Abstract
Background
The staphylococci have increasingly been associated with infections worldwide and anti-microbial resistance has made these versatile pathogens more recalcitrant in the hospital setting.
Objectives
This study sought to investigate the occurrence and distribution of Staphylococcus species as well as determine the prevalence of methicillin resistant Staphylococcus aureus (MRSA) and methicillin resistant coagulase negative staphylococci (MRCoNS) among clinical samples from University of Benin Teaching Hospital (UBTH) in Benin City.
Methods
Ninety one (91) clinical isolates comprising S. aureus and Coagulase Negative staphylococci (CoNS) were recovered from routine clinical specimens and anti-microbial susceptibility tests were carried out. Polymerase Chain Reaction (PCR) was thereafter carried out on these isolates to detect mecA gene.
Results
Staphylococcus species had its highest prevalence from infected wounds of patients (28.8%) while urine samples showed the least (5.4%). The highest level of resistance was to ceftazidime (S. aureus - 68%, CoNS - 75.6%) while the least resistance was observed for meropenem (S. aureus- 26%, CoNS- 46.3%). Using phenotypic method (with 1µg oxacillin antibiotic disc), the distribution of MRSA and MRCoNS was 44.0% and 46.3% respectively. PCR analysis showed that 38.0% of S. aureus and 41.5% of the CoNS had mecA gene respectively; wound swabs showed the highest prevalence with 30.5% of staphylococcal isolates being mecA gene positive. There was also no significant association between the Staphylococcal isolates and their isolation rate, isolation site and mecA gene distribution (p > 0.05).
Conclusion
This study draws attention on the increase in the prevalence of mecA gene (39.6%) and an increase in multidrug resistant staphylococci when compared to previous studies in our country; it recommends laboratory guidance and periodic review to stem the tide of resistance.
Keywords: Staphylococcus aureus (S. aureus), coagulase-negative staphylococci (CoNS), mecA gene, methicillin-resistance
Introduction
Staphylococcus aureus is a non-motile, aerobic or facultative anaerobic Gram positive coccus that inhabits the nasal membranes and skin of warm blooded animals and man, in whom it could cause a range of infections from mild, such as skin infections and food poisoning, to life threatening, such as pneumonia, sepsis, osteomyelitis and infectious endocarditis1. The CoNS are part of the normal flora of human skin2, these organisms have relatively low virulence but are increasingly recognized as agents of clinically significant infection of the bloodstream and other sites3. Risk factors for CoNS infection include foreign bodies (such as indwelling prosthetic devices or intravascular catheters) and immune compromise3.
The introduction of methicillin in 1960 as an alternative antibiotic for the treatment of beta-lactamase (penicillinase) producing S. aureus was greeted with resistance almost immediately4,5. Since that time, strains of S. aureus resistant to methicillin emerged and became a major clinical problem within hospitals in Europe in the 1960s6, in the United States in the 1970s7, and in several countries of the world8. Resistance to methicillin and semisynthetic penicillins has been observed in more than 80% of coagulase-negative Staphylococcal isolates9. MRSAs and MRCoNS have therefore emerged as the most important cause of hospital-acquired infections (HAI) and community-acquired infections (CAI), resulting in increased morbidity and mortality in the hospital settings10,11.
The specific genetic mechanism of its resistance in MRSAs has been identified as a mobile genetic element (staphylococcal cassette chromosome mec) integrated into the S. aureus chromosome, within which the mecA gene encodes a specific methicillin-resistant transpeptidase known as penicillin-binding protein 2a (PBP2a)12. This protein has a low affinity for beta-lactam antimicrobial drugs, thus bacteria expressing this protein are resistant to all types of these drugs13, and in recent times; aminoglycosides and quinolones13.
In Nigeria, the occurrence of MRSA was first documented by researchers in 1987 with its prevalence pegged at 50.6%14. Subsequent studies have seen a variation in its prevalence15,16,17. Literature is growing on CoNS and its incrimination in clinical infections in Nigeria18,19; several factors affect the reliability of the disk diffusion technique for the detection of methicilin resistance among the staphylococci17,20. Though molecular methods have emerged as the gold standard, there is still little data on the distribution of MRSA and MRCoNS in our locality.
This study therefore aims at evaluating the prevalence and distribution of MRSAs and MRCoNS among staphylococci isolated from clinical samples by determining the distribution of mecA gene among these organisms in Benin City using PCR based method.
