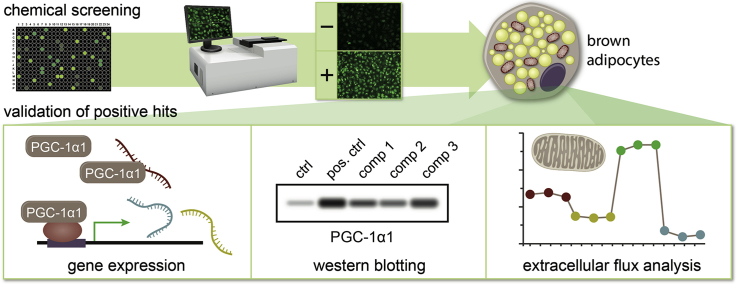
Abstract
Objective
The peroxisome proliferator-activated receptor-γ coactivator-1α1 (PGC-1α1) regulates genes involved in energy metabolism. Increasing adipose tissue energy expenditure through PGC-1α1 activation is potentially beneficial for systemic metabolism. Pharmacological PGC-1α1 activators could be valuable tools in the fight against obesity and metabolic disease. Finding such compounds has been challenging partly because PGC-1α1 is a transcriptional coactivator with no known ligand-binding properties. While, PGC-1α1 activation is regulated by several mechanisms, protein stabilization is a crucial limiting step due to its short half-life under unstimulated conditions.
Methods
We designed a cell-based high-throughput screening system to identify PGC-1α1 protein stabilizers. Positive hits were tested for their ability to induce endogenous PGC-1α1 protein accumulation and activate target gene expression in brown adipocytes. Select compounds were analyzed for their effects on global gene expression and cellular respiration in adipocytes.
Results
Among 7,040 compounds screened, we highlight four small molecules with high activity as measured by: PGC-1α1 protein accumulation, target gene expression, and uncoupled mitochondrial respiration in brown adipocytes.
Conclusions
We identify compounds that induce PGC-1α1 protein accumulation and show that this increases uncoupled respiration in brown adipocytes. This screening platform establishes the foundation for a new class of therapeutics with potential use in obesity and associated disorders.
Keywords: Small molecule screening, PGC-1a, PGC-1alpha, PGC-1alpha1, Protein stabilization, UCP1, Mitochondrial respiration, Brown adipose tissue
Abbreviations: C/EBP, CCAAT-enhancer-binding proteins; FCCP, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone; HEK, human embryonic kidney; HNF, hepatocyte nuclear factor; HPRT, hypoxanthine-guanine phosphoribosyltransferase; IDP, intrinsically disordered protein; MEF2, myocyte enhancer factor 2; NRF2, nuclear respiratory factors 2; PPAR, peroxisome proliferator-activated receptor; ROS, reactive oxygen species; PGC, peroxisome proliferator-activated receptor-γ coactivator; PTM, post-translational modifications; UCP1, uncoupling protein 1; SREBP1, sterol regulatory element-binding protein 1; XBP1, X-box binding protein 1
Graphical abstract
Highlights
-
•
A high-throughput platform to identify PGC-1α1 activators.
-
•
PGC-1α1 protein stabilizers work as activators in brown adipocytes.
-
•
Small molecule PGC-1α1 activators induce Ucp1 expression and cellular respiration.
1. Introduction
Proteins of the peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) family of transcriptional coactivators regulate genes involved in energy metabolism and are expressed in energy-demanding tissues like fat, skeletal muscle, liver, and brain [1]. Interestingly, a single PGC-1α gene can be differently regulated by alternative promoter usage and alternative splicing to generate discrete PGC-1α variants with different biological activities [2], [3], [4]. For example, PGC-1α1, the founding member of the family [5] is a strong regulator of mitochondrial biogenesis and oxidative metabolism, whereas PGC-1α4 regulates muscle mass and strength [2], [3], [5], [6]. In skeletal muscle, induction of PGC-1α1 promotes a more oxidative phenotype, efficient fuel handling, and increases resistance to fatigue [7]. In addition, skeletal muscle PGC-1α1 activates local kynurenine detoxification, thereby positively impacting on mental health [8], [9]. In the brain, PGC-1α1 has been implicated in reactive oxygen species (ROS) detoxification and protection from neurodegenerative disease [10]. However, it is in brown adipose tissue that PGC-1α1 shows the highest expression and has some of its most well-established roles. Initially discovered for its ability to induce mitochondrial biogenesis and adaptive thermogenesis in brown adipocytes, PGC-1α1 has been shown to coordinate the expression of thermogenic genes, of which uncoupling protein 1 (Ucp1) is of central importance [5], [6], [11]. This mechanism is also inducible in white adipose depots, through an adipocyte population called beige, brite, or recruitable (hereon referred to as beige adipocytes) [12], [13], [14], [15]. Moreover, PGC-1α1 expression correlates with typical markers of beige-selective genes in human brown adipose tissue [16]. Therefore, elevated PGC-1α1 levels in adipose tissue could have beneficial effects on systemic metabolism, making PGC-1α1 an interesting target in the treatment of obesity and type 2 diabetes [17], [18]. Genetic deletion of PGC-1α1 (knockout) in adipose tissue perturbs the mitochondrial and thermogenic gene programs rendering mice carrying the knockout alleles more sensitive to high-fat diet-induced insulin resistance [19]. In addition, diabetic humans have reduced PGC-1α levels in adipose tissue, which may contribute to the pathogenesis of metabolic disease [20], [21]. For these reasons, there are great therapeutic promises in finding ways to activate PGC-1α1 and its target genes.
PGC-1α1 activation can be achieved through several mechanisms including gene expression, protein stabilization, and post-translational modifications. Previous attempts to screen for regulators of PGC-1α1 action have focused on identifying inducers of PGC-1α1 gene transcription (potential activators) or of PGC-1α1 acetylation (potential inhibitors) [22], [23], [24], [25], [26]. However, the short protein half-life makes protein stabilization a limiting step in the activation process of PGC-1α1 and downstream target gene transcription [4], [27], [28], [29], [30], [31], [32]. Indeed, PGC-1α1 levels are tightly controlled by several E3 ubiquitin-ligases that target the protein for ubiquitin-proteasome-mediated degradation [27], [29], [30], [33]. Additionally, it has been suggested that PGC-1α1 is an intrinsically disordered protein, which makes it susceptible to default degradation by the 20S proteasome [34].
Here, we report the development of a cell-based high-throughput screening system that allows for the identification of agents that activate PGC-1α1 by increasing protein stability. Using this system, we were able to identify several small molecule PGC-1α1 activators that are biologically active in brown adipocytes. Treatment of adipocytes with select compounds leads to PGC-1α1 protein accumulation, increased Ucp1 expression, and higher mitochondrial respiration rates.
2. Experimental procedures
2.1. Plasmids and cell lines
2.1.1. Establishment of the screening cell line
To generate a pEGFP-C1-mousePGC-1α1 plasmid (containing a neomycin selection cassette), the cDNA for mouse PGC-1α1 (mPGC-1α1) was excised from a previously published pcDNA3.1-Flag-mPGC-1α1 plasmid [2] using XhoI and NotI. The pEGFP-C1 was opened with BglII and the cDNA for mouse PGC-1α1 was subsequently blunt-end ligated downstream of EGFP. To efficiently produce EGFP-PGC-1α1 fusion protein the A in the start codon for PGC-1α1 was point mutated to a G (generating valine instead of methionine) using QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, #200523).
To generate the screening cell line (293-EGFPmPGC-1α1), human embryonic kidney (HEK) 293-T cells were transfected with the pEGFP-C1-mPGC-1α1 plasmid using Lipofectamine® 2000 (Thermo Fisher Scientific). Stable cell lines were generated by culturing cells in the presence of 500 μg/ml G418 (also termed Geneticin®, Thermo Fisher Scientific, #11811). Single cell-derived colonies were obtained using a dilution protocol. Cells were subsequently cultured according to standard protocols for HEK 293 cells in DMEM high glucose (Gibco, #31966021) containing 10% Fetal Bovine Serum (FBS, Sigma–Aldrich, #F7524) and penicillin/streptomycin (pen/strep) (Gibco, #15140122) in the presence of 500 μg/ml G418. Cells were grown at 37 °C in 5% CO2.
2.1.2. Brown adipocyte cell line
Validation of compound screening was done in an immortalized brown preadipocyte line of mouse origin and was a kind gift from Dr. Bruce Spiegelman (Dana-Farber Cancer Institute and Harvard Medical School, Boston, USA) and has been described previously [35]. Preadipocytes were cultured in DMEM high glucose (Gibco, #31966021) containing 20% FBS (Sigma–Aldrich, #F7524), 20 mM Hepes (Gibco, #15630056), and pen/strep (Gibco, #15140122). For differentiation, cells were seeded and grown to confluence in differentiation medium (DMEM high glucose supplemented with 10% FBS, 20 nM insulin, and 1 nM triiodothyronine, T3). Cells were subsequently induced to differentiate with an induction medium (i.e. differentiation medium supplemented with 0.125 mM indomethacin, 0.5 μM dexamethasone and 0.5 mM isobutyl methylxanthine) for 2 days, after which, medium was changed to differentiation medium for 3–4 additional days. Fully differentiated brown adipocytes were exposed to 10 μM of compound or 0.1% DMSO for 8 h. Isoproterenol (10 μM) was included as a positive control (as previously reported [35]). MG132 treated cells were included to control for general proteasome inhibition. Following treatment, cells were used for cellular respirometry or harvested for isolation of protein and RNA. Cells were grown at 37 °C in 5% CO2.
