Abstract
Cell alignment in muscle, nervous tissue, and cartilage is requisite for proper tissue function; however, cell sheeting techniques using the thermosensitive polymer poly(N-isopropyl acrylamide) (PNIPAAm) can only produce anisotropic cell sheets with delicate and resource-intensive modifications. We hypothesized that electrospinning, a relatively simple and inexpensive technique to generate aligned polymer fibers, could be used to fabricate anisotropic PNIPAAm and poly(caprolactone) (PCL) blended surfaces that both support cell viability and permit cell sheet detachment via PNIPAAm dissolution. Aligned electrospun PNIPAAm/PCL fibers (0%, 25%, 50%, 75%, 90%, and 100% PNIPAAm) were electrospun and characterized. Fibers ranged in diameter from 1–3 μm, and all fibers had an orientation index greater than 0.65. Fourier transform infrared spectroscopy was used to confirm the relative content of PNIPAAm and PCL. For advancing water contact angle and mass loss studies, only high PNIPAAm-content fibers (75% and greater) exhibited, temperature-dependent properties like 100% PNIPAAm fibers, whereas 25% and 50% PNIPAAm fibers behaved similarly to PCL-only fibers. 3T3 fibroblasts seeded on all PNIPAAm/PCL fibers had high cell viability and spreading except for the 100% PNIPAAm fibers. Cell sheet detachment by incubation with cold medium was successful only for 90% PNIPAAm fibers, which had a sufficient amount of PCL to allow cell attachment and spreading but not enough to prevent detachment upon PNIPAAm dissolution. This study demonstrates the feasibility of using anisotropic electrospun PNIPAAm/PCL fibers to generate aligned cell sheets that can potentially better recapitulate anisotropic architecture to achieve proper tissue function.
Introduction
Cell alignment, which can influence developmental and physiological processes, is driven by biophysical cues, particularly matrix nanotopography.1 To better mimic native cell microenvironments in vitro, techniques to generate surface anisotropy, from electrospinning fibers2,3 to micropatterning to photolithography, have been widely developed and applied to anisotropic tissues, namely muscle and nervous tissue. Both micropatterning and photolithography can be used to design surfaces with high resolution but are resource-intensive, requiring microprinters and clean rooms.4 Electrospinning, on the other hand, is a relatively simple and inexpensive technique to fabricate polymer nano- and micro-fibers that can be seeded with cells.5,6 Electrospun fibers are extracellular matrix-mimicking in that they provide a 3-dimensional fibrous micro-environment. Aligned electrospun fibers have been successful as scaffolds for generating nervous and beating cardiac tissues that can be implanted into animal models.7,8 These transplants, however, are rather limited in that they are only 1–2 cell layers thick, which can severely limit function depending on tissue type, and that the material component may elicit a host response upon implantation.9
Cell sheeting is a technique to generate biomaterial-free, tissue-like constructs for transplant. Teruo Okano pioneered the re-purposing of thermosensitive poly(N-isopropyl-acrylamide) (PNIPAAm) as a surface coating to enable cell sheeting in vitro.10 PNIPAAm undergoes a rapid coil-to-globule transition at its lower critical solution temperature (LCST) of 32 °C that determines how the hydrophilic and hydrophobic domains interact with water. Below 32 °C, PNIPAAm readily dissolves in water; above 32 °C, PNIPAAm's hydrophilic domains are sequestered and PNIPAAm precipitates in aqueous solutions.11 Thus, for cells grown on PNIPAAm-grafted tissue culture plates, cell sheet detachment is possible when the incubation temperature is lowered below the LCST: PNIPAAm expands, forcing the cell sheet to detach without perturbing cell–cell and cell–ECM adhesions. Using this technique, cells sheets have been generated for transplantation to the heart, cornea, and kidney.12,13 Yet generating aligned cell sheets has been challenging. Although grafting hydrophilic domains to PNIPAAm-grafted plates spatially controls cell attachment, leading to cell alignment, this approach required chemical synthesis and photolithography patterning.14,15
Here, we describe the use of electrospun PNIPAAm and poly (ε-caprolactone) (PCL) blended fibers (denoted PNIPAAm/PCL fibers) to generate anisotropic cell sheets. PCL is a common polymer for electrospinning, and electrospun PCL fibers have been successfully used as cell culture scaffolds.16–18 Due to PNIPAAm's thermo-sensitive nature, we hypothesized that fibers would need to contain enough PNIPAAm to enable detachment of cell sheets yet contain a sufficient amount of PCL to ensure proper cell attachment. While PNIPAAm has been successfully electrospun,19 it has primarily been used for drug delivery20,21 and as sacrificial fibers to negatively pattern hydrogels.22 Only one previous report described the use of electrospun PNIPAAm fibers for cell culture, which required chemical modification of PNIPAAm to enable gelatin grafting.23 Our method does not require chemical modification or resource-intensive techniques, thus saving time and expense, and has the potential to generate tissue-specific, aligned cell sheets for transplant studies.
