosis. Thus, circulating fragments from unmodified or post-translationally modified proteins generated in a tumor microenvironment have the potential to serve as diagnostic or prognostic markers.
Another peptide peak was identified as a fragment of Complement C4A/B, a hydrolytic fragment of complement C4. C4A/B contributes to the propagation of activation pathways that lead to the formation of a membrane-associated complex that attacks foreign antigens [37]. In recent studies, a possible role for complement activation in tumor growth and oncogenic capabilities has been identified [38,39], particularly in hypopharyngeal squamous cells [40,41], colorectal cancer [42], lung cancer [43], and breast cancer [44]. It is possible that higher levels of complement components and activation fragments in plasma are the result of cancer cells that are resistant to complement-mediated cytotoxicity, and this leads to a sustained release of complement fragments. In the present study, the higher levels of C4A/B that were detected in the PTC samples suggest that this complement protein can potentially serve as a biomarker for diagnosing PTC.
Conclusions
In this study, peptidome patterns from WCX-MB-purified serum samples were directly profiled with MALDI-TOF-MS, and a classification model was generated that effectively distinguished PTC patients from benign tumors and healthy controls. Moreover, nano-LC/ESI-MS/MS identified fibrinogen α chain and complement component 4A/B as potential biomarkers for distinguishing PTC. Our future studies will be aimed at developing antibodies against the candidate markers identified and to verify their efficacy in a large preoperative cohort.
Supplementary Table
Supplementary Table 1.
Coefficient of variation (CV) values determined for the ion intensities of ten different peaks identified in ten pooled PTC serum samples by MALDI-TOF MS.
| Mass | CV (%) |
|---|---|
| 1546.069 | 24.18 |
| 2167.17 | 18.26 |
| 2663.308 | 23.78 |
| 3886.029 | 23.86 |
| 2955.75 | 22.27 |
| 4056.434 | 14.30 |
| 4093.956 | 18.35 |
| 5338.196 | 22.06 |
| 7759.919 | 18.26 |
| 9276.545 | 24.99 |
Footnotes
Source of support: Departmental sources
References
- 1.Visciano C, Prevete N, Liotti F, et al. Tumor-associated mast cells in thyroid cancer. Int J Endocrinol. 2015;2015:705169. doi: 10.1155/2015/705169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.SEER Cancer Statistics Review, 1975–2014. http://seer.cancer.gov/statfacts/html/thyro.html.
- 3.Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer. 2009;115:3801–7. doi: 10.1002/cncr.24416. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Wang W. Increasing incidence of thyroid cancer in Shanghai, China, 1983–2007. Asia Pacific J Public Health. 2015;27:NP223–29. doi: 10.1177/1010539512436874. [DOI] [PubMed] [Google Scholar]
- 5.Hundahl SA, Cady B, Cunningham MP, et al. Initial results from a prospective cohort study of 5583 cases of thyroid carcinoma treated in the United States during 1996. Cancer. 2000;89:202–17. doi: 10.1002/1097-0142(20000701)89:1<202::aid-cncr27>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 6.Lim H, Devesa SS, Sosa JA, et al. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. AMA. 2017;317:1338–48. doi: 10.1001/jama.2017.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orosco RK, Hussain T, Brumund KT, et al. Analysis of age and disease status as predictors of thyroid cancer-specific mortality using the Surveillance, Epidemiology, and End Results database. Thyroid. 2015;25:125–32. doi: 10.1089/thy.2014.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samaan NA, Schulz PN, Hickey RC, et al. The results of various modalities of treatment of well differentiated thyroid carcinomas: A retrospective review of 1599 patient. J Clin Endocrinol Metab. 1992;75:714–20. doi: 10.1210/jcem.75.3.1517360. [DOI] [PubMed] [Google Scholar]
