Abstract
The Gram-positive bacterial cell wall is a large supramolecular structure and its assembly requires coordination of complex biosynthetic pathways. In the step that merges the two major biosynthetic pathways in Staphylococcus aureus cell wall assembly, conserved protein ligases attach wall teichoic acids to peptidoglycan, but the order of biosynthetic events is a longstanding question. Here, we use a chemical approach to define which of the possible peptidoglycan intermediates are substrates for wallteichoic acid ligases, thereby establishing the order of cell wall assembly. We have developed a strategy to make defined glycan chain-length polymers of either uncrosslinked or crosslinked peptidoglycan, and we find that wall teichoic acid ligases cannot transfer wall teichoic acid precursors to the crosslinked substrates. A 1.9Å crystal structure of a LytR-CpsA-Psr (LCP) family ligase in complex with a wall teichoic acid precursor defines the location of the peptidoglycan binding site as a long, narrow groove, and suggests that the basis for selectivity is steric exclusion of crosslinked peptidoglycan. Consistent with this hypothesis, we have found that chitin oligomers are good substrates for transfer, showing that LCPs do not discriminate crosslinked from uncrosslinked peptidoglycan substrates by recognizing features of the uncrosslinked stem peptide. We conclude that wall teichoic acids are coupled to uncrosslinked peptidoglycan chains at an early stage of peptidoglycan synthesis and may create marks that define the proper spacing of subsequent crosslinks.
Graphical abstract
The bacterial cell wall is a complex macromolecule and the target of many antibiotics. Peptidoglycan (PG), strands of glycan polymers joined by peptide crossbridges, forms the foundation of the cell wall in all bacteria.1 In Gram-positive bacteria the peptidoglycan is covalently modified with additional glycopolymers that comprise up to 50% of the cell wall by mass.2 These glycopolymers are frequently wall teichoic acid (WTA) glycopolymers, which play important roles in cell growth and division.2 In pathogens such as Staphylococcus aureus (S. aureus), wall teichoic acid is essential for host colonization and virulence.3 Importantly, inhibiting wall teichoic acid biosynthesis re-sensitizes methicillin-resistant S. aureus (MRSA) to beta-lactams.4 Underlying all of these effects of wall teichoic acids on bacterial physiology is their central role in regulating bacterial cell wall biosynthesis.2,5 Understanding how and when wall teichoic acids are attached to peptidoglycan is fundamental to understanding cell physiology and has implications for developing novel antimicrobials.
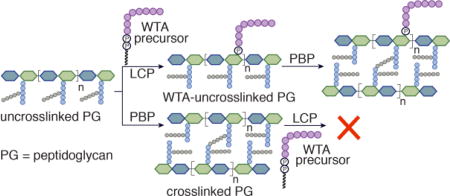
In the final steps of peptidoglycan biosynthesis, S. aureus translocates the monomer building block Lipid II to the cell surface6 where it is polymerized into linear glycan strands that are crosslinked by penicillin-binding proteins (PBPs) to adjacent glycan strands via their stem peptides.7 Concurrently, WTA precursors are biosynthesized in the cytoplasm,2 translocated to the cell surface,8 and transferred to a peptidoglycan substrate.9 Studies in cells have tried to address whether WTA attachment to peptidoglycan occurs before or after glycan strands are crosslinked (Figure 1 and Figure S1), but have not reached a definitive conclusion.10 Here, using a chemical approach, we show that WTA precursors can only be attached to uncrosslinked peptidoglycan, implying that transfer occurs at an early stage of cell wall biosynthesis.