Materials & methods
Fabrication PNIPAAm/PCL fibers
For PNIPAAm-only fibers, PNIPAAm (300 000 Da, Scientific Polymer Products, Ontario, NY) was dissolved 20% (wt/v) in methanol (Fisher Chemical, Pittsburgh, PA), as previously described.22 For PCL-only fibers, PCL (80 000 Da, Sigma-Aldrich, St Louis, MO) was dissolved 10% (wt/v) in hexafluoro-isopropanol (Sigma-Aldrich). For PNIPAAm/PCL fibers, PNIPAAm and PCL (at ratios of 9 :1, 3: 1, 1:1, and 1: 3, respectively) were dissolved 12–18% (wt/v) in a 1 : 3 mixture of methanol and chloroform (Sigma-Aldrich). All polymer solutions were dissolved by continuous stirring until clear and homogenous. To electrospin, a syringe pump (New Era Pump Systems, Inc.) was used to dispense the polymer solutions from a 10 mL syringe with a 25 G blunted stainless steel needle at 2.0 mL h−1 for the PCL-only solution and 1.0 mL h−1 for all PNIPAAm-containing solutions. A high voltage supply (Gamma High Voltage, Ormond Beach, FL) was used to apply a charge of 5–15 kV (optimal charge determine for each solution, Table S1†) to the needle to initiate jet formation. Fibers were deposited on a rotating grounded 7.6 cm diameter aluminum collector. To obtain aligned fibers, the collector was rotated at 2500–3200 rotations per minute (RPM, approximately 10.0–12.8 m s−1). The working distance from the needle and to collector was set at 11 cm. PNIPAAm/PCL fibers are referred to by the percent PNIPAAm content (i.e., 75% PNIPAAm fibers consist of 75% PNIPAAm and 25% PCL). Similarly, “high PCL-content” refers to 0%, 25%, and 50% PNIPAAm and “high PNIPAAm-content” refers to 75%, 90%, and 100% PNIPAAm.
Characterization of PNIPAAm/PCL fibers
Fiber orientation and diameter
PNIPAAm/PCL fibers were sputter coated with 12 nm platinum/palladium and imaged using a Zeiss Supra 40VP scanning electron microscope (SEM, 5 kV). SEM images (n = 9) were analyzed using NIH ImageJ software, specifically the OrientationJ24,25 and DiameterJ26 plug-ins to determine fiber orientation and diameter, respectively. The fiber orientation index, S, was calculated from angle distribution histograms using the following equation:27
where α is the difference between an individual fiber angle and the mean angle of all fibers. S varies from 0 to 1, for perfectly random and perfectly aligned fibers, respectively.
Fourier transform infrared spectroscopy
Fourier-transform infrared (FTIR) attenuated total reflectance (ATR) was used to verify fiber polymer composition. Spectra of dry PNIPAAm/PCL fibers was collected over a range of wavelengths (400 cm−1 to 3000 cm−1) at a resolution of 2 cm−1 using a Thermo Scientific Nicolet iS10 FT-IR spectrometer (Waltham, MA). Background spectra was collected prior to each individual sample.
PNIPAAm mass loss
PNIPAAm/PCL fibers were cut (approximately 1 cm × 1 cm, n = 3) and weighed before being immersed in ultrapure water. To ensure complete PNIPAAm dissolution, fibers were rinsed 3 times in 2 mL of water over 24 hours. Fibers were then dried under vacuum for 48 hours before measuring their final weight. Percent mass lost was determined by subtracting the final weight from the original weight. Fibers were imaged before and after rinsing, and original and post-dissolution areas were calculated in ImageJ. Percent contraction was determined by dividing the final area by the original area.
Advancing contact angle measurement
Advancing contact angles were measured using a FTA-200 goniometer (First Ten Angstroms, Portsmouth, VA) to determine the relative hydrophobicity of dry (non-wetted) and wetted PNIPAAm/PCL fibers. Fibers were cut into squares (approximately 1.75 cm × 1.75 cm, n = 3). Wetted PNIPAAm/PCL fiber squares were secured in CellCrown inserts and rinsed in water warmed to 37 °C for 24 hours. Fibers were then dried in a vacuum oven at 35–55°, above the LCST of PNIPAAm and below the melting temperature. PNIPAAm/PCL fibers were placed on a heating platform to maintain the temperature between 32–60 °C, as measured by an infrared thermometer. Briefly, a drop of purified water was deposited at 0.8 μL per second from a 10 mL syringe on the PNIPAAm/PCL fibers, and high resolution images were subsequently captured. When possible, the contact angle was determined in the sessile drop session mode in the instrument-associated software. Otherwise, advancing contact angle was calculated by manually defining the location of the fiber plane and the drop's curvature.
Cell studies
All cell studies were performed using NIH 3T3 fibroblasts purchased from American Type Culture Collection (Manassas, VA), passages 15–25. 3T3 fibroblasts were maintained in 10% fetal bovine serum (ThermoFisher) and 1% penicillin/streptomycin (ThermoFisher) in low glucose Dulbecco's Modified Eagle's Medium with l-glutamine (Sigma-Aldrich). 3T3 fibroblasts were passaged using trypsin/EDTA (Sigma-Aldrich).
PNIPAAm/PCL fiber sterilization and protein coating
PNIPAAm/PCL fibers and parafilm were cut into squares (1.75 cm × 1.75 cm or 2.5 cm × 2.5 cm) and sterilized by ultraviolet light, 30 minutes each side. Fibers were secured using 12-well or 24-well CellCrown inserts (Scaffdex, Tampere, Finland) and parafilm (Bemis, Oshkosh, WI)—effectively making stand-alone PNIPAAm/PCL fiber bottom wells (Fig. S1f)—to improve handling and prevent PNIPAAm fiber contraction upon wetting. Prior to coating, fibers were wetted with Dulbecco's Phosphate Buffered Saline (DPBS). Fibers were then coated with a 1:50 dilution of Growth-factor Reduced (GFR)-Matrigel (Corning, Corning, NY) in DMEM. Fibers were also pre-treated with fetal bovine serum (FBS, ThermoFisher) for at least 10 minutes immediately prior to cell seeding, as recommended by Haraguchi et al.28 All solutions were warmed to 37 °C and fibers were kept on a hot plate in the tissue culture hood to ensure that PNIPAAm remained above its LCST
Cell viability on PNIPAAm/PCL fibers
3T3 fibroblasts were seeded onto PNIPAAm/PCL fibers (n = 3) at 120 000 cells per cm2. This cell density was selected to ensure that the absorbance for the colorimetric assay was in the linear range. 24 hours post-seeding, cells were rinsed and treated with the tetrazolium dye 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxy-methoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS, CellTiter 96® AQueous One Solution Cell Proliferation Assay, Promega, Madison, WI). Solution absorbance was measured at 490 nm on a Biotek Cytation 3 Cell Imaging Multi-Mode Reader. Each condition was normalized by solution absorbance of 3T3 fibroblast-seeded PCL-only controls to determine relative cell attachment.