2.2. High-throughput screening, image acquisition and analysis
The compound library used in this study was a collection of compounds from the Enamine Ltd Drug-Like Set and Pharmacological Diversity Set, which constitutes a total of 28,160 compounds. Compounds in the Drug-Like Set are selected using diversity sorting from the combined file of high-throughput screening and historical collections and strictly conform to rules of Lipinski and Veber [36], [37], and do not bear undesired reactive functional groups. Compounds from the Pharmacological Diversity Set are particularly recommended for research on new targets because only biologically relevant chemical space has been searched. Moreover, the design is based on the use of predicted pharmacological properties of compounds. Each compound from Enamine collection was profiled by over 3000 activities. Compounds predicted to be toxic are excluded; the others are clustered by their activities.
293-EGFPmPGC-1α1 cells, stably expressing the EGFP-PGC-1α1 fusion protein, were seeded with laminin (L2020, Sigma–Aldrich) 0.2 μg/cm2 in 384-well optical bottom plates (BD Falcon Optilux #353962 plates) using a Multidrop. Prior to the experiment 50 nl of the library compounds were transferred using an ECHO 550 (Labcyte Inc.) acoustic liquid handler to destination plates and stored at −20 °C. Twenty-four hours after seeding the cells the compound plates were thawed and 20 μl of cell culture medium was added to the plates using a Multidrop. Compound plates were shaken for 1 h at RT after which 20 μl compound solution was transferred to the cell plate using a PerkinElmer Janus 384 MTD system. Cells were incubated in a final concentration 10 μM of each compound, 0.1% DMSO (=negative control) or 10 μM of a proteasome inhibitor, MG132 (=positive control) for 8 h in 37 °C, 5% CO2. After 8 h treatment, cells were fixed in 4% paraformaldehyde and stained with Hoechst 33342 (nuclear staining). Fluorescence was detected in a fully automated system using Operetta® High Content Imaging System (PerkinElmer). Images were captured using a 10× LWD objective in wide field fluorescence mode in 2 fluorescent channels: Ex.360-340/Em.410-480 for Hoechst 33342 and Ex.460-490/Em.500-550 for EGFP. Columbus 2.4 (PerkinElmer) analysis software was used to allocate and segment the cells. The Columbus Find Nuclei Building Block feature was applied to the Hoechst channel images to identify the nuclei that were then used as a seeding point to help Find Cytoplasm Building Block identify cell cytoplasm using EGFP channel. Additionally, Find Spots Building Block was applied to identify EGFP aggregations. Calculate Intensity Properties Building Blocks were used to determine the mean, sum and median EGFP intensity in whole cells as well as in nuclear and cytoplasm regions. The results were exported as mean and median values per well. Z′ factor values for each plate for different quantification parameters were calculated and plotted using Columbus tertiary analysis.
2.3. Identification of active compounds
The values for the following parameters were exported to a text file and then converted into % activation based on negative (0.1% DMSO – 0%) and positive (MG132 – 100%) controls for each plate: Intensity Cell Sum − Median per Well, Intensity Nucleus Sum − Median per Well, Intensity Cytoplasm Sum − Median per Well, Number of Spots − Median per Well. Percent activity was calculated according to the formula:
where MIN refers to the negative controls (DMSO-treated cells) and MAX refers to the positive controls (MG132-treated cells). In addition, Number of Cells was monitored to estimate compound toxicity. The normalized results were imported to TIBCO Spotfire (PerkinElmer) analysis and visualization software. After excluding wells containing less than 1000 cells (2 compounds) and auto fluorescent compounds (1 compound), the mean and standard deviation of % activation (Intensity Cell Sum − Median per Well) for all tested compounds were calculated. The threshold value of mean [sample] + 2 ∗ stdev [sample] was applied to select active compounds. Identified active compounds were then classified according to their EGFP intensity increase in different cell compartments. Raw values of Intensity Cell Sum − Median per Well, Intensity Nucleus Sum − Median per Well, Intensity Cytoplasm Sum − Median per Well, Number of Spots − Median per Well parameters were subjected to hierarchical clustering using TiBCO Spotfire tool (hierarchical agglomerative method) with Euclidean distance and Complete linkage as clustering and Z-score as normalization methods, to generate a heat map and group active compounds.
2.4. Chemical structure analysis
The chemical structures of the primary hits were compared with the structures of previously known compounds in Bioactivity assay data available through PubChem. Compound structures were also run against Pan-assay interference compounds (PAINS) patterns.
2.5. Immunoblotting
Protein extracts were prepared in a modified FLAG lysis buffer (50 mM Tris–HCl pH 7.4, 180 mM NaCl, 1% Triton X-100, 15% glycerol, 1 mM EDTA) supplemented with 1 mM dithiotreitol (DTT) and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Protein extracts were resolved by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and transferred into polyvinylidene difluoride membranes. Membranes were stained by Ponceau S to evaluate transfer efficiency and to control for equal loading. Immunoblotting was performed using a PGC-1α-specific antibody (4C1.3, Calbiochem, Cat. No. ST1202) and a α-tubulin-specific antibody (T6199, Sigma–Aldrich). Protein bands were quantified using a Chemidoc MP system (Biorad) and normalized to α-tubulin.
2.6. Protein stability and half-life
Protein stability was assessed by treating cells (HEK 293 or immortalized brown adipocytes) as described before with 10 μM compound, MG132 or isoproterenol (positive control) followed by protein extraction. The half-life of ectopically expressed Flag-PGC-1α1 and EGFP-PGC-1α1 was determined in HEK 293 cells transfected with either plasmid for 24 h and subsequently treated with 100 μg/ml cycloheximide for 0, 15, 30, 45, 60 or 90 min, after which cells were harvested for protein.
2.7. Cellular respirometry by extracellular flux analysis (Seahorse™)
Mitochondrial respiration was assessed in fully differentiated brown adipocytes treated as described above. Oxygen consumption was measured using the Seahorse XF 24 analyzer (Agilent) as an indication of mitochondrial respiration. The analysis was performed in DMEM pH 7.4, 25 mM glucose, and 1 mM pyruvate as substrate. The oxygen consumption rate (OCR) was measured at baseline and followed by sequential stimulation with oligomycin (1 μM), carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP, 1 μM), and antimycin A (2 μM). Results were presented as OCR pmoles/min/30,000 cells. Subtracting antimycin A-insensitive OCR (non-mitochondrial respiration) from baseline OCR reveals basal mitochondrial respiration. ATP-linked respiration is represented by oligomycin-sensitive OCR. Oligomycin-insensitive rates represent uncoupled respiration (proton leak), and FCCP rates represent maximal respiratory capacity. Experiments were repeated at least 4 times.
2.8. Gene expression analysis
RNA was extracted using Isol RNA lysis reagent (5 PRIME), DNase treated (Invitrogen), and reverse transcribed (Applied Biosystems). Gene expression was measured using Power SYBR Green PCR Master Mix in a Viia 7 Real-Time PCR System (Applied Biosystems) and normalized to hypoxanthine-guanine phosphoribosyltransferase (Hprt). Expression values shown are normalized for Hprt and expressed relative to experimental controls. Primer sequences are listed in Table S1.
Microarray analysis of gene expression was performed using Affymetrix® Mouse Gene 2.1 ST array. Microarray and basic bioinformatics analysis of data, including normalization, annotations, experimental group comparisons with fold change, and p-value calculations, were performed at the Bioinformatics and Gene expression Analysis (BEA) core facility of the Karolinska Institutet. Microarray data were further processed using Ingenuity Pathway Analysis, David (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resource, Venny (http://bioinfogp.cnb.csic.es/tools/venny/) and Venn generator (http://jura.wi.mit.edu/bioc/tools/venn.php) to generate proportional sized Venn diagrams. To predict common regulators of gene expression, microarray data were further analyzed by DiRE (distant regulatory elements of co-regulated genes). The array data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE107630.
2.9. Statistical analysis
Statistical analysis was performed using Graphpad Prism software. Data presented are mean ± SEM. One-way ANOVA was used to analyze statistical differences between multiple groups. Statistical significance was defined as p < 0.05.