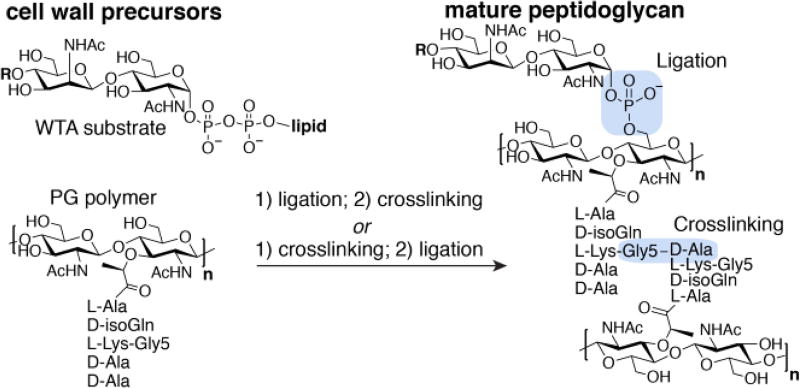
Figure 1.
The order of assembly of the Gram-positive cell wall has not been established. R represents the WTA polymer chain, and n represents the number of peptidoglycan repeats.
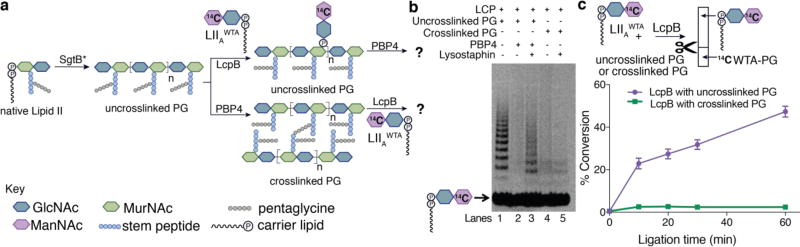
To compare WTA transfer to crosslinked and uncrosslinked peptidoglycan, we needed defined peptidoglycan fragments that could be resolved by polyacrylamide gel electrophoresis (PAGE). We previously reported that all three S. aureus LCP (LytR-CpsA-Psr) proteins can transfer a truncated radiolabeled wall teichoic acid precursor, LIIAWTA, to synthetic, uncrosslinked peptidoglycan oligomers,12b,13 resulting in a radiolabeled ladder of modified peptidoglycan fragments, but we were unable to test transfer to the corresponding crosslinked oligomers due to lack of access to substrates containing the pentaglycine branch required for crosslinking by S. aureus penicillin-binding proteins. We recently developed a strategy to obtain native Lipid II from S. aureus, making the preparation of crosslinked S. aureus peptidoglycan possible (Figure S2).11 To prepare short peptidoglycan oligomers, we incubated this Lipid II with a mutant transglycosylase, SgtBY181D (SgtB*), that releases peptidoglycan prematurely during polymerization (Figure 2a).12 The oligomers produced range from approximately two to ten disaccharide units and can be separated to disaccharide resolution. Here, we prepared the required crosslinked substrates by incubating native Lipid II with SgtB* to make peptidoglycan oligomers that were then crosslinked using S. aureus PBP4 (Figure 2a). We next tested (1) whether WTA-modified peptidoglycan could be crosslinked by a PBP, and (2) whether crosslinked peptidoglycan could be modified with WTA (Figure 2a). When WTA-modified, uncrosslinked peptidoglycan was treated with S. aureus PBP4, we observed disappearance of the radiolabeled ladder of modified fragments because the polymers became too large to enter the gel11a,14 (compare lanes 1 and 2, Figure 2b). Consistent with this, treatment with lysostaphin, which cleaves the pentaglycine-bridged crosslinks, restored the radiolabeled ladder (lane 3).11b,13b,15 In contrast, when crosslinked peptidoglycan (25% crosslinked; Table S1) was first incubated with the WTA ligase LcpB and then treated with lysostaphin, we did not observe the appearance of a radiolabeled ladder (lanes 4 and 5; compare lane 5 to lane 3, Figure 2b). These results showed that uncrosslinked PG modified with WTA is a good substrate for crosslinking, whereas crosslinked PG is a poor substrate for WTA transfer.
Figure 2.