Cell alignment on PNIPAAm/PCL fibers
3T3 fibroblasts were seeded onto PNIPAAm/PCL fibers at 240 000 cells per cm2. 24 hours post-seeding, cells on PNIPAAm/PCL fibers were rinsed and fixed in 4% (v/v) paraformaldehyde for 15 minutes. Cells were permeabilized with 0.2% (v/v) Triton X-100 for 10 minutes and then stained with 6.6 μM rhodamine phalloidin (ThermoFisher). Following actin staining, cell nuclei were stained with 300 nM 4′,6-diamidino-2-phenyl-indole (DAPI, ThermoFisher). All solutions were warmed to and fibers were incubated at 37 °C, above PNIPAAm's LCST, as described previously.14 Cell-seeded fibers were dissembled from CellCrown inserts, placed on slides, and imaged on a Biotek Cytation 3 Cell Imaging Multi-Mode Reader incubated to 37 °C. Actin images were analyzed using NIH ImageJ software and OrientationJ plug-in to determine cell alignment.
Cell sheet detachment from PNIPAAm/PCL fibers
3T3 fibro-blasts were seeded onto PNIPAAm/PCL fibers at 630 000 cells per cm2 and allowed to grow for 4 days. Medium was changed every day. Prior to detachment, cell were stained with 5 μM calcein, AM (ThermoFisher) and nuclear stain Hoescht 33342 (ThermoFisher) for 30 minutes. Detachment was initiated by rinsing 5 times over 10-15 minutes with cold (approximately 4 °C) medium to dissolve and remove PNIPAAm. Cell sheets were rinsed from the CellCrown insert with additional medium and imaged on an Olympus IX83 fluorescent microscope.
Statistical methods
All data are presented as mean ± standard deviation. Statistical significance was calculated by performing one-way ANOVA analysis followed by Tukey's multiple comparison in GraphPad, Prism Software. Differences are considered significant for p < 0.05.
Results and discussion
Characterization of PNIPAAm/PCL fibers
To generate PNIPAAm/PCL co-fibers, a solvent combination— methanol and chloroform—that dissolves both PNIPAAm and PCL was identified by referring to previous reports on PCL solubility in electrospinning solvents.29 SEM imaging confirmed fiber formation (Fig. 1A). Eletrospinning PCL-only and PNIPAAm-only fibers in methanol and chloroform was attempted; however, fiber formation and alignment was poor compared to the PNIPAAm/PCL blended fibers. Consequently, PCL-only and PNIPAAm-only fibers were electrospun using HFP and methanol, respectively, following previous reports.22,30 Interestingly, only the 100% PNIPAAm fibers exhibited a flat, ribbon-like morphology, as was previously reported for PNIPAAm fibers electrospun by Rockwood et al.19 Average fiber diameters ranged from 1 to 3 μm, with PCL-only fibers being the smallest diameter and PNIPAAm-only fibers having the largest diameter (Fig. 1B). Orientation analysis confirmed fiber alignment, with all conditions having an orientation index greater than 0.65, with 0% and 25% PNIPAAm fibers having significantly greater orientation indices than fibers containing 50% or more PNIPAAm (Fig. 1C). Differences in diameter and fiber orientation between PCL-only, PNIPAAm-only, and the PNIPAAm/PCL fibers may be attributable to the solvent of choice, which affects solution viscosity—a parameter known to largely determine fiber diameter.31 Furthermore, the solution viscosity is also influenced by the relative amounts of PNIPAAm and PCL, as their respective molecular weights, effective chain lengths, and solubility in methanol and chloroform differ. We observed that higher PCL-content fibers could be electrospun from solutions for which the combined polymer concentration of the solution was lower (Table S1†). For example, 90% PNIPAAm fibers were electrospun from a solution of 18% (wt/v) of 9 :1 PNIPAAm: PCL whereas 25% PNIPAAm fibers were electrospun from a solution of 12% (wt/v) of 1: 3 PNIPAAm : PCL.
Fig. 1.
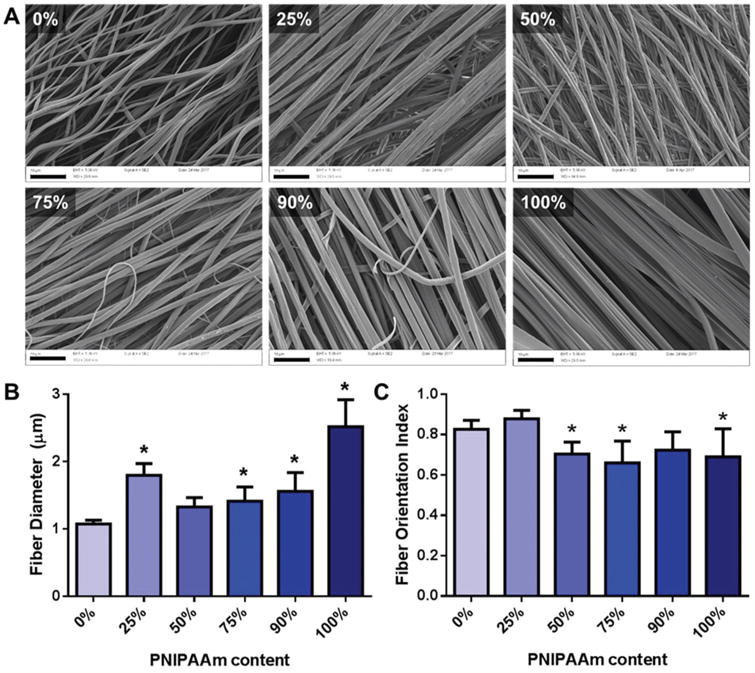
PNIPAAm/PCL fibers. (A) SEM images of electrospun PNIPAAm/PCL fibers. Percentage in upper left indicates PNIPAAm content. Scalebars (black bars, bottom left) are 10 μm. (B) Average fiber diameter determined using DiameterJ. (C) Fiber orientation index determined using OrientationJ. *p < 0.05 compared to 0% PNIPAAm fibers.