3. Results
3.1. A cell-based screening system for PGC-1α1 stabilizers
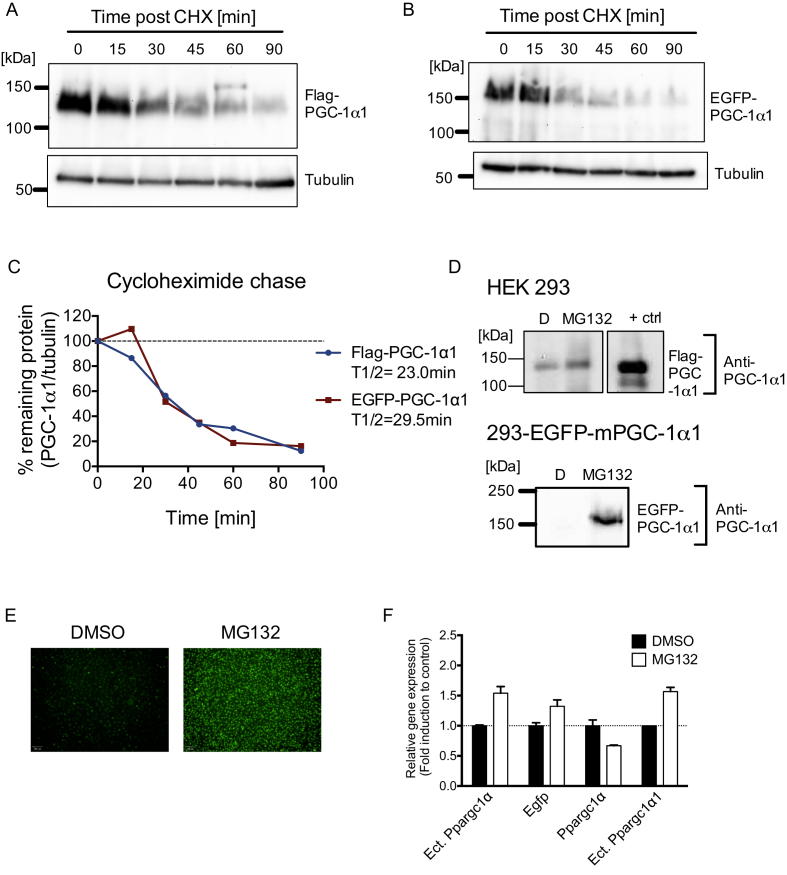
PGC-1α1 has been shown to have a short half-life, which is conserved even upon ectopic expression of the protein in different cells [4], [30]. Using HEK 293 cells, we determined that adding a N-terminal tag (either Flag or EGFP, Enhanced Green Fluorescent Protein) to PGC-1α1 does not significantly affect protein stability (Figure 1A–C). However, to achieve robust expression of the EGFP-PGC-1α1 chimera in HEK 293 cells, it was important to mutate the start codon of PGC-1α1 (Supplementary Figure 1) to prevent expression of the untagged protein. We next generated a HEK 293 stable cell line that expresses EGFP-mPGC-1α1 under a CMV promoter (293-EGFPmPGC-1α1 cell line), at levels that are efficiently degraded by the endogenous protein degradation machinery, but still robustly accumulate upon inhibition of the ubiquitin-proteasome system. Indeed, treatment of the 293-EGFPmPGC-1α1 stable cell line with a proteasome inhibitor (MG132) induced accumulation of the EGFP-PGC-1α1 fusion protein, detectable by both immunoblotting (Figure 1D) and by fluorescence microscopy (Figure 1E). To further validate that MG132-induced EGFP-PGC-1α1 accumulation was due to protein stabilization, and not to effects on the CMV promoter (i.e. increased transcription), we analyzed expression of the EGFP-PGC-1α1 transcript by targeted qRT-PCR using specific probes (Figure 1F). The small increase in ectopic PGC-1α1 gene expression (measured by both isoform specific and unspecific primers) would not account for the robust stabilization of PGC-1α1 protein (Figure 1F,D).
Figure 1.
Developing a platform to screen for activators of PGC-1α1 stabilization. HEK 293 cells were transiently transfected with Flag-PGC-1α1 (A) or EGFP-PGC-1α1 (B) and treated with an inhibitor of translation (cycloheximide, CHX) for various times. PGC-1α1 protein levels at different time points were used to calculate the half-life for Flag-PGC-1α1 and EGFP-PGC-1α1 (A–C). The 293-EGFPmPGC-1α1 stable cell line expressing a EGFP-PGC-1α1 fusion protein was stimulated with 10 μM MG132 to block proteasomal degradation. EGFP-PGC-1α1 was detected by western blot (D) or fluorescence microscopy (E). (F) Gene expression of total PGC-1α (detected as Exon2) or isoform PGC-1α1 in 293-EGFPmPGC-1α1 following an 8 h MG132 (10 μM) treatment. Unless stated otherwise transcripts detected are endogenous (human). Ectopic (ect.) transcripts are of mouse origin. Results shown in panel F are from a representative experiment and displayed as mean ± SEM.
3.2. Identification of small molecule PGC-1α1 stabilizers/activators
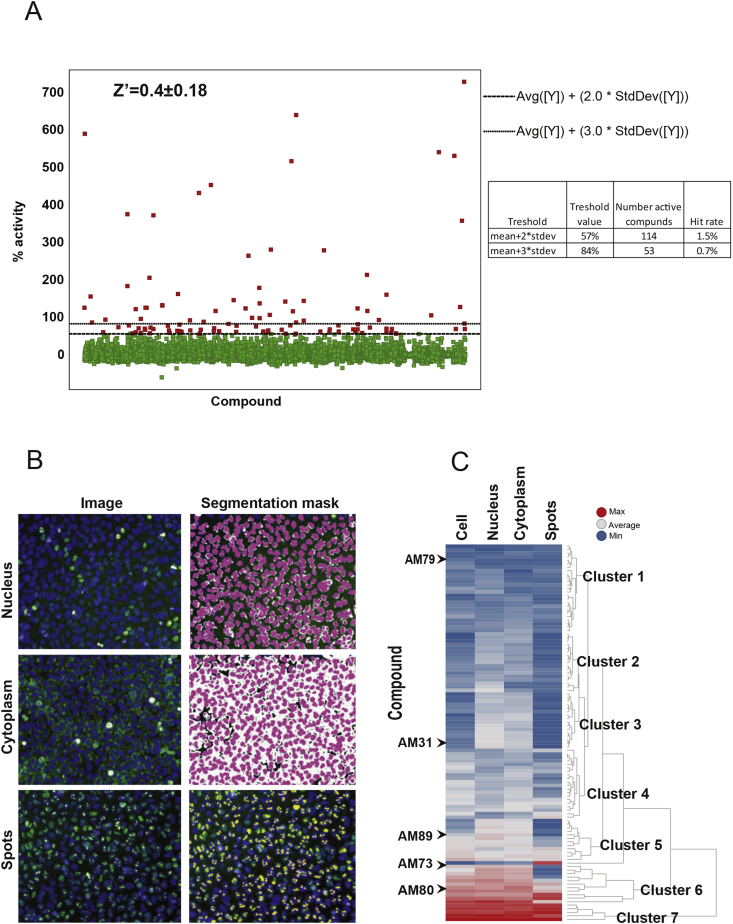
To identify compounds that stabilize PGC-1α1 protein and activate its target genes, we adapted the 293-EGFPmPGC-1α1 cells to grow in 384-well optical-bottom plates, which are suitable for automated cell plating, compound treatment, and fluorescence microscope analysis. For the selection of compounds to screen, we chose a subset of the drug-like and pharmacological diversity sets from Enamine Ltd, which consists of approximately 7040 small molecule compounds (approximately 25% of the library). Cells were then exposed to compounds (or controls) for 8 h to minimize any unspecific activation of the CMV promoter that drives EGFP-PGC-1α1 expression. Changes in fluorescence intensity and localization were detected and quantified using a fully automated high content microscopy system. The well median of integrated intensity measured with whole cell boundary (Cell sum intensity median per well) was used as a preferred parameter + nuclear intensity, giving reasonable Z′ factor values (Z value) and consistent assay window, to calculate percent activation. The cut-off of 57%, equal to the mean of % activation + 2 ∗ standard deviation (SD) was applied to identify active compounds which generated 114 primary hits (Figure 2A). Remaining compounds were classified according to the subcellular localization of the EGFP-PGC-1α1 signal (whole cell, nuclear, cytoplasmic or spots) (Figure 2B). Analysis of the screening output data by similarity in cellular distribution of the EGFP-PGC-1α1 signal (cell sum, nucleus, cytoplasm or punctate) induced by the screening compounds generated 7 clusters (Figure 2C). Clusters included compounds that induced a strong signal in all cellular compartments (clusters 4 and 6) including a nuclear punctate distribution (cluster 7), to compounds that induced a modest subcellular localization (clusters 2, 3 and 5) and weak induction (cluster 1).
Figure 2.
Screening results and cut-off selection. (A) Distribution of EGFP-PGC-1α1 signal and selection of the cut-off value after screening of 7040 compounds (approximately 25% of the library). The mean and standard deviation of % activation (Intensity Cell Sum − Median per Well) for all tested compounds were calculated according to (1 − ((x − mean [MIN])/(mean[MAX] − mean[MIN])) * 100%), where MIN refers to the negative controls (DMSO-treated cells) and MAX refers to the positive controls (MG132-treated cells). A threshold value of mean + 2 ∗ stdev was applied to select active compounds. The table on the right panel shows the number of hits and hit rates with two different cut-offs. (B) Segmentation masks were used to identify where the EGFP-PGC-1α1 signal was located either; nucleus, cytoplasm or aggregation. Example of fluorescence picture on the left and corresponding segmentation mask to the right. (C) Clustering of hits according to similarities in cellular distribution of EGFP-PGC-1α1 signal. Clustering of hits is shown on the right side and five selected compounds chosen for further investigation are shown under their respective cluster.