Substrate preferences define the order of cell wall assembly as 1) formation of uncrosslinked PG polymer; 2) transfer of WTA to PG; 3) strand crosslinking. (a) Scheme shows the order in which enzymatic reactions are performed to test substrate preferences of a prototypical WTA ligase, LcpB.11a,12b An SgtB mutant (SgtB*) that makes short oligomers was used for PAGE analysis (b) and wild-type SgtB was used for paper chromatography (c). (b) PAGE autoradiograph shows that uncrosslinked PG modified with WTA (lane 1) can be crosslinked (lanes 2 and 3); however, crosslinked PG cannot be modified with WTA as no bands were observed after lysostaphin treatment (lane 5). (c) Schematic of paper strip assay and a corresponding time course confirms that crosslinked PG is a poor substrate for transfer.
To quantitatively compare WTA transfer to uncrosslinked and crosslinked peptidoglycan polymers, we developed a paper chromatography assay that separates PG polymers from the radiolabeled WTA substrate, LIIAWTA (Figure 2c). Using wild-type SgtB, we prepared long, uncrosslinked peptidoglycan, split the reaction mixture, and incubated part of it with PBP4 to produce crosslinked peptidoglycan.16 The uncrosslinked and crosslinked substrates were then incubated with radiolabeled LIIAWTA and LcpB, and the reaction mixtures were separated.17 Strips were cut to isolate WTA-modified peptidoglycan polymers (retained at the baseline) from radiolabeled LIIAWTA starting material (Figure 2c, schematic). The WTA substrate was readily incorporated into uncrosslinked polymer, but minimal transfer to crosslinked polymer was detected. To test whether LcpB’s preference for uncrosslinked peptidoglycan depends on the penicillin-binding protein used, we also tested the essential S. aureus enzyme, PBP2, which both polymerizes Lipid II and crosslinks the resulting polymers. To prepare uncrosslinked polymers for the comparison, we used PBP2 variant, PBP2S398G, which contains a mutation in its transpeptidase domain that prevents crosslinking (Figure S4).11a,11b LC/MS analysis showed that 17% of available sites were crosslinked by wild-type PBP2 (Table S1). We again found that only uncrosslinked peptidoglycan was a substrate for transfer (Figure S4), showing that the results do not depend on the peptidoglycan polymerase or transpeptidase used.
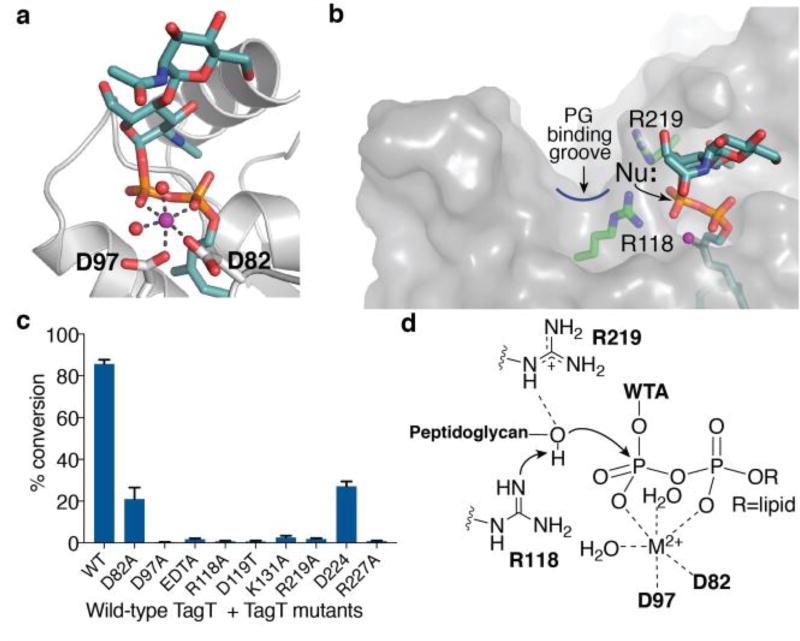
We took a structural approach to better understand the substrate preferences of LCP proteins. Efforts to crystallize the S. aureus LCP proteins were unsuccessful, so we focused on the B. subtilis TagT, which was previously crystallized with octaprenyl pyrophosphate.18 We first verified that TagT ligates LIIAWTA to synthetic, uncrosslinked peptidoglycan oligomers (Figure S5) and then crystallized it with two WTA precursors, LIWTA (containing a monosaccharide) or LIIAWTA (containing a disaccharide), yielding 1.8Å and 1.9Å structures, respectively (Figure 3a; Table S3). Most features of the structures are similar, but the orientations of the saccharide moieties and pyrophosphate are different, and only the structure with the disaccharide (LIIAWTA) contains a divalent cation in the active site (Figure S6). This finding is notable because LCPs are metal ion-dependent transferases (Figure S7)12b,18–19 and we have