FTIR spectroscopy confirmed that relative polymer compositions of PNIPAAm/PCL fibers followed the starting PNIPAAm concentration (Fig. 2), as PCL-specific peaks increased with PCL-content and PNIPAAm-specific peaks increased with PNIPAAm content. The PCL-only (0% PNIPAAm) fibers show a strong peak at 1727 cm−1 indicating carbonyl stretching, which is reduced as PNIPAAm content increases and is absent for the 100% PNIPAAm fibers. Similarly, PNIPAAm-only fibers (100% PNIPAAm) show strong peaks at 1626 and 1559 cm−1 for amide group vibrations that become less strong as PNIPAAm content decreases and are completely absent for the 0% PNIPAAm fibers. Additional absorbance peaks for each polymer are listed in Table 1.32–34
Fig. 2.
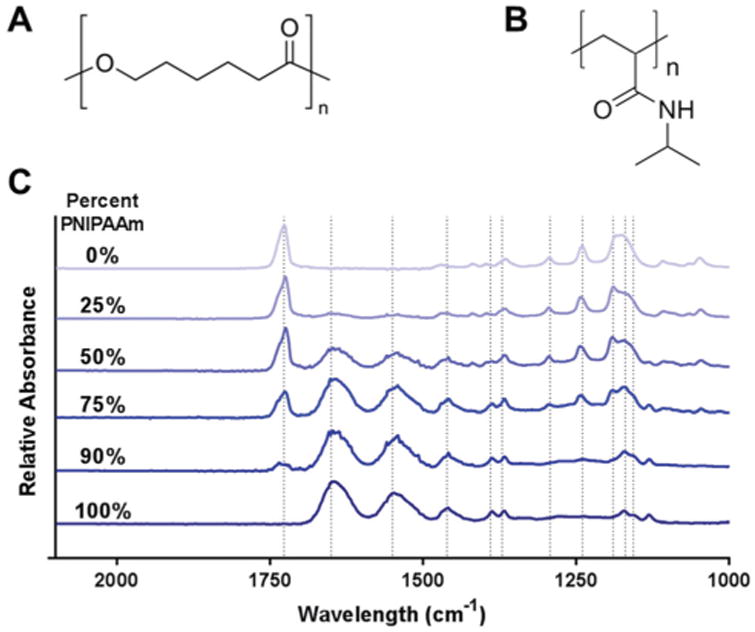
Chemical structures of (A) PCL and (B) PNIPAAm. (C) FTIR spectra of PNIPAAm/PCL fibers. Dashed lines indicate absorption peaks.
Table 1. Absorption peaks and assignments for PNIPAAm and PCL.
| Position (cm−1) | Polymer | Assignment |
|---|---|---|
| 1727 | PCL | Carbonyl stretching |
| 1626 | PNIPAAm | Amide I |
| 1559 | PNIPAAm | Amide II |
| 1462 | PNIPAAm | CH3 asymmetrical deformation |
| 1390, 1371 | PNIPAAm | CH3 symmetrical deformation |
| 1293 | PCL | C–C, C–O stretching |
| 1240 | PCL | C–O–C asymmetric stretching |
| 1190 | PCL | OC–O stretching |
| 1170 | PCL | COC symmetric stretching |
| 1157 | PCL | C–O, C–C stretching |
To further confirm relative PNIPAAm content and evaluate the potential of PNIPAAm/PCL fibers for cell sheet detachment via PNIPAAm dissolution, we immersed PNIPAAm/PCL fibers in room temperature (approximately 20 °C) water to dissolve out PNIPAAm (Fig. 3). Fibers with a considerable amount of PCL showed little mass loss (less than 5% the original weight) whereas high-content PNIPAAm fibers lost more than 50% their original mass (Fig. 3A). Observations of PNIPAAm/PCL fiber area and axial length changes before and after wetting supported the mass loss data: high PNIPAAm-content fiber area contracted more than 55% whereas high PCL-content fibers did not contract but instead slightly swelled (Fig. 3B). Furthermore, 75% and 90% PNIPAAm fiber contraction was uniaxial, perpendicular to fiber orientation (Fig. 3C and S2†). As expected, 100% PNIPAAm fibers completely dissolved, preventing measurements of mass loss and changes in area and axes length. For high PCL-content fibers, the percent mass lost does not match the starting percent PNIPAAm content, indicating that the PCL protects PNIPAAm from dissolving.
Fig. 3.