3.3. Chemical structure analysis
To determine if we could gain any mechanistic insight about the selected compounds or, for example, predict potential toxicity, we compared the chemical structures of our primary hits with the structures of previously known compounds in Bioactivity assay data available through PubChem. Among the 114 primary hits we were able to identify 4 structures tested in previous bioactivity assays and 2 of them (AM72 and AM90) were declared as active but not good hits in their respective assay. However, these 2 compounds were not selected as candidates in our validation assays based on the gene expression and protein stabilization results. Compound structures were also searched for PAINS patterns. The chemical structures of any detected PAINS were then individually analyzed to further evaluate their potential usefulness in further validation process.
3.4. Primary and secondary validation of PGC-1α1 activators using brown adipocytes
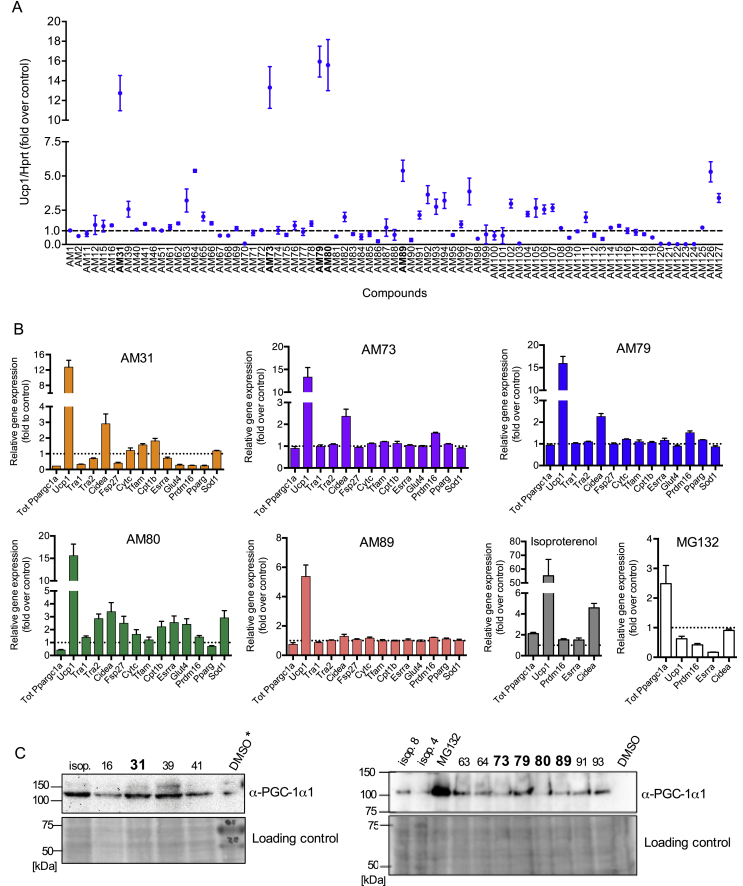
From the primary 114 hits we selected 79 compounds with the highest EGFP-mPGC-1α1 intensities and representing different EGFP-mPGC-1α1 distribution patterns (nucleus, cytosolic and spots) for further validation assays. Primary validation of the selected 79 compounds was performed using a mouse brown preadipocyte cell line, which can be differentiated to mature brown adipocytes with endogenous PGC-1α1 expression. Differentiated brown adipocytes are responsive to beta-adrenergic stimulation (using, for example, isoproterenol) to activate PGC-1α1-driven gene networks [35]. Fully differentiated brown adipocytes were exposed to each compound (10 μM final concentration), or DMSO (negative control), or 10 μM isoproterenol (positive control) for 8 h. MG132 was included to compare compound effects to a general proteasome inhibitor. Primary compound validation was then performed by quantitative real-time PCR (qRT-PCR) analysis of PGC-1α1 target gene activation (Figure 3A). To this end, we determined the expression of known PGC-1α1 target genes involved in browning/thermogenesis, mitochondrial biogenesis, oxidative phosphorylation, and anti-oxidant defenses [5], [6], [10], [38], [39]. This approach was used to exclude compounds with general toxic effects for the cells, including general proteasome inhibitors, which could impact PGC-1α1-induced gene expression and biological effects [40]. Among the genes we evaluated, Ucp1 showed the highest induction after compound treatment (Figure 3A). Among all the validated compounds, 19 compounds (approximately 23% of the selected compounds) induced Ucp1 expression by at least 2-fold with the highest observed induction at 15-fold for compound AM80 (Figure 3A). Treatment of brown adipocytes with AM31, AM73, AM79, and AM80 increased Ucp1 12- to 15-fold, while for AM89 the increase was 5-fold (Figure 3B). In comparison, the positive control isoproterenol induced Ucp1 55-fold. Treatment of brown adipocytes with the general proteasome inhibitor MG132 resulted in a repression of gene expression for most PGC-1α1 target genes (Figure 3B), despite the modest increase in total PGC-1α mRNA expression. AM31, AM73, AM79, and AM80 also induced Cidea expression (Figure 3B). Of the tested compounds, AM80 showed the broadest activity on increasing PGC-1α1 target gene expression in brown adipocytes.
Figure 3.
Hit validation in brown adipocytes identified five compounds that stabilize PGC-1α1 protein and activate Ucp1 transcription. Fully differentiated brown adipocytes were treated for 8 h with a compound (10 μM), a positive control, isoproterenol (10 μM) 4 h or 8 h, MG132 8 h (10 μM), or with a negative control D = DMSO and harvested for RNA and protein. ∗indicates that in that lane a protein molecular size marker was loaded together with the DMSO-treated negative control extract. (A) Ucp1 expression in brown adipocytes for 79 validated compounds. (B) Gene expression panels for five select compounds and for controls, isoproterenol and MG132. (C) PGC-1α1 protein stabilization by western blot for a selection of validated compounds. Compounds that were chosen for further analysis are shown in bold. Isoproterenol 4 h (i4) or 8 h (i8). m, protein molecular size marker. Lower panel of (C) shows Ponceau S stained PVDF membranes. Results in A and B are mean ± SEM of a single experiment.
Secondary hit validation was performed by testing each compound for its ability to stabilize endogenous PGC-1α1 protein in brown adipocytes (Figure 3C and Supplementary Figures 3–6). To determine the maximal PGC-1α1 protein accumulation in brown adipocytes, MG132 was used to block proteasome degradation, and isoproterenol was used as a biologically active positive control. As expected PGC-1α1 stabilization by MG132 treatment was higher than stabilization following treatment with isoproterenol (Figure 3C). Of the tested compounds, 45 induced PGC-1α1 protein accumulation as determined by immunoblotting using whole cell extracts from mature brown adipocytes treated with each compound, and anti-PGC-1α antibodies (Figure 3C and Supplementary Figures 3–6). In summary, 22 of the validated compounds induced Ucp1 expression (>2-fold) and 17 compounds induced both Ucp1 expression and PGC-1α1-protein accumulation (Supplementary Figures 3–6). Of these 17 compounds, we selected five that stabilized PGC-1α1 protein and induced a robust increase in Ucp1 expression (>5-fold). The selected compounds were AM31, AM73, AM79, AM80, and AM89. In fact, when compared to the positive and negative controls, these compounds were able to stabilize endogenous PGC-1α1 to levels comparable to cold-induced brown adipose tissue [2], [41].
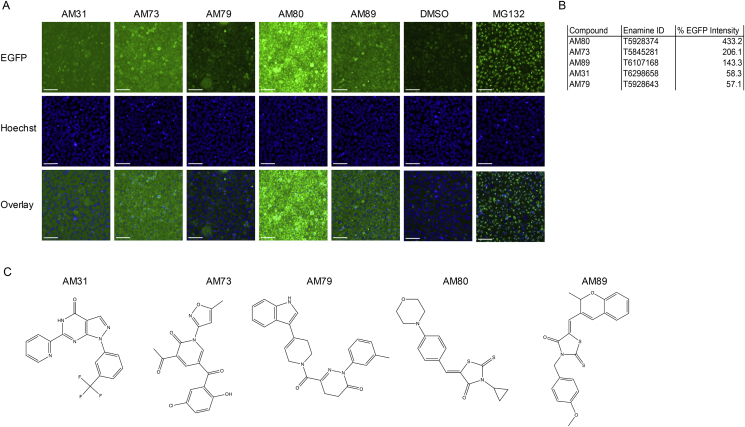
Screening images, EGFP-PGC-1α1 intensities, Enamine IDs and chemical structures for the five selected compounds are shown in Figure 4 A-C. High-resolution images for the selected compounds are shown in Supplementary Figures S7–11.
Figure 4.
Compound structures and screening results (A) Screening images including EGFP, Hoechst and overlay of five selected compounds. Enamine codes, working names and EGFP intensities for the five selected compounds (B). (C) Compound structures for AM31, AM73, AM79, AM80, and AM89. Scale bars indicate 100 μM. Original un-cropped screening images for the five selected compounds are found in Supplementary Figures 7–11.