previously shown that WTA precursors must contain at least a disaccharide to serve as substrates for transfer.12b Although the N-acetyl glucosamine sugar of the LIWTA substrate makes several contacts with TagT, the corresponding GlcNAc of LIIAWTA is oriented differently due to changes in the glycosidic linkage and pyrophosphate bonds (Figure S6). These changes evidently prevent steric clashes that would otherwise arise between the ManNAc sugar and the protein. Therefore, the second sugar in the WTA precursor orients the substrate so that the pyrophosphate is in a conformation that can bind a divalent cation (Figure 3), making it competent for reaction.
Figure 3.
Crystal structure of TagT and LIIAWTA defines the LCP active site for WTA transfer. a) A close-up view of the LIIAWTA substrate showing the pyrophosphate coordinated by Mg2+ (purple sphere), two waters (red spheres) and aspartate oxygen atoms of D82 and D97. b) In-line attack of the nucleophile on the pyrophosphate defines the location of the PG nucleophile. c) Mutations of conserved charged residues in the active site reduce or abolish TagT activity. All points are the mean ± s.e.m. (n=3). d) Proposed mechanism for deprotonation of the nucleophile.
The orientation of the metal-bound pyrophosphate defines the trajectory of nucleophilic attack by the peptidoglycan substrate (Figure 3b). The Mg2+ ion is coordinated by two oxygens on the adjacent LIIAWTA pyrophosphate, two waters, and one carboxylate oxygen from each of two strictly conserved aspartate residues (D97 and D82, Figure 3a). Aspartate to alanine substitutions at these positions either greatly (D82A) or completely (D97A) inhibited TagT ligase activity (Figure 3c), as did alanine substitutions at analogous positions in LcpB (Figure S8). Because the nucleophile approaches a phosphate in-line with the leaving group in a phosphoryl transfer reaction,20 and the bonds formed in this phosphoryl transfer reaction are known, we can infer that the peptidoglycan substrate binds in a long, narrow groove proximal to the anomeric phosphate of LIIAWTA (Figure 3b). This groove and the adjacent WTA binding pocket contain a number of strictly conserved residues, which were all found to be important for ligase activity (Figure 3c, Figure S9).21 Notably, three of these residues are arginines. R227 appears to play a role in stabilizing the pyrophosphoryl-oxygens of the WTA substrate (Figure S9). R219 makes no polar contacts to the WTA substrate, and its guanidinium nitrogens are positioned above the proposed trajectory of the peptidoglycan nucleophile. Likewise, R118 is adjacent to the proposed nucleophile, with one guanidinium nitrogen 3.0Å from the anomeric phosphate and the second nitrogen proximal to the proposed trajectory of the nucleophilic MurNAc hydroxyl (Figure 3b). Although uncommon, arginines can act as general bases in catalytic reactions.22 Catalytic arginines in solvent-exposed pockets are often near other arginines, which may tune the pKa, and may also be adjacent to a carboxylate that facilitates proton transfer. These features are found in the LCP structure.23 Based on our analysis, we propose that R118 acts as a base to deprotonate the C6-hydroxyl of MurNAc, while R219 plays a supporting role in coordinating the nucleophile (Figure 3d).