- 9.Durante C, Montesano T, Torlontano M, et al. Papillary thyroid cancer: Time course of recurrences during postsurgery surveillance. J Clin Endocrinol Metab. 2013;98:636–42. doi: 10.1210/jc.2012-3401. [DOI] [PubMed] [Google Scholar]
- 10.Yassa L, Cibas ES, Benson CB, et al. Long-term assessment of a multidisciplinary approach to thyroid nodule diagnostic evaluation. Cancer. 2007;111:508–16. doi: 10.1002/cncr.23116. [DOI] [PubMed] [Google Scholar]
- 11.Wong CK, Wheeler MH. Thyroid nodules: rational management. World J Surg. 2000;24:934–41. doi: 10.1007/s002680010175. [DOI] [PubMed] [Google Scholar]
- 12.Raab SS, Vrbin CM, Grzybicki DM, et al. Errors in thyroid gland fine needle aspiration. Am J Clin Pathol. 2006;125:873–82. doi: 10.1309/7RQE-37K6-439T-4PB4. [DOI] [PubMed] [Google Scholar]
- 13.Baier ND, Hahn PF, Gervais DA, et al. Fine-needle aspiration biopsy of thyroid nodules: Experience in a cohort of 944 patients. Am J Roentgenol. 2009;193:1175–79. doi: 10.2214/AJR.08.1840. [DOI] [PubMed] [Google Scholar]
- 14.Zhong LC, Lu F, Ma F, et al. Ultrasound-guided fine-needle aspiration of thyroid nodules: does the size limit its efficiency? Int J Clin Exp Pathol. 2015;8:3155–59. [PMC free article] [PubMed] [Google Scholar]
- 15.Güney G1, Şahiner İT. Malignancy rates of thyroid cytology: Cyst fluid benign or non-diagnostic? Med Sci Monit. 2017;23:3556–61. doi: 10.12659/MSM.905718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saussez S, Glinoer D, Chantrain G, et al. Serum galectin-1 and galectin-3 levels in benign and malignant nodular thyroid disease. Thyroid. 2008;18:705–12. doi: 10.1089/thy.2007.0361. [DOI] [PubMed] [Google Scholar]
- 17.Stanciu AE, Serdarevic N, Hurduc AE, Stanciu MM. IL-4, IL-10 and high sensitivity-CRP as potential serum biomarkers of persistent/recurrent disease in papillary thyroid carcinoma with/without Hashimoto’s thyroiditis. Scand J Clin Lab Invest. 2015;75:539–48. doi: 10.3109/00365513.2015.1057895. [DOI] [PubMed] [Google Scholar]
- 18.Tabur S, Korkmaz H, Özkaya M, et al. Serum calprotectin: A new potential biomarker for thyroid papillary carcinoma. Tumour Biol. 2015;36:7549–56. doi: 10.1007/s13277-015-3468-1. [DOI] [PubMed] [Google Scholar]
- 19.Makki FM, Taylor SM, Shahnavaz A, et al. Serum biomarkers of papillary thyroid cancer. J Otolaryngol Head Neck Surg. 2013;42:16. doi: 10.1186/1916-0216-42-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 21.Whiteaker JR, Zhao L, Zhang HY, et al. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albrethsen J. Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem. 2007;53:852–58. doi: 10.1373/clinchem.2006.082644. [DOI] [PubMed] [Google Scholar]
- 23.Cottrell JS. Protein identification using MS/MS data. J Proteomics. 2011;74:1842–51. doi: 10.1016/j.jprot.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Yin X, Subramanian S, Hwang SJ, et al. Protein biomarkers of new-onset cardiovascular disease. Arterioscler Thromb Vasc Biol. 2014;34:939–45. doi: 10.1161/ATVBAHA.113.302918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horala A, Swiatly A, Matysiak J, et al. Diagnostic value of serum angiogenesis markers in ovarian cancer using multiplex immunoassay. Int J Mol Sci. 2017;18:123. doi: 10.3390/ijms18010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ClinProTools 3.0 Software for biomarker detection and evaluation User Manual. Bruker Daltonics; Bremen, Germany: 2011. Basics on data preparation model generation and spectra classification; pp. 51–96. [Google Scholar]
- 27.Bleeker S, Moll H, Steyerberg E, et al. External validation is necessary in prediction research: A clinical example. J Clin Epidemiol. 2003;56:826–32. doi: 10.1016/s0895-4356(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 28.Ioannidis JP. A roadmap for successful applications of clinical proteomics. Proteomics Clin Appl. 2011;5:241–47. doi: 10.1002/prca.201000096. [DOI] [PubMed] [Google Scholar]
- 29.Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–99. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- 30.Simpson-Haidaris PJ, Rybarczyk B. Tumors and fibrinogen. The role of fibrinogen as an extracellular matrix protein. Ann N Y Acad Sci. 2001;936:406–25. [PubMed] [Google Scholar]