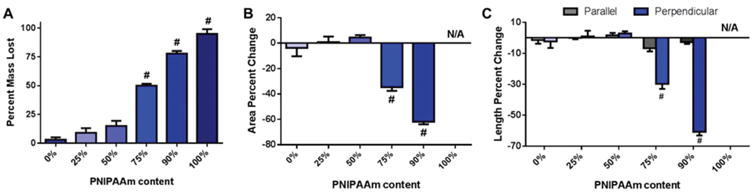
PNIPAAm dissolution from PNIPAAm/PCL fibers (A) PNIPAAm/PCL mass loss in water. PNIPAAm/PCL fiber (B) area percent change and (C) axes (relative to fiber direction) length percent change following PNIPAAm dissolution. # p < 0.05 compared to all other groups.
Relative hydrophobicity of the PNIPAAm/PCL fibers was determined by measuring advancing water contact angle above 32 °C, showing that dry and wetted 100% PNIPAAm fibers (θadv = 88.0° and 55.3°, respectively) were significantly less hydrophobic than 0% PNIPAAm fibers (θadv = 120.7° and 107.1°, Fig. 4 and S3†). In fact, all dry, high PCL-content fibers (0%, 25%, 50% PNIPAAm) were all significantly more hydro-phobic than 100% PNIPAAm fibers with θadv > 120°. Because PNIPAAm undergoes a coil-to-globule transition at its LCST, it does not become truly hydrophobic above its LCST; rather, the hydrophobic domains are exposed to the aqueous solution, enabling protein adsorption.11 Evaluations of relatively thick and thin layers of PNIPAAm-grafted surfaces found that θadv decreased with thickness, meaning thicker PNIPAAm surfaces were less hydrophobic.13 Because biomaterial hydrophobicity is an indicator of the degree of protein adsorption, low hydrophobicity may impair cell attachment and spreading.
Fig. 4.
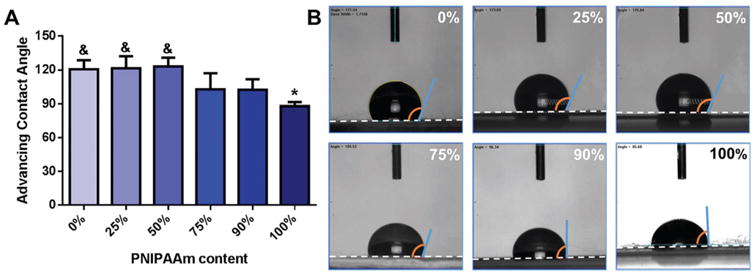
(A) Advancing water contact angle on PNIPAAm/PCL fibers. (B) Representative images of water droplet on fibers. White dashed lines indicate fiber edge. * p < 0.05 compared to 0% PNIPAAm fibers. & p < 0.05 compared to 100% PNIPAAm fibers.
The mass loss, area contraction, and advancing water contact angle data are largely consistent in that the high PCL-content fibers behave similarly and that significant differences are observed for high PNIPAAm-content fibers. Significant mass loss and area contraction from PNIPAAm dissolution starts to occur with 75% PNIPAAm. A possible explanation for these data is that the PNIPAAm/PCL fibers may have a core-sheath architecture, with a PNIPAAm core and PCL sheath. This has been previously observed for PNIPAAm and PCL blended fibers electrospun in dimethylformamide (DMF) and chloroform, although with a PNIPAAm sheath around a PCL core.35 The authors proposed a thermodynamic argument: because DMF was a better solvent for PNIPAAm than for PCL and because DMF had a much lower boiling point than chloroform, the DMF evaporated first, leaving PNIPAAm on the exterior. In our case, methanol's boiling point (64.7 °C) is slightly higher than chloroform's boiling point (61.2 °C), and methanol is a good solvent for PNIPAAm but a bad solvent for PCL. Applying the same thermodynamic argument, the PCL should be dissolved in the chloroform portion, which would evaporate first to leaving PCL on the exterior of the fibers. For high PCL-content fibers, the PCL may form an entire sheath around the PNIPAAm; however, as PNIPAAm content increases, there would not be enough PCL to protect PNIPAAm from dissolution, as observed by the mass loss and contact angle data.
Cell viability and alignment on PNIPAAm/PCL fibers
After confirming the relative PNIPAAm content, we assessed the behavior of NIH 3T3 fibroblasts on PNIPAAm/PCL fibers. Given PNIPAAm's relative low hydrophobicity as indicated by advancing water contact angle, we coated PNIPAAm/PCL fibers with a 1 : 50 GFR-Matrigel dilution and pre-treated fibers with FBS prior to seeding, as recommended by Haraguchi et al.;28 other groups have combined PNIPAAm with gelatin,23 chitosan,36 fibronectin,14,37 collagen,38 poly-l-lysine, and laminin39 to improve cell adhesion.
MTS assay 24-hours post-seeding demonstrated that cells attached and were viable on PNIPAAm/PCL fibers as compared to the PCL-only (0% PNIPAAm) control (Fig. 5). However, cells were significantly less viable (60% relative to PCL-only control) on 100% PNIPAAm fibers. To visualize cytoskeletal actin, cells were stained with rhodamine phalloidin 24-hours post-seeding on PNIPAAm/PCL fibers. Fibers containing PCL (0%, 25%, 50%, 75%, and 90% PNIPAAm) showed robust spreading and significant cell alignment in a preferred direction (Fig. 4B and C). On 100% PNIPAAm fibers, cells showed notably less spreading and grew in clusters, which is in line with the relatively poor cell viability observed on these fibers. As PNIPAAm is clearly not toxic to cells, as indicted by comparable cell viability on 0%, 25%, 50%, 75%, and 90% PNIPAAm fibers, it is likely that cell attachment was affected by poor adhesion protein adsorption due to its decreased hydrophobicity. This is consistent with our advancing water contact results and previous reports that protein adsorption onto PNIPAAm, especially thick PNIPAAm films (>15–20 nm), is severely limited.37,39 Understandably, coating with cell adhesion proteins28 and grafting gelatin to PNIPAAm23 has been used to improve cell attachment to and spreading on PNIPAAm surfaces. Representative histograms of cell angle show that cells had relatively high alignment on high-content PCL fibers (Fig. S4†). Although less aligned, cells on 75% and 90% PNIPAAm fibers had a preferred angle to which the cells aligned. The actin ridges that appeared on 75% and 90% PNIPAAm fibers is likely due to PNIPAAm contracture as the fibers had to be removed from the CellCrown inserts for imaging. This phenomenon affected the orientation analysis as these ridges, which are perpendicular to cell orientation angle, dampened the preferred cell angle peak.