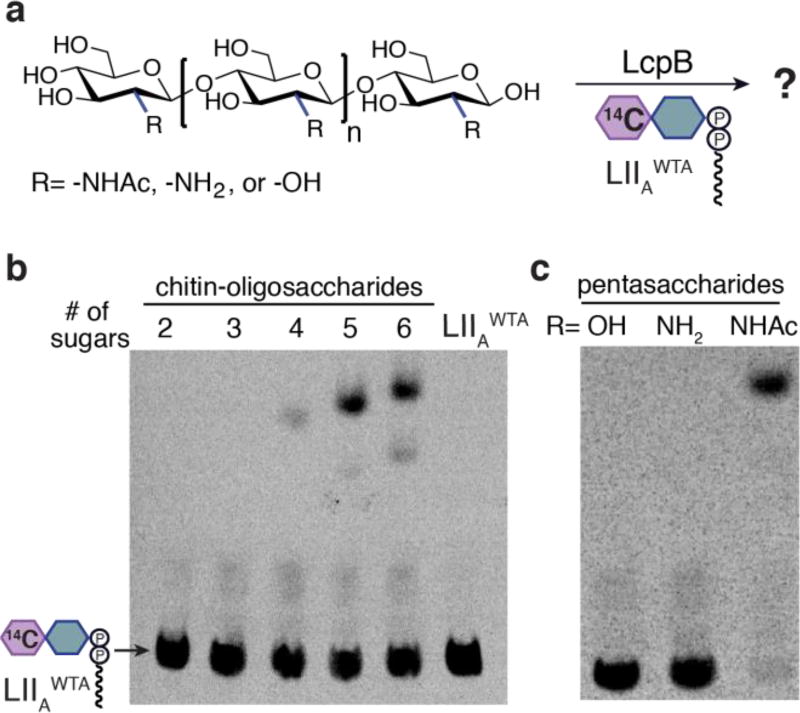
The peptidoglycan binding groove identified in the TagT-LIIAWTA structure is narrow, suggesting that crosslinked peptidoglycan is sterically excluded. Nonetheless, we considered the possibility that LCP enzymes might recognize specific chemical features in the stem peptides of uncrosslinked peptidoglycan, which differ in crosslinked peptidoglycan. To assess whether any feature of the stem peptides found in uncrosslinked PG is required, we tested chitin oligosaccharides of increasing length. These oligosaccharides have the same linkage stereochemistry as peptidoglycan, and also contain N-acetyl groups on every monosaccharide, but they lack the stem peptide and the lactic acid moiety present on the C3 position of every other sugar in peptidoglycan. We found that LcpB and TagT transferred WTA onto chitin oligosaccharides containing five or more sugars; shorter oligosaccharides reacted slowly or not at all (Figure 4; Figures S10–12). The corresponding deacetylated oligosaccharides could not be labelled with LIIAWTA, nor could cellulose-based oligomers (β(1,4)-linked glucose). Hence, the stem peptide of peptidoglycan is not required for WTA ligation, but the C2-N-acetyl groups play crucial roles in substrate recognition.
Figure 4.
LCP enzymes can use substrates lacking a stem peptide. (a) Oligosaccharides tested as substrates for LcpB (shown) and TagT (Figure S10). (b) PAGE autoradiography showing that oligosaccharides containing five or six GlcNAcs (n=3 or 4) are substrates (5 minute reactions). (c) Pentasaccharides lacking C2-N-acetyl are not substrates (20 minute reactions).