3.5. PGC-1α1 activators increase mitochondrial respiration in brown adipocytes
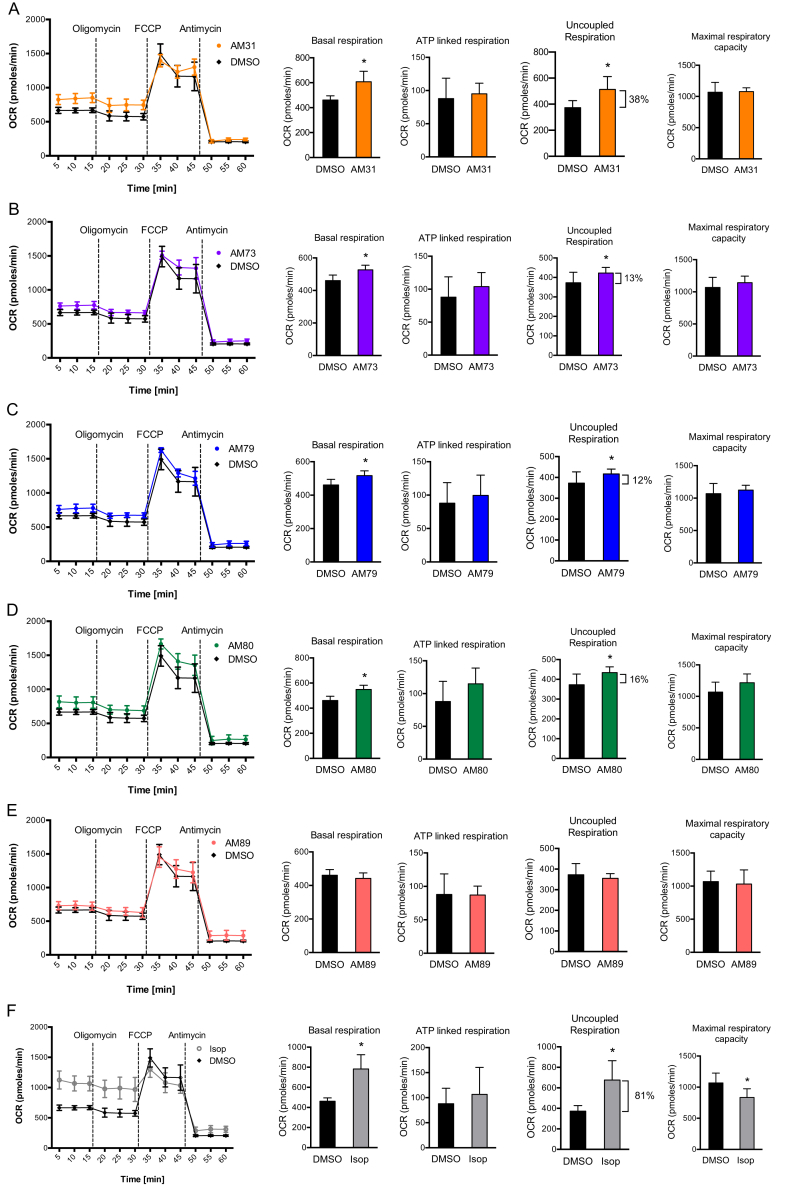
We next measured cellular respiration in brown fat cells treated with AM31, AM73, AM79, AM80, or AM89. In line with the observed increases in Ucp1 expression (Figure 3C), we observed an increase in basal mitochondrial respiration (corrected for non-mitochondrial respiration) for all compounds except AM89 (Figure 5A–F). This increase in basal respiration was maintained even after addition of oligomycin (an inhibitor of ATP synthase), indicating an increase in uncoupled respiration rather than ATP turnover. Only compound AM80 showed a small increased maximal respiration (after addition of FCCP).
Figure 5.
Compound treatment increased basal mitochondrial respiration trough increased proton leak in brown adipocytes. Brown adipocytes were treated for 8 h with 10 μM of AM31 (A), AM73 (B), AM79 (C), AM80 (D), AM89 (E) or 10 μM of isoproterenol (F) after which oxygen consumption rates (OCRs) were measured before and after addition of oligomycin (blocks ATP synthesis), FCCP (=uncouples respiration, leads to maximal respiration) and antimycin (=inhibitor of complex III and of mitochondrial respiration). Data shown is the average ± SEM of four independent experiments. For statistical analysis, each condition (basal/oligomycin/FCCP/antimycin) was analyzed individually. Different groups were analyzed by one-way ANOVA. ∗ indicates a p-value < 0.5.
3.6. Global gene expression analysis of adipocytes treated with PGC-1α1 activators highlights effects on energy metabolism
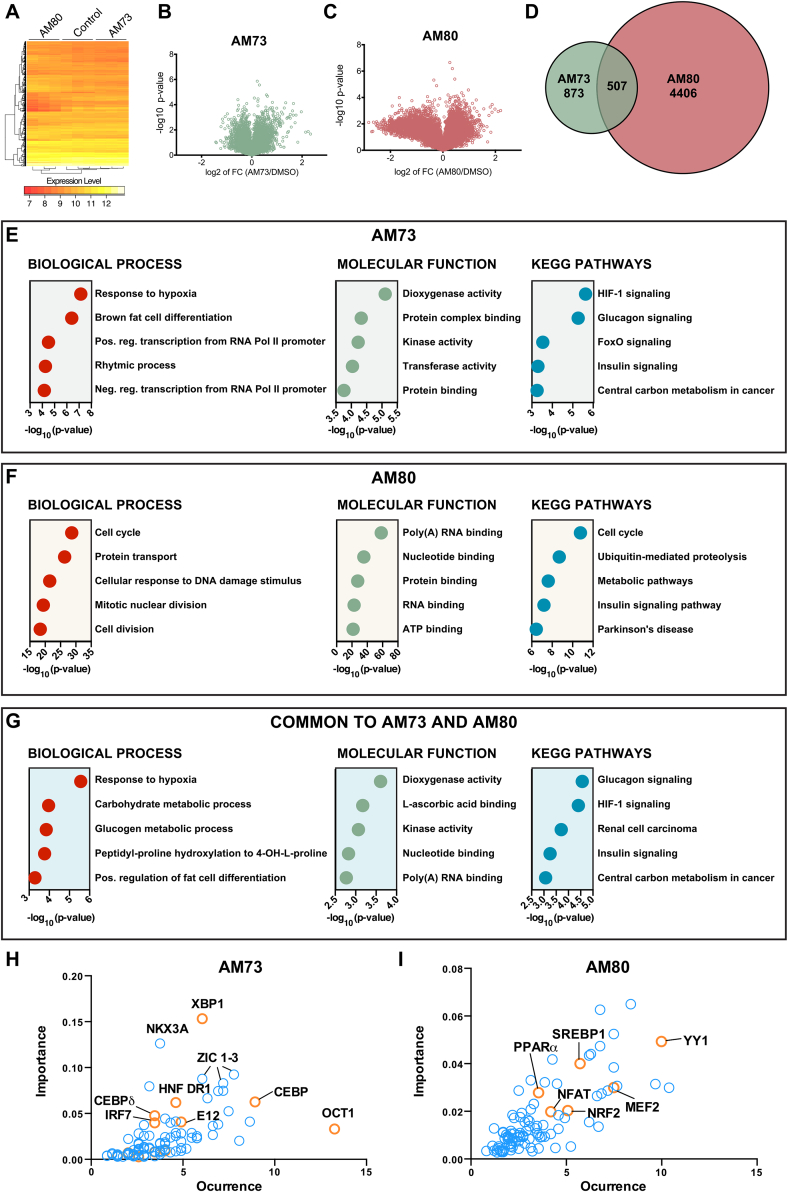
Since PGC-1α1 activation can be achieved through several pathways we decided to explore the mechanisms of action for select compounds. Consequently, we performed a global analysis of gene expression in brown adipocytes treated with AM73 or AM80 for 8 h (data deposited at Gene Expression Omnibus under accession number GSE107630). We selected these two compounds based on their overall profile as PGC-1α1 activators. This included the EGFP-PGC-1α1 intensities from the screening cell line (Figure 4B), the ability to stabilize endogenous PGC-1α1, and to activate PGC-1α1 target genes in brown adipocytes (Figure 3A–C). The general distribution of target gene expression for AM73 and AM80 in brown adipocytes is represented in a heat map and in volcano plots (Figure 6A–C). In this analysis, we identified 1380 genes regulated by AM73 and 4913 regulated by AM80, of which 507 genes were common to both compounds (Figure 6D). To further analyze these results, we looked at the biological processes, molecular functions, and KEGG pathways regulated by either compound alone or by both (Figure 6E–G). Interestingly, AM73-regulated pathways comprised typical PGC-1α1-controlled pathways such as hypoxia, brown fat cell differentiation, carbohydrate metabolism, and insulin signaling (Figure 6E). These pathways were also shared between AM73 and AM80 (Figure 6G). To further analyze the transcriptional networks regulating the genes differentially expressed upon AM73/AM80 treatment, we performed a DiRE (distant regulatory elements of co-regulated genes) analysis. In line with our observations, and further supporting a functional activation of PGC-1α1, we found that regulatory regions surrounding co-regulated genes tended to contain transcription factor binding sites for many previously known PGC-1α1 binding partners, including CCAAT-enhancer-binding proteins (C/EBP), hepatocyte nuclear factor (HNF), X-box binding protein 1 (XBP1), peroxisome proliferator-activated receptor α (PPARα), nuclear respiratory factors 2 (NRF2), myocyte enhancer factor 2 (MEF2), sterol regulatory element-binding protein 1 (SREBP1) (Figure 6H–I). Complete lists of regulatory elements as predicted by DiRE are found in Supplementary Tables 2 and 3.