Fig. 5.
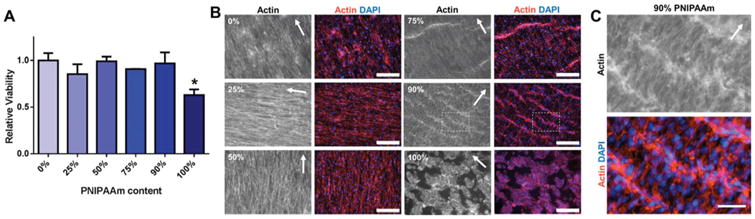
Cell viability and cell alignment on PNIPAAm/PCL fibers. (A) Cell viability relative to 0% PNIPAAm fibers determined by MTS assay. * p < 0.05 compared to 0% PNIPAAm fibers. (B) Representative images of fibroblasts seeded on PNIPAAm/PCL fibers with actin (left) and actin/DAPI overlays (right). Scalebars are 200 μm. (C) Insets from 90% PNIPAAm images, as indicated by white dashed-box in (B). Scalebar is 50 μm. * p < 0.05 compared to 0% PNIPAAm fibers.
Cell sheet detachment
We attempted detachment of aligned fibroblast cell sheets by incubating cell-seeded PNIPAAm/PCL fibers in cold medium. Following previous reports on cell sheeting,14,28 we seeded cells at an ultra-high density—630 000 cells per cm2—to ensure sufficient cell–cell adhesion and ECM deposition. Detachment was initiated by rinsing cell-seeded PNIPAAm/PCL fibers with cold medium to dissolve the PNIPAAm. Cell sheet detachment from PNIPAAm/PCL fibers was only successful for 90% PNIPAAm fibers (Fig. 6A), which occurred rapidly (less than 15 minutes). For lower PNIPAAm-content fibers, too much PCL remained preventing cell detachment. Cell sheet detachment from 100% PNIPAAm fibers was unsuccessful because the cells did not form a complete monolayer, as indicated by our cell morphology data (Fig. 5B). Cell sheets exhibited slight curling at the edges, evidenced by the thickening around the edges. Calcein staining confirmed that cell sheets were viable, intact, and consisted of aligned cells (Fig. 6B and C). Compared to previous reports of anisotropic cell sheeting from anisotropic PNIPAAm surfaces,14,23 relatively little contraction of the cell sheet was observed. This may be due to the presence of residual PCL, which can be observed in phase contrast images of the cell sheets (Fig. 6D and E).
Fig. 6.
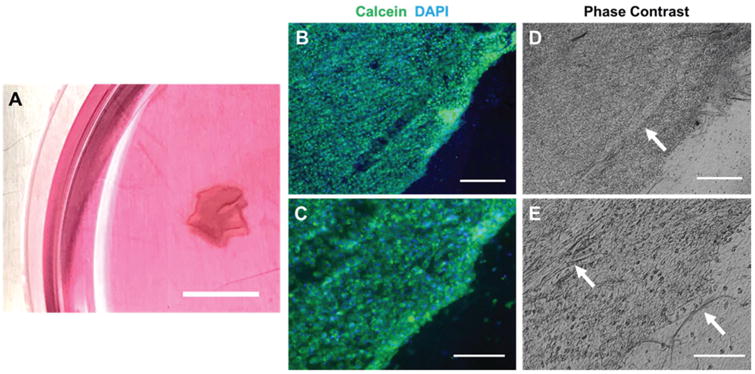
Cell sheet detachment. (A) Cell sheets detached from 90% PNIPAAm fibers using room temperature medium; scalebar is 1 cm. (B, C) Cell sheet viability was confirmed with calcein, AM live-staining. (D, E) Corresponding phase contrast images of (B, C); white arrows indicate residual PCL. Scalebars are (B, D) 400 μm and (C, E) 200 μm.
Our data demonstrate that electrospun PNIPAAm/PCL fibers can be used to culture aligned cells but only 90% PNIPAAm fibers can be used for cell detachment. This follows our original hypothesis that PNIPAAm and PCL have complementary roles and must be present in sufficient amounts. PCL encourages cell attachment, but too much PCL precludes cell sheet detachment. Likewise, PNIPAAm's low hydrophobicity limits for cell attachment but is necessary for cell sheet detachment. The cell sheets generated by our method did show some contraction, which is unfavorable for cell sheet stacking. Gelatin hydrogel plungers have been previously used to prevent cell sheet contracture and could easily be used in conjunction with our PNIPAAm/PCL fibers.28 We used 3T3 fibroblasts as a proof of principle to demonstrate that PNIPAAm/PCL fibers can generate cell sheets. Our system can be used with other cell types to possibly generate more complex tissue structures, such as blood vessels with better cellular architecture of the tunica media. Thus, electrospun PNIPAAm/PCL fibers, which are simple and relatively inexpensive to produce, have the potential to be used to generate anisotropic cell sheets that can either enable analyses that are typically precluded by the use of plates or biomaterial scaffolds or be used to create tissue-like constructs for in vivo transplantation.40,41
Conclusions
Here, we developed a simple, inexpensive, and low-resource system to generate cell sheets. PNIPAAm and PCL were successfully electrospun to generate aligned PNIPAAm/PCL blended fibers on which 3T3 fibroblasts could be cultured. Cell viability and cell alignment was observed on PCL and PNIPAAm/PCL fibers whereas cell viability and cell alignment was impaired on 100% PNIPAAm fibers. Detachment of viable cell sheets by incubation with room temperature medium was successful for 90% PNIPAAm fibers; cell sheets did not detach from fibers containing less PNIPAAm and cells did not form a contiguous monolayer on 100% PNIPAAm. In summary, our findings demonstrate that anisotropic cell sheets can be generated simply using electrospun PNIPAAm/PCL fibers—without chemical synthesis or resource-intensive techniques.