Taken together, the results presented here establish the order of the final steps of S. aureus cell wall assembly, and likely other Gram-positive organisms that contain LCPs, and also provide a mechanistic rationale for the sequence of events. Using comparable uncrosslinked and crosslinked peptidoglycan fragments assembled in vitro from native S. aureus Lipid II, we have shown that WTA precursors can only be transferred to uncrosslinked strands. Because Lipid II itself is not a substrate for transfer,12b these findings imply that uncrosslinked PG is first made and then modified with WTA before crosslinking. As the stem peptide is not required for recognition, WTA ligases do not discriminate between uncrosslinked and crosslinked substrates by recognizing a feature found only in the former. Instead, the basis for selectivity appears to be steric exclusion of crosslinked peptidoglycan from the long, narrow peptidoglycan binding groove. The ability to use chitin as an alternative substrate to uncrosslinked peptidoglycan will facilitate development of assays for LCP inhibitors, which may serve as beta-lactam potentiators to treat MRSA.4,11a
The order of cell wall assembly, with wall teichoic acid attachment occurring prior to crosslinking, suggests a role for these peptidoglycan modifications in regulating crosslinking. It has been observed previously that S. aureus PBP4 is mislocalized in the absence of wall teichoic acid,24 resulting in decreased crosslinking; a scaffolding model in which wall teichoic acids anchor PBPs has been proposed.4a It is also possible that wall teichoic acid marks have physical effects on peptidoglycan polymer conformation that affect crosslinking rates. Access to defined peptidoglycan substrates with and without WTA modifications now makes it possible to address these models.
Supplementary Material
Acknowledgments
This research was supported by GM076710 and U19 AI109764 to D.K. and S.W, as well as GM066174 to D.K. This work also used NE-CAT beamlines (GM103403), a Pilatus detector (RR029205), and an Eiger detector (OD021527) at the APS (DE-AC02-06CH11357). This work also acknowledges Veerasak Srisuknimit for helpful discussions with crosslinking reactions.
Footnotes
ASSOCIATED CONTENT
Supplemental figures and tables, experimental procedures, compound analysis, protein purification and crystallization protocols. This material is available free of charge on the ACS Publications website.
The authors declare no competing financial interests.
References
- 1.Silhavy TJ, Kahne D, Walker S. Cold Spring Harb. Perspect. Biol. 2010;2:a000414–a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown S, Maria JPS, Walker S. Annu. Rev. Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. Nat. Med. 2004;10:243. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]; (b) Wanner S, Schade J, Keinhorster D, Weller N, George SE, Kull L, Bauer J, Grau T, Winstel V, Stoy H, Kretschmer D, Kolata J, Wolz C, Broker BM, Weidenmaier C. Nat. Microbiol. 2017;2:16257. doi: 10.1038/nmicrobiol.2016.257. [DOI] [PubMed] [Google Scholar]; (c) Collins LV, Kristian SA, Weidenmaier C, Faigle M, van Kessel KPM, van Strijp JAG, Gotz F, Neumeister B, Peschel A. J. Infect. Dis. 2002;186:214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]; (d) Weidenmaier C, Kokai-Kun JF, Kulauzovic E, Kohler T, Thumm G, Stoll H, Gotz F, Peschel A. Int. J. Med. Microbiol. 2008;298:505–513. doi: 10.1016/j.ijmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 4.(a) Campbell J, Singh AK, Maria JPS, Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S. ACS Chem. Biol. 2011;6:106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Farha MA, Leung A, Sewell EW, D'Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED. ACS Chem. Biol. 2013;8:226–233. doi: 10.1021/cb300413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weidenmaier C, Peschel A. Nat. Rev. Microbiol. 2008;6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 6.Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vollmer W, Blanot D, de Pedro MA. FEMS Microbiol. Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 8.Lazarevic V, Karamata D. Mol. Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 9.(a) Neuhaus FC, Baddiley J. Microbiol. Mol. Biol. Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yokoyama K, Miyashita T, Araki Y, Ito E. Eur. J. Biochem. 1986;161:479–489. doi: 10.1111/j.1432-1033.1986.tb10469.x. [DOI] [PubMed] [Google Scholar]