Materials and methods
Sources of clinical specimens
The staphylococci isolates used in this study were obtained by serial sampling. Ninety one (91) consecutive non-repetitive clinical isolates comprising S. aureus and CoNS were recovered from routine clinical specimens sent to the diagnostic laboratory of Medical Microbiology department, University of Benin Teaching Hospital, Benin city (UBTH).The clinical samples were; wound swabs (25), endocervical swabs (4), high vaginal swabs (3), eye swabs (4), ear swabs (9), urethral swabs (3), seminal fluid (10), blood (17), urine (12), aspirates (2) and urinary catheter tips (2). These specimens were collected from patients admitted in wards (in-patients), and from patients attending out-patient clinics.
Isolation and identification
The isolates were identified to be S. aureus following Gram stain that showed Gram positive cocci and biochemical tests which showed positive results for catalase and coagulase (slide and/or tube) as described in standard Medical Microbiology laboratory manual21. Similarly isolates identified as CoNS were Gram positive cocci and catalase positive but were negative for both slide and tube coagulase tests. The isolates were thereafter stored at 4°C on Mueller Hinton agar slants for further work.
Antimicrobial susceptibility testing
In vitro anti-bacterial susceptibility tests were performed on each isolate using anti-bacterial drugs. Plates were prepared with Mueller Hinton's agar for use in the Kirby-Bauer method. Broth cultures were adjusted to turbidity standard, equivalent to McFarland 0.5. This was then used for disc diffusion according to Clinical and Laboratory Standard Institute (CLSI) criteria, using the following discs; gentamicin (10 µg), ciprofloxacin (5 µg), ofloxacin (5 µg), amoxicillin-clavulanate (30 µg), cefuroxime (30 µg), ceftriaxone (30 µg) ceftazidime (30 µg), cloxacillin (5 µg), meropenem (10 µg) (all from Abtek Biologicals Ltd, Liverpool, U.K). These were then incubated at 37°C for 18 hrs. Sensitivity pattern was determined by measuring the zones of inhibition with a calibrated ruler and comparing with the control organism. Interpretative criteria for susceptibility tests were according to CLSI22.
An oxacillin disc (1 µg) was used to detect methicillin resistance. Sensitivity was read after incubation for 24 hrs at 35° C. Isolates were regarded as sensitive or resistant according to CLSI criteria23.
Multiple antibiotic resistance (MAR) index was calculated for each isolate using the formular;
Chromosomal DNA Extraction
DNA extraction was carried out as previously described24; Isolates were harvested into 1.0 ml of sterile water, vortexed to mix, and centrifuged at 10,000 r.p.m for 5 min. The supernatant was discarded and the pellets were washed again with sterile water. After this, 200µl of sterile water was added to the pellets, the pellets were vortexed to homogenize, and boiled in a dry bath at 100°C for 10 minutes. This was followed by vortexing and centrifugation at 12,000 r.p.m for 5 min. The supernatant containing the DNA were transferred to another tube and stored at −20°C. The concentration and purity of the extracted DNA was estimated using a nanodrop spectrophotometer.
PCR amplification of mecA gene.
The mecA gene was amplified using the primer set mecA1( AAAATCGATGGTAAAGGTTGGC) and mecA2 (5'AGTTCTGCAGTACCGGATTTTGC3') as described by Del Vechio et al (1995)25. PCR was performed in a 20 µl of a reaction mixture containing 1X PCR Buffer (Solis Biodyne), 1.5 mM MgCl2, 200 µM of each dNTP (Solis Biodyne), 20 pMol of each primer, 2.5 units of TaqDNA polymerase (Solis Biodyne), 10–200 ng of extracted DNA, and sterile distilled water was used to make up the reaction mixture. Thermal cycling was conducted in an Eppendorf Thermal Cycler Nexus series for an initial denaturation of 95°C for 5 min, followed by 30 consecutive cycles of 95°C for 30 sec; 55°C for 30 sec, and 72°C for 1 min. This was followed by a final extension step of 72°C for 10 min. The amplification product was separated on 1.5% agarose gel electrophoresis, visualized by ethidium bromide staining and photographed under Ultraviolet illumination. 100bp DNA ladder (Solis Biodyne) was used as DNA molecular weight standard. A single band with a molecular weight of 533bp signifies the presence of the mecA gene. S. aureus ATCC 43300 served as the positive control strain.