Figure 6.
Global gene expression analysis identified classical browning and thermogenic pathways regulated by AM73 and AM80. RNA from brown adipocytes treated with 10 μM AM73 or AM80 for 8 h was used for global gene expression analysis by microarray. All data were normalized for a negative control (DMSO). Distribution of genes affected by AM73 or AM80 treatment shown as heat map (A), Volcano plots (B–C) and overlapping genes (D). Biological, molecular and KEGG pathways affected by AM73 (E), AM80 (F), or both (G). The gene regulatory regions for genes affected by either treatment was analyzed by DiRE (distant regulatory regions of co-regulated genes) to identify putative factors regulating the gene expression changes, H–I.
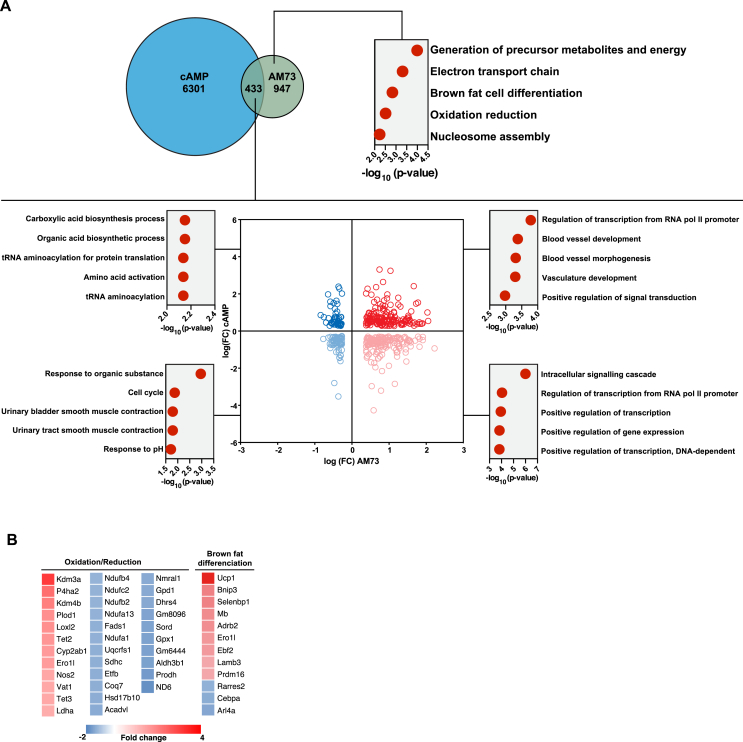
To compare the gene expression changes following AM73/AM80 treatment with those observed for other known browning agents, we used publicly available data from cAMP-treated brown adipocytes (GEO GSE5041 [35] obtained with the same cells used in this study). Cyclic AMP is a second messenger of beta-adrenergic receptor stimulation, a classical inducer of thermogenic programs in brown fat cells [42]. Among 6734 genes regulated by cAMP, 433 were co-regulated by AM73, corresponding to approximately 31% of AM73 targets (Figure 7A). AM80 and cAMP shared 39% of their target genes (Supplementary Figure 12). The pathways regulated by both cAMP and AM73 were further divided (Figure 7A) as induced by both cAMP and AM73 (top right), reduced by both cAMP and AM73 (bottom left), induced by cAMP and reduced by AM73 (top left) or reduced by cAMP and induced by AM73 (bottom right). For example, blood vessel development was induced by both cAMP and AM73. Interestingly, oxidation/reduction and brown fat cell differentiation were among the pathways regulated by AM73 alone (Figure 7A–B). A similar analysis was performed for cAMP and AM80, and is shown in Supplementary Figure S12.
Figure 7.
Comparison of global gene expression changes following cAMP or AM73 treatment of murine brown adipocytes. RNA from brown adipocytes treated with 10 μM AM73 for 8 h was used for global gene expression analysis by microarray. Gene lists for AM73-treated brown adipocytes was compared to previously published results from the same cell line treated with cAMP for 4 h. Overlap of hits between cAMP 4 h versus AM73 (A). Top right panel of A shows Gene Ontology (GO) pathways significantly changed by AM73 alone (excluding genes shared with cAMP). The lower panel shows pathways changed by both cAMP and AM73; both up (upper right) or both down (lower left); or in opposite direction; up with cAMP 4 h and down with AM73 (upper left) or down with cAMP 4 h and up with AM73/AM80 (lower right). (B) Gene lists for the AM73-regulated pathways brown fat cell differentiation and oxidation reduction.
4. Discussion
PGC-1α1 exerts its role as a transcriptional coactivator by binding to a variety of transcription factors (such as nuclear receptors) as well as to other transcription-associated protein complexes such as the Mediator, histone-modifying enzymes, and the splicing machinery [43], [44], [45]. PGC-1α1 does not contain a DNA-binding domain but works as a docking platform for other proteins to promote assembly of the necessary gene regulatory protein complexes [1]. In many cases, the binding partners for PGC-1α1 are expressed in the target cells leaving PGC-1α1 as the limiting factor for target gene activation [19], [35]. The reason for that is the fact that activation of PGC-1α1 is a highly regulated process, that relies heavily on diverting the protein from ubiquitin-proteasome-mediated degradation. In this context, and to explore the possibility of using an unbiased approach to identify new strategies to target PGC-1α1, we designed a cell-based screening system that targets PGC-1α1 activity through increased protein stability. Other previously published PGC-1α1 screening systems have aimed at finding transcriptional activators or inducers of PGC-1α1-acetylation that could lead to PGC-1α1 inhibition in the liver [22], [23], [24], [25].
Using our screening platform, we tested 7040 small molecule compounds and identified 114 primary hits (representing a 1.51% hit rate). Based on the EGFP-PGC-1α1 signal intensity and subcellular distribution pattern we selected 79 compounds for validation assays. Of 79 validated compounds 17 compounds were confirmed true positive hits by PGC-1α1 protein accumulation and target gene activation in a biologically relevant system. To obtain as much information as possible from the screening results, we decided to use more relaxed criteria (i.e. mean + 2 ∗ stdev instead of mean + 3 ∗ stdev, commonly used in high throughput screening) for selecting compounds to be tested in the hit validation stage. However, to screen larger compound sets with this system, elimination of false positives could be further enhanced by a combination of higher cut-off limit (e.g. mean + 3 ∗ stdev) and another round of secondary screening in the same screening cell line. Nevertheless, our platform proved to be stable and reproducible and would easily be transferable to a larger format.
The validation process relied primarily on the analysis of PGC-1α1 target genes involved in non-shivering thermogenesis, using a brown adipocyte cell line that expresses PGC-1α1 endogenously [35]. This was done to preclude any compounds that although stabilizing PGC-1α1 did not lead to target gene activation (due to, for example, cell toxicity). With this approach, we validated 79 compounds and showed that 45 compounds stabilized PGC-1α1, 17 of which also induced Ucp1 expression by at least 2-fold.
One major concern of screening for PGC-1α1 protein stabilizers is identifying candidates that affect general protein turnover, such as proteasome inhibitors. To address this, we performed hit validation in two steps looking at both PGC-1α1 protein stability and target gene activation. Using this approach, we were able to monitor the biological activity of PGC-1α1 thereby increasing the chances of identifying compounds that target PGC-1α1-related pathways. Moreover, determining PGC-1α1 target gene expression in brown adipocytes treated with the proteasome inhibitor MG132 resulted in reduced expression for most genes we analyzed. This gene expression profile is clearly different from the ones we determined for our selected compounds, indicating that none of the tested compounds are general proteasome inhibitors. One additional concern is that our PGC-1α1 activators may inhibit E3 ubiquitin-ligases that are not specific to PGC-1α1. Although such compounds could still be usable, they could also affect the stability of other proteins with possible unwanted effects.
However, the overlap between PGC-1α1 stabilizers and inducers of target gene expression was not complete. For example, several compounds stabilized PGC-1α1 protein but only induced small (<1.5 fold) or no changes in the target gene expression measured in our validation assays. It is possible that these compounds induced stabilization of PGC-1α1 protein by post-translational modifications (PTMs) that target PGC-1α1 to other, yet unknown target genes. In contrast only 5 compounds induced Ucp1 expression without affecting PGC-1α1 protein stability. It is likely that these compounds induced Ucp1 expression through PGC-1α1-independent mechanisms, or that they induce PTMs that lead to more efficient target gene activation without increasing total PGC-1α1 protein.
Along with the changes in gene expression and PGC-1α1 protein accumulation in brown adipocytes, compound AM31, AM73, AM79, and AM80 increased basal respiration measured as oxygen consumption rate. This change was maintained after blocking ATP synthase (following oligomycin addition), suggesting that the effect on basal mitochondrial respiration was primarily due to increased uncoupled respiration, likely related to the increase in Ucp1 expression. The maximal respiratory capacity was slightly increased with compound AM80, which could indicate more mitochondria and/or mitochondrial protein.