Supplementary Material
Acknowledgments
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1610403 and National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under award number EB007507. The authors thank Dr Nicholas Peppas for use of the Thermo Scientific Nicolet iS10 FT-IR spectrometer, the University of Texas Institute of Cell and Molecular Biology for use of the Zeiss Supra SV40 SEM, and the University of Texas Center for Nano- and Molecular Science for use of the FTA-200 goniometer, Will Goth for the design and construction of the electrospinning rotating collector, and Daegi Lee for assistance with image processing.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c7bm00324b
References
- 1.Kim DH, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. J Cell Biol. 2012;197:351–360. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim SH, Liu XY, Song H, Yarema KJ, Mao HQ. The effect of nanofiber-guided cell alignment on the preferential differentiation of neural stem cells. Biomaterials. 2010;31:9031–9039. doi: 10.1016/j.biomaterials.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levorson EJ, et al. Fabrication and characterization of multiscale electrospun scaffolds for cartilage regeneration. Biomed Mater. 2013;8:014103. doi: 10.1088/1748-6041/8/1/014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falconnet D, Csucs G, Michelle Grandin H, Textor M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials. 2006;27:3044–3063. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Doshi J, Reneker DH. Electrospinning process and applications of electrospun fibers. Conference Record of the 1993 IEEE Industry Applications Society Annual Meeting, 1993. 1993;3:1698–1703. doi:10.1109/ IAS.1993.299067. [Google Scholar]
- 6.Pham QP, Sharma U, Mikos AG. Electrospinning of Polymeric Nanofibers for Tissue Engineering Applications: A Review. Tissue Eng. 2006;12:1197–1211. doi: 10.1089/ten.2006.12.1197. [DOI] [PubMed] [Google Scholar]
- 7.Kai D, et al. Stem cell-loaded nanofibrous patch promotes the regeneration of infarcted myocardium with functional improvement in rat model. Acta Biomater. 2014;10:2727–2738. doi: 10.1016/j.actbio.2014.02.030. [DOI] [PubMed] [Google Scholar]
- 8.Wang TY, Forsythe JS, Nisbet DR, Parish CL. Promoting engraftment of transplanted neural stem cells/ progenitors using biofunctionalised electrospun scaffolds. Biomaterials. 2012;33:9188–9197. doi: 10.1016/j.biomaterials.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Anderson JM, et al. Biomaterials Science: an Introduction to Materials in Medicine, Host reactions to biomaterials and their evaluation. Academic Press; 1996. [Google Scholar]
- 10.Yamada N, et al. Thermo-responsive polymeric surfaces; control of attachment and detachment of cultured cells. Makromol Chem, Rapid Commun. 1990;11:571–576. [Google Scholar]
- 11.Pelton R. Poly(N-isopropylacrylamide) (PNIPAM) is never hydrophobic. J Colloid Interface Sci. 2010;348:673–674. doi: 10.1016/j.jcis.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, et al. Cell sheet engineering: Recreating tissues without biodegradable scaffolds. Biomaterials. 2005;26:6415–6422. doi: 10.1016/j.biomaterials.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 13.Yamato M, et al. Temperature-responsive cell culture surfaces for regenerative medicine with cell sheet engineering. Prog Polym Sci. 2007;32:1123–1133. [Google Scholar]
- 14.Takahashi H, Nakayama M, Shimizu T, Yamato M, Okano T. Anisotropic cell sheets for constructing three-dimensional tissue with well-organized cell orientation. Biomaterials. 2011;32:8830–8838. doi: 10.1016/j.biomaterials.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi H, Nakayama M, Itoga K, Yamato M, Okano T. Micropatterned Thermoresponsive Polymer Brush Surfaces for Fabricating Cell Sheets with Well-Controlled Orientational Structures. Biomacromolecules. 2011;12:1414–1418. doi: 10.1021/bm2000956. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, et al. Three-dimensional poly-(ε-caprolactone) nano-fibrous scaffolds directly promote the cardiomyocyte differentiation of murine-induced pluripotent stem cells through Wnt/β-catenin signaling. BMC Cell Biol. 2015;16:22. doi: 10.1186/s12860-015-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cipitria A, et al. Design, fabrication and characterization of PCL electrospun scaffolds—a review. J Mater Chem. 2011;21:9419–9453. [Google Scholar]
- 18.Croisier F, et al. Mechanical testing of electrospun PCL fibers. Acta Biomater. 2012;8:218–224. doi: 10.1016/j.actbio.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Rockwood DN, Chase DB, Akins RE, Jr, Rabolt JF. Characterization of electrospun poly(N-isopropyl acrylamide) fibers. Polymer. 2008;49:4025–4032. [Google Scholar]
- 20.Tran T, et al. Controllable and switchable drug delivery of ibuprofen from temperature responsive composite nano-fibers. Nano Convergence. 2015;2:15. [Google Scholar]