- 10.(a) Bracha R, Davidson R, Mirelman D. J Bacteriol. 1978;134:412–417. doi: 10.1128/jb.134.2.412-417.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mauck J, Glaser L. J. Biol. Chem. 1972;247:1180–1187. [PubMed] [Google Scholar]; (c) Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Gotz F. Mol. Microbiol. 2010;75:864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- 11.(a) Qiao Y, Srisuknimit V, Rubino F, Schaefer K, Ruiz N, Walker S, Kahne D. Nat. Chem. Biol. 2017;13:793–798. doi: 10.1038/nchembio.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Srisuknimit V, Qiao Y, Schaefer K, Kahne D, Walker S. J. Am. Chem. Soc. 2017;139:9791–9794. doi: 10.1021/jacs.7b04881. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Welsh MA, Taguchi A, Schaefer K, Van Tyne D, Lebreton F, Gilmore MS, Kahne D, Walker S. J. Am. Chem. Soc. 2017;139:17727–17730. doi: 10.1021/jacs.7b10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Rebets Y, Lupoli T, Qiao Y, Schirner K, Villet R, Hooper D, Kahne D, Walker S. ACS Chem. Biol. 2014;9:459–467. doi: 10.1021/cb4006744. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schaefer K, Matano LM, Qiao Y, Kahne D, Walker S. Nat. Chem. Biol. 2017;13:396–401. doi: 10.1038/nchembio.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Gale RT, Sewell EW, Garrett TA, Brown ED. Chem. Sci. 2014;5:3823–3830. [Google Scholar]; (b) Lee W, Schaefer K, Qiao Y, Srisuknimit V, Steinmetz H, Muller R, Kahne D, Walker S. J. Am. Chem. Soc. 2016;138:100–103. doi: 10.1021/jacs.5b11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helassa N, Vollmer W, Breukink E, Vernet T, Zapun A. FEBS J. 2012;279:2071–2081. doi: 10.1111/j.1742-4658.2012.08592.x. [DOI] [PubMed] [Google Scholar]
- 15.Thumm G, Gotz F. Mol. Microbiol. 1997;23:1251–1265. doi: 10.1046/j.1365-2958.1997.2911657.x. [DOI] [PubMed] [Google Scholar]
- 16.(a) Qiao Y, Lebar MD, Schirner K, Schaefer K, Tsukamoto H, Kahne D, Walker S. J. Am. Chem. Soc. 2014;136:14678–14681. doi: 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lebar MD, Lupoli TJ, Tsukamoto H, May JM, Walker S, Kahne D. J. Am. Chem. Soc. 2013;135:4632–4635. doi: 10.1021/ja312510m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye XY, Lo MC, Brunner L, Walker D, Kahne D, Walker S. J. Am. Chem. Soc. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 18.Eberhardt A, Hoyland CN, Vollmer D, Bisle S, Cleverley RM, Johnsborg O, Havarstein LS, Lewis RJ, Vollmer W. Microb. Drug Resist. 2012;18:240–255. doi: 10.1089/mdr.2011.0232. [DOI] [PubMed] [Google Scholar]
- 19.(a) Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. EMBO J. 2011;30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gale RT, Li FKK, Sun T, Strynadka NCJ, Brown ED. Cell Chem. Biol. 2017;24:1537–1546. doi: 10.1016/j.chembiol.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Lassila JK, Zalatan JG, Herschlag D. Annu. Rev. Biochem. 2011;80:669–702. doi: 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubscher J, Luthy L, Berger-Bachi B, Stutzmann Meier P. BMC Genomics. 2008;9:617. doi: 10.1186/1471-2164-9-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillen Schlippe YV, Hedstrom L. Arch. Biochem. Biophys. 2005;433:266–278. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 23.(a) Fitch CA, Platzer G, Okon M, Garcia-Moreno BE, McIntosh LP. Protein Sci. 2015;24:752–761. doi: 10.1002/pro.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xu B, Jacobs MI, Kostko O, Ahmed M. Chemphyschem. 2017;18:1503–1506. doi: 10.1002/cphc.201700197. [DOI] [PubMed] [Google Scholar]
- 24.Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SR. Proc. Natl. Acad. Sci. U S A. 2010;107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.