Since this chemical compound library subset was selected for structural diversity, thus enhancing the chances of targeting different components of the PGC-1α1 degradation pathway, very little information is available for each hit. We next performed global gene expression analysis of brown adipocytes treated with either AM73 or AM80 (selected for their EGFP intensities and induction of Ucp1 expression) in order to (a) better understand the mechanisms of PGC-1α1 stabilization, and (b) compare the browning/thermogenic signature of these compounds with other known browning agents. By looking at the microarray data and the genes highlighted in the analysis, we can conclude that AM73, and to some extent AM80, regulates pathways involved in brown fat differentiation, response to hypoxia, and carbohydrate metabolism. In our prediction of transcription factors controlling the gene expression changes, we confirmed several known PGC-1α1-binding partners, including C/EBP, XBP1, HNF, PPARα, NRF2, MEF2, and SREBP1. All of these transcription factors are expressed in adipocytes and it is likely that PGC-1α1 is the limiting factor controlling the transcription of their respective target genes. In line with this, PGC-1α1 ablation in either adipose tissue or adipocytes doesn't affect the expression levels of adipogenesis-regulating transcription factors like PPARγ and C/EBPα [19], [35]. However, the absence of PGC-1α1 affects mainly the induction of genes such as Ucp1, Cytc, and Esrra by, for example, β-adrenergic stimuli (e.g. cAMP).
Interestingly, when compared to each other or to cAMP, AM73 and AM80 seem to also have specific gene signatures. This could indicate distinct PGC-1α1 activating mechanisms not related to the classical cAMP-induced pathways. For example, oxidation/reduction and brown fat cell differentiation were pathways regulated by AM73 alone. These pathways included typical browning genes, e. g. Ucp1, Prdm16, and Ebf2, as well as Kdm3a (also termed Jmjd1a or Jhdm2a), an H3K9-specific demethylase that regulates Ppara and Ucp1 expression [46]. Also induced by AM73 and included in the oxidation/reduction pathway are Kdm4b (lysine demethylase 4B) and Tet2 (tet methylcytosine dioxygenase 2). Kdm4b and Tet2 are both involved in adipogenesis by regulation of C/EBPβ activity and the PPARγ locus, respectively. Overall, the global gene expression profile for AM73 indicated a regulation of pathways involved in browning and thermogenesis distinct from β-adrenergic signaling.
Together with the effect of AM73 on Ucp1 expression and uncoupled respiration, the global gene expression changes suggested activation of thermogenic processes in brown adipocytes. Previous studies show that overexpression of human PGC-1α1 induces brown fat features in human white adipocyte [11]. It is therefore tempting to speculate that AM73 could also be used to activate thermogenic processes in white adipocytes. Moreover, it is conceivable that co-treatment with classical browning factors, e.g. low-dose isoproterenol, could accelerate the thermogenic effects of the compounds in white adipocytes. Finally, a careful titration of each compound, in combination or not with other browning agents, could potentially result in stronger effects than the ones we report here.
The exact mechanisms of action for the compounds are still unclear. Several reports have shown that PGC-1α1 stability can be controlled by PTMs such as phosphorylation [47], [48], [49], [50], [51], [52], acetylation [53] and deacetylation [54], [55], methylation [56], SUMOylation [57] or O-GlcNAcylation [28]. Many of these PTMs impact the interaction with binding partners or increase the association of PGC-1α1 with E3 ligases, eg Skp1/Cullin/F-box-cell division control 4 (SCFCdc4) and Ring-finger-containing protein 34 (RNF34), thereby targeting the protein for proteasomal degradation [27], [29]. PGC-1α1 stability is also proposed to be governed by the N-terminal pathway [30]. Yet, our N-terminal Flag- and EGFP-PGC-1α1 fusion proteins did not significantly differ in their stability compared to previous observations [30], [58], suggesting that other proteasome targeting pathways might be more dominant. Additionally, the compounds could act on the protein itself. PGC-1α1 has been suggested to be an intrinsically disordered protein. These are proteins that lack a stable three-dimensional structure and rapidly cycle between different conformations [59]. Interaction with small compounds could theoretically stabilize a conformation making the protein more stable and less susceptible for degradation [60].
One concern with systemic PGC-1α1 activation with a small molecule such as the compounds that we identify in this study is that higher PGC-1α1 activity in the liver could induce glucose output, an undesirable effect in an obesity or type 2 diabetes setting. However, the specificity of PGC-1α1 action in different tissues critically depends on the transcription factors present in those cells and on the post-translational status of PGC-1α1 itself. For example, in hepatocytes PGC-1α1 modification through phosphorylation by S6kinase attenuates its ability to enhance gluconeogenic gene transcription but does not interfere with the PGC-1α1-driven activation of mitochondrial and fatty acid oxidation pathways [50]. Moreover, AMPK activation in skeletal muscle drives the phosphorylation and subsequent activation of PGC-1α1 whereas AMPK activation in hepatocytes blocks fasting-induced PGC-1α1 trough TORC2 and CREB [49]. These tissue-specific regulatory pathways clearly demonstrate that the outcome of PGC-1α1 activation will depend not only on the stability of the protein but also on the post-translational modifications of PGC-1α1 itself as well as the presence of specific transcription factors. Although this complicates the prediction of activating PGC-1α1 systemically, it also highlights the possibility of activating a specific gene network (e.g. mitochondrial genes and fatty acid oxidation) while leaving other pathways unaffected (e.g. gluconeogenesis). In addition, further development of these compounds through medical chemistry could avoid targeting the liver, and therefore reduce any unwanted effects on hepatic glucose output.
In addition to metabolic disease, PGC-1α1 activation could potentially be useful in other diseases as well. These include neurodegenerative disorders, mood, and especially muscle atrophy, for which there are still no effective treatments [7], [8], [61]. Muscle-specific expression of PGC-1α1 leads to the enrichment of type I fibers in transgenic mice and to a more oxidative phenotype [7] that protects these muscles from atrophy [61]. PGC-1α1 also drives and maintains the function of synapses in hippocampal dendritic spines [62] and neuromuscular junctions [63]. Furthermore, higher expression of PGC-1α1 in muscle protects the brain from high levels of kynurenine, a tryptophan metabolite that induced stress responses in the brain and has been linked to depression [8].
In summary, by developing a platform to screen for inducers of PGC-1α1 protein accumulation and activity, we have identified four compounds with the ability to stabilize PGC-1α1 protein, activate Ucp1 expression and induce uncoupled respiration in brown adipocytes. The specific platform development allows for easy adaptation to other screens, such as siRNA or other biologicals. This study sets the foundation for a novel generation of therapeutics based on the activation of PGC-1α1 that could be of use in metabolic disease and other pathologies.
Acknowledgments
This work was supported by grants from Karolinska Institutet (J.L.R), The Swedish Research Council (J.L.R.), The Novo Nordisk Foundation (J.L.R.), The Strategic Research Programs in Diabetes (J.L.R.) and in Regenerative Medicine (J.L.R.) at Karolinska Institutet, The Swedish Diabetes Association (J.L.R.). During part of this study A.T.P-K and J.C.C. were recipients of postdoctoral fellowships from the Swedish Society for Medical Research and D.M.S.F. and V.M.-R. were recipients of postdoctoral fellowship from the Wenner-Gren Foundation. We would like to thank the core facility at Novum, BEA, Bioinformatics and Expression Analysis, which is supported by the board of research at the Karolinska Institute and the research committee at the Karolinska hospital.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.01.017.