- 21.Yu H, Jia Y, Chen G, Zhang Y. Fabrication of core/ sheath PCL/PEG–PNIPAAm fibers as thermosensitive release carriers by a new technique combining blend electrospinning and ultraviolet-induced graft polymerization. Mater Lett. 2016;164:505–508. [Google Scholar]
- 22.Lee JB, et al. Development of 3D Microvascular Networks Within Gelatin Hydrogels Using Thermoresponsive Sacrificial Microfibers. Adv Healthcare Mater. 2016;5:781–785. doi: 10.1002/adhm.201500792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Wang L, Wang P, Yang Y, Wang F. Fabrication of Thermoresponsive Nanofibers for Cell Sorting and Aligned Cell Sheet Engineering. J Nanosci Nanotechnol. 2016;16:5520–5527. doi: 10.1166/jnn.2016.11738. [DOI] [PubMed] [Google Scholar]
- 24.Rezakhaniha R, et al. Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech Model Mechanobiol. 2012;11:461–473. doi: 10.1007/s10237-011-0325-z. [DOI] [PubMed] [Google Scholar]
- 25.Püspöki Z, Storath M, Sage D, Unser M. Transforms and Operators for Directional Bioimage Analysis: A Survey. Adv Anat, Embryol Cell Biol. 2016;219:69–93. doi: 10.1007/978-3-319-28549-8_3. [DOI] [PubMed] [Google Scholar]
- 26.Hotaling NA, Bharti K, Kriel H, Simon CG, Jr, Diameter J. A validated open source nanofiber diameter measurement tool. Biomaterials. 2015;61:327–338. doi: 10.1016/j.biomaterials.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferdman AG, Yannas IV. Scattering of Light from Histologic Sections: A New Method for the Analysis of Connective Tissue. J Invest Dermatol. 1993;100:710–716. doi: 10.1111/1523-1747.ep12472364. [DOI] [PubMed] [Google Scholar]
- 28.Haraguchi Y, et al. Fabrication of functional three-dimensional tissues by stacking cell sheets in vitro. Nat Protoc. 2012;7:850–858. doi: 10.1038/nprot.2012.027. [DOI] [PubMed] [Google Scholar]
- 29.Bordes C, et al. Determination of poly(ε-caprolactone) solubility parameters: Application to solvent substitution in a microencapsulation process. Int J Pharm. 2010;383:236–243. doi: 10.1016/j.ijpharm.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 30.Nam J, Huang Y, Agarwal S, Lannutti J. Materials selection and residual solvent retention in biodegradable electrospun fibers. J Appl Polym Sci. 2008;107:1547–1554. [Google Scholar]
- 31.Thompson CJ, Chase GG, Yarin AL, Reneker DH. Effects of parameters on nanofiber diameter determined from electrospinning model. Polymer. 2007;48:6913–6922. [Google Scholar]
- 32.Beattie DA, et al. In situ particle film ATR FTIR spectroscopy of poly (N -isopropyl acrylamide) (PNIPAM) adsorption onto talc. Phys Chem Chem Phys. 2014;16:25143–25151. doi: 10.1039/c4cp03161j. [DOI] [PubMed] [Google Scholar]
- 33.Elzein T, Nasser-Eddine M, Delaite C, Bistac S, Dumas P. FTIR study of polycaprolactone chain organization at interfaces. J Colloid Interface Sci. 2004;273:381–387. doi: 10.1016/j.jcis.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Dybal J, Trchová M, Schmidt P. The role of water in structural changes of poly(N-isopropylacrylamide) and poly (N-isopropylmethacrylamide) studied by FTIR, Raman spectroscopy and quantum chemical calculations. Vib Spectrosc. 2009;51:44–51. [Google Scholar]
- 35.Chen M, et al. Thermo-Responsive Core-Sheath Electrospun Nanofibers from Poly (N-isopropylacrylamide)/ Polycaprolactone Blends. Chem Mater. 2010;22:4214–4221. [Google Scholar]
- 36.Wang J, Chen L, Zhao Y, Guo G, Zhang R. Cell adhesion and accelerated detachment on the surface of temperature-sensitive chitosan and poly(N-isopropyl-acrylamide) hydrogels. J Mater Sci: Mater Med. 2009;20:583–590. doi: 10.1007/s10856-008-3593-0. [DOI] [PubMed] [Google Scholar]
- 37.Akiyama Y, Kikuchi A, Yamato M, Okano T. Ultrathin poly(N-isopropylacrylamide) grafted layer on polystyrene surfaces for cell adhesion/detachment control. Langmuir. 2004;20:5506–5511. doi: 10.1021/la036139f. [DOI] [PubMed] [Google Scholar]
- 38.Moran MT, Carroll WM, Selezneva I, Gorelov A, Rochev Y. Cell growth and detachment from protein-coated PNIPAAm-based copolymers. J Biomed Mater Res. 2007;81A:870–876. doi: 10.1002/jbm.a.31089. [DOI] [PubMed] [Google Scholar]
- 39.Moran MT, Carroll WM, Gorelov A, Rochev Y. Intact endothelial cell sheet harvesting from thermo-responsive surfaces coated with cell adhesion promoters. J R Soc, Interface. 2007;4:1151–1157. doi: 10.1098/rsif.2007.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu T, et al. Fabrication of pulsatile cardiac tissue grafts using a novel 3-dimensional cell sheet manipulation technique and temperature-responsive cell culture surfaces. Circ Res. 2002;90:e40. doi: 10.1161/hh0302.105722. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, et al. Reconstruction of functional tissues with cell sheet engineering. Biomaterials. 2007;28:5033–5043. doi: 10.1016/j.biomaterials.2007.07.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