- 31.Son HJ, Park JW, Chang HJ, et al. Preoperative plasma hyperfbrinogenemia is predictive of poor prognosis in patients with nonmetastatic colon cancer. Ann Surg Oncol. 2013;20:2908–13. doi: 10.1245/s10434-013-2968-8. [DOI] [PubMed] [Google Scholar]
- 32.Sandanayake NS, Camuzeaux S, Sinclair J, et al. Identification of potential serum peptide biomarkers of biliary tract cancer using MALDI MS profiling. BMC Clin Pathol. 2014;14:7. doi: 10.1186/1472-6890-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Li B, Guo T, et al. Application value of mass spectrometry in the differentiation of benign and malignant liver tumors. Med Sci Monit. 2017;23:1636–44. doi: 10.12659/MSM.901064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebert MP, Niemeyer D, Deininger SO, et al. Identification and confirmation of increased fibrinopeptide a serum protein levels in gastric cancer sera by magnet bead assisted MALDI-TOF mass spectrometry. J Proteome Res. 2006;5:2152–58. doi: 10.1021/pr060011c. [DOI] [PubMed] [Google Scholar]
- 35.Jiang HG, Li J, Shi SB, et al. Value of fibrinogen and D-dimer in predicting recurrence and metastasis after radical surgery for non-small cell lung cancer. Med Oncol. 2014;31(7):1–9. doi: 10.1007/s12032-014-0022-8. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Gao J, Ming B, et al. The pretreatment platelet and plasma fibrinogen level correlate with tumor progression and metastasis in patients with pancreatic cancer. Platelets. 2014;25(5):382–87. doi: 10.3109/09537104.2013.827782. [DOI] [PubMed] [Google Scholar]
- 37.Schifferli JA, Ng YC, Peters DK. The role of complement and its receptor in the elimination of immune complexes. N Engl J Med. 1986;315:488–95. doi: 10.1056/NEJM198608213150805. [DOI] [PubMed] [Google Scholar]
- 38.Pio R, Ajona D, Lambris JD. Complement inhibition in cancer therapy. Semen Immunol. 2013;25:54–64. doi: 10.1016/j.smim.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutkowski MJ, Sughrue ME, Kane AJ, et al. Cancer and the complement cascade. Mol Cancer Res. 2010;8:1453–65. doi: 10.1158/1541-7786.MCR-10-0225. [DOI] [PubMed] [Google Scholar]
- 40.Tian WD, Li JZ, Hu SW, et al. Proteomic identification of alpha-2-HS-glycoprotein as a plasma biomarker of hypopharyngeal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;18:9021–31. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao J, Fan YX, Yang Y, et al. Identification of potential plasma biomarkers for esophageal squamous cell carcinoma by a proteomic method. Int J Clin Exp Pathol. 2015;8:1535–44. [PMC free article] [PubMed] [Google Scholar]
- 42.Bertuzzi M, Marelli C, Bagnati R, et al. Plasma clusterin as a candidate pre-diagnosis marker of colorectal cancer risk in the Florence cohort of the European Prospective Investigation into Cancer and Nutrition: A pilot study. BMC Cancer. 2015;15:56. doi: 10.1186/s12885-015-1058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ajona D, Pajares MJ, Corrales L, et al. Investigation of complement activation product C4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst. 2013;105:1385–93. doi: 10.1093/jnci/djt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Broek, Sparidans RW, Schellens JH, Beijinen JH. Quantitative assay for six potential breast cancer biomarker peptides in human serum by liquid chromatography coupled to tandem mass spectrometry. J Chromatogr B. 2010;878:590–602. doi: 10.1016/j.jchromb.2010.01.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Coefficient of variation (CV) values determined for the ion intensities of ten different peaks identified in ten pooled PTC serum samples by MALDI-TOF MS.
| Mass | CV (%) |
|---|---|
| 1546.069 | 24.18 |
| 2167.17 | 18.26 |
| 2663.308 | 23.78 |
| 3886.029 | 23.86 |
| 2955.75 | 22.27 |
| 4056.434 | 14.30 |
| 4093.956 | 18.35 |
| 5338.196 | 22.06 |
| 7759.919 | 18.26 |
| 9276.545 | 24.99 |