Conflict of interest
None.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Correia J.C., Ferreira D.M., Ruas J.L. Intercellular: local and systemic actions of skeletal muscle PGC-1s. Trends in Endocrinology and Metabolism. 2015;26:305–314. doi: 10.1016/j.tem.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Ruas J.L., White J.P., Rao R.R., Kleiner S., Brannan K.T., Harrison B.C. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151:1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Redondo V., Pettersson A.T., Ruas J.L. The hitchhiker's guide to PGC-1alpha isoform structure and biological functions. Diabetologia. 2015;58:1969–1977. doi: 10.1007/s00125-015-3671-z. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Redondo V., Jannig P.R., Correia J.C., Ferreira D.M., Cervenka I., Lindvall J.M. Peroxisome proliferator-activated receptor gamma coactivator-1alpha isoforms selectively regulate multiple splicing events on target genes. Journal of Biological Chemistry. 2016 doi: 10.1074/jbc.M115.705822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 6.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 7.Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 8.Agudelo L.Z., Femenia T., Orhan F., Porsmyr-Palmertz M., Goiny M., Martinez-Redondo V. Skeletal muscle PGC-1alpha1 modulates kynurenine metabolism and mediates resilience to stress-induced depression. Cell. 2014;159:33–45. doi: 10.1016/j.cell.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 9.Cervenka I., Agudelo L.Z., Ruas J.L. Kynurenines: Tryptophan's metabolites in exercise, inflammation, and mental health. Science. 2017;357 doi: 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- 10.St-Pierre J., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jager S. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 11.Tiraby C., Tavernier G., Lefort C., Larrouy D., Bouillaud F., Ricquier D. Acquirement of brown fat cell features by human white adipocytes. Journal of Biological Chemistry. 2003;278:33370–33376. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- 12.Kajimura S., Spiegelman B.M., Seale P. Brown and beige fat: physiological roles beyond heat generation. Cell Metabolism. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrovic N., Walden T.B., Shabalina I.G., Timmons J.A., Cannon B., Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. Journal of Biological Chemistry. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walden T.B., Hansen I.R., Timmons J.A., Cannon B., Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. American Journal of Physiology. Endocrinology and Metabolism. 2012;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- 15.Wu J., Bostrom P., Sparks L.M., Ye L., Choi J.H., Giang A.H. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharp L.Z., Shinoda K., Ohno H., Scheel D.W., Tomoda E., Ruiz L. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS ONE. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komen J.C., Thorburn D.R. Turn up the power – pharmacological activation of mitochondrial biogenesis in mouse models. British Journal of Pharmacology. 2014;171:1818–1836. doi: 10.1111/bph.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svensson K., Handschin C. Modulation of PGC-1alpha activity as a treatment for metabolic and muscle-related diseases. Drug Discovery Today. 2014;19:1024–1029. doi: 10.1016/j.drudis.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Kleiner S., Mepani R.J., Laznik D., Ye L., Jurczak M.J., Jornayvaz F.R. Development of insulin resistance in mice lacking PGC-1alpha in adipose tissues. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9635–9640. doi: 10.1073/pnas.1207287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Enerback S., Smith U. Reduced expression of FOXC2 and brown adipogenic genes in human subjects with insulin resistance. Obesity Research. 2003;11:1182–1191. doi: 10.1038/oby.2003.163. [DOI] [PubMed] [Google Scholar]
- 21.Semple R.K., Crowley V.C., Sewter C.P., Laudes M., Christodoulides C., Considine R.V. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1alpha is reduced in the adipose tissue of morbidly obese subjects. International Journal of Obesity and Related Metabolic Disorders. 2004;28:176–179. doi: 10.1038/sj.ijo.0802482. [DOI] [PubMed] [Google Scholar]
- 22.Arany Z., Wagner B.K., Ma Y., Chinsomboon J., Laznik D., Spiegelman B.M. Gene expression-based screening identifies microtubule inhibitors as inducers of PGC-1alpha and oxidative phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4721–4726. doi: 10.1073/pnas.0800979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharabi K., Lin H., Tavares C.D., Dominy J.E., Camporez J.P., Perry R.J. Selective chemical inhibition of PGC-1alpha gluconeogenic activity ameliorates type 2 diabetes. Cell. 2017;169:148–160. doi: 10.1016/j.cell.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L.N., Zhou H.Y., Fu Y.Y., Li Y.Y., Wu F., Gu M. Novel small-molecule PGC-1alpha transcriptional regulator with beneficial effects on diabetic db/db mice. Diabetes. 2013;62:1297–1307. doi: 10.2337/db12-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz M., Courilleau D., Jullian J.C., Fortin D., Ventura-Clapier R., Blondeau J.P. A cardiac-specific robotized cellular assay identified families of human ligands as inducers of PGC-1alpha expression and mitochondrial biogenesis. PLoS ONE. 2012;7:e46753. doi: 10.1371/journal.pone.0046753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y., Dominy J.E., Choi Y.J., Jurczak M., Tolliday N., Camporez J.P. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature. 2014;510:547–551. doi: 10.1038/nature13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson B.L., Hock M.B., Ekholm-Reed S., Wohlschlegel J.A., Dev K.K., Kralli A. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes & Development. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan H.B., Han X., Li M.D., Singh J.P., Qian K., Azarhoush S. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metabolism. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei P., Pan D., Mao C., Wang Y.X. RNF34 is a cold-regulated E3 ubiquitin ligase for PGC-1alpha and modulates brown fat cell metabolism. Molecular and Cellular Biology. 2012;32:266–275. doi: 10.1128/MCB.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trausch-Azar J., Leone T.C., Kelly D.P., Schwartz A.L. Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1{alpha} via the N-terminal pathway. Journal of Biological Chemistry. 2010;285:40192–40200. doi: 10.1074/jbc.M110.131615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trausch-Azar J.S., Abed M., Orian A., Schwartz A.L. Isoform-specific SCF(Fbw7) ubiquitination mediates differential regulation of PGC-1alpha. Journal of Cellular Physiology. 2015;230:842–852. doi: 10.1002/jcp.24812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano M., Tokudome S., Shimizu N., Yoshikawa N., Ogawa C., Shirakawa K. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor gamma coactivator 1alpha. Journal of Biological Chemistry. 2007;282:25970–25980. doi: 10.1074/jbc.M703634200. [DOI] [PubMed] [Google Scholar]
- 33.Sen N., Satija Y.K., Das S. PGC-1alpha, a key modulator of p53, promotes cell survival upon metabolic stress. Molecular Cell. 2011;44:621–634. doi: 10.1016/j.molcel.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 34.Adamovich Y., Shlomai A., Tsvetkov P., Umansky K.B., Reuven N., Estall J.L. The protein level of PGC-1alpha, a key metabolic regulator, is controlled by NADH-NQO1. Molecular and Cellular Biology. 2013;33:2603–2613. doi: 10.1128/MCB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 37.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. Journal of Medicinal Chemistry. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 38.Hallberg M., Morganstein D.L., Kiskinis E., Shah K., Kralli A., Dilworth S.M. A functional interaction between RIP140 and PGC-1alpha regulates the expression of the lipid droplet protein CIDEA. Molecular and Cellular Biology. 2008;28:6785–6795. doi: 10.1128/MCB.00504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael L.F., Wu Z., Cheatham R.B., Puigserver P., Adelmant G., Lehman J.J. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou W., Slingerland J.M. Links between oestrogen receptor activation and proteolysis: relevance to hormone-regulated cancer therapy. Nature Reviews. Cancer. 2014;14:26–38. doi: 10.1038/nrc3622. [DOI] [PubMed] [Google Scholar]
- 41.Chang J.S., Fernand V., Zhang Y., Shin J., Jun H.J., Joshi Y. NT-PGC-1alpha protein is sufficient to link beta3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis. Journal of Biological Chemistry. 2012;287:9100–9111. doi: 10.1074/jbc.M111.320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connolly E., Nanberg E., Nedergaard J. Norepinephrine-induced Na+ influx in brown adipocytes is cyclic AMP-mediated. Journal of Biological Chemistry. 1986;261:14377–14385. [PubMed] [Google Scholar]
- 43.Wallberg A.E., Yamamura S., Malik S., Spiegelman B.M., Roeder R.G. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Molecular Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 44.Li S., Liu C., Li N., Hao T., Han T., Hill D.E. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metabolism. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monsalve M., Wu Z., Adelmant G., Puigserver P., Fan M., Spiegelman B.M. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Molecular Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 46.Tateishi K., Okada Y., Kallin E.M., Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puigserver P., Rhee J., Lin J., Wu Z., Yoon J.C., Zhang C.Y. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Molecular Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 48.Li X., Monks B., Ge Q., Birnbaum M.J. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 49.Jager S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lustig Y., Ruas J.L., Estall J.L., Lo J.C., Devarakonda S., Laznik D. Separation of the gluconeogenic and mitochondrial functions of PGC-1{alpha} through S6 kinase. Genes & Development. 2011;25:1232–1244. doi: 10.1101/gad.2054711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodgers J.T., Haas W., Gygi S.P., Puigserver P. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metabolism. 2010;11:23–34. doi: 10.1016/j.cmet.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson R.M., Barger J.L., Edwards M.G., Braun K.H., O'Connor C.E., Prolla T.A. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lerin C., Rodgers J.T., Kalume D.E., Kim S.H., Pandey A., Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metabolism. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 54.Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} Journal of Biological Chemistry. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 55.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 56.Teyssier C., Ma H., Emter R., Kralli A., Stallcup M.R. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes & Development. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rytinki M.M., Palvimo J.J. SUMOylation attenuates the function of PGC-1alpha. Journal of Biological Chemistry. 2009;284:26184–26193. doi: 10.1074/jbc.M109.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinez-Redondo V., Jannig P.R., Correia J.C., Ferreira D.M., Cervenka I., Lindvall J.M. Peroxisome proliferator-activated receptor gamma Coactivator-1 alpha isoforms selectively regulate multiple splicing events on target genes. Journal of Biological Chemistry. 2016;291:15169–15184. doi: 10.1074/jbc.M115.705822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 60.Neira J.L., Bintz J., Arruebo M., Rizzuti B., Bonacci T., Vega S. Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Scientific Reports. 2017;7:39732. doi: 10.1038/srep39732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandri M., Lin J., Handschin C., Yang W., Arany Z.P., Lecker S.H. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng A., Wan R., Yang J.L., Kamimura N., Son T.G., Ouyang X. Involvement of PGC-1alpha in the formation and maintenance of neuronal dendritic spines. Nature Communications. 2012;3:1250. doi: 10.1038/ncomms2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mills R., Taylor-Weiner H., Correia J.C., Agudelo L.Z., Allodi I., Kolonelou C. Neurturin is a PGC-1alpha1-controlled myokine that promotes motor neuron recruitment and neuromuscular junction formation. Molecular Metabolism. 2017 doi: 10.1016/j.molmet.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.