Calculation of sensitivity and specificity
Calculations of sensitivity were made by dividing the number of strains detected as resistant by the susceptibility test method by the total number of strains that were mecA positive. Specificity was calculated by dividing the number of strains detected as susceptible by the test method by the total number of strains that were mecA negative.
Statistical analysis
Statistical analysis was by the Chi (X2) square test using INSTAT® software. A p value of < 0.05 was deemed statistically significant.
Result
A total of 91 clinical isolates of Staphylococcus species (S. aureus - 50 (54.9%), CoNS - 41 (45.1%)) were recovered from clinical samples during the period of study. Although the isolation rate of Staphylococcus species was higher from males (52.7%), gender was however not a risk factor for Staphylococcal infection (X2 = 0.2637, P > 0.05). There was no significant association in the occurrence of Staphylococci and the isolation sites (X2 = 0.5481, P> 0.05).
The isolation rate of Staphylococcus species from clinical samples was highest from infected wounds of patients (28.8%) while urine samples showed the least isolation rate (5.4%) as seen in Table 1.
Table 1.
Distribution of Staphylococcus species recovered from clinical samples.
| Clinical Sample | No of Samples | No of Culture Positive samples |
Distribution in clinical samples |
|
| S. aureus | CoNS | |||
| Wound Swab | 87 | 62 | 13 (15.0) | 12 (13.8) |
| Endocervical swab |
35 | 9 | 1 (2.9) | 3 (8.6) |
| High vaginal swab |
39 | 13 | 2 (5.1) | 1 (2.6) |
| Eye swab | 22 | 5 | 2 (9.1) | 2 (9.1) |
| Ear swab | 58 | 22 | 6 (10.3) | 3 (5.2) |
| Urethral swab | 19 | 3 | 2 (10.5) | 1 (5.3) |
| Seminal fluid | 36 | 11 | 9 (25.0) | 1 (2.8) |
| Blood | 65 | 26 | 7 (10.8) | 10 (15.4) |
| Urine | 223 | 89 | 6 (2.7) | 6 (2.7) |
| Aspirates | 21 | 4 | 1 (4.8) | 1 (4.8) |
| Catheter tip | 10 | 5 | 1 (6.3) | 1 (6.3) |
Number in parentheses = value in percentage. CoNS- Coagulase negative Staphylococci
The anti-microbial susceptibility pattern showed varying degrees of resistance. The highest level of resistance was toceftazidime (S. aureus - 68%, CoNS - 75.6%). The antibiotic with the least resistance was meropenem (S. aureus-26%, CoNS- 46.3%). The antibiogram is well detailed in Table 2.
Table 2.
Antimicrobial susceptibility pattern of Staphylococcus species recovered from Clinical samples.
| Antibiotics | S. aureus | CoNS | ||
| n = 50 | n = 41 | |||
| S | R | S | R | |
| Oxacillin | 28 (56.0) | 22 (44.0) | 22 (53.7) | 19 (46.3) |
| Meropenem | 37 (74.0) | 13 (26.0) | 22 (53.7) | 19 (46.3) |
| Ceftriaxone | 27 (54.0) | 23 (46.0) | 16 (39.0) | 25 (61.0) |
| Cefuroxime | 31 (62.0) | 19 (3.8) | 19 (46.3) | 22 (53.7) |
| Ofloxacin | 33 (66.0) | 17 (34.0) | 19 (46.3) | 22 (53.7) |
| Amoxicillin-clavulanate | 30 (60.0) | 20 (40.0) | 21 (51.2) | 20 (48.5) |
| Cloxacillin | 26 (52.0) | 24 (48.0) | 15 (36.6) | 26 (63.4) |
| Ceftazidime | 16 (32.0) | 34 (68.0) | 10 (24.4) | 31 (75.6) |
| Gentamicin | 28 (56.0) | 22 (44.0) | 19 (46.3) | 22 (53.7) |
| Erythromycin | 28 (56.0) | 22 (44.0) | 15 (36.6) | 26 (63.4) |
CoNS- Coagulase negative Staphylococci. S – Sensitive, R – Resistant. Number in parentheses = value in percentage.
Using phenotypic method (with 1µg oxacillin antibiotic disc), the distribution of MRSA and MRCoNS was 44.0% and 46.3% respectively. Multiple antibiotic resistance (MAR) index was calculated for each of the isolates. It was observed that majority of isolates had MAR indices of above 0.2 (S. aureus — 60%, CoNS — 80.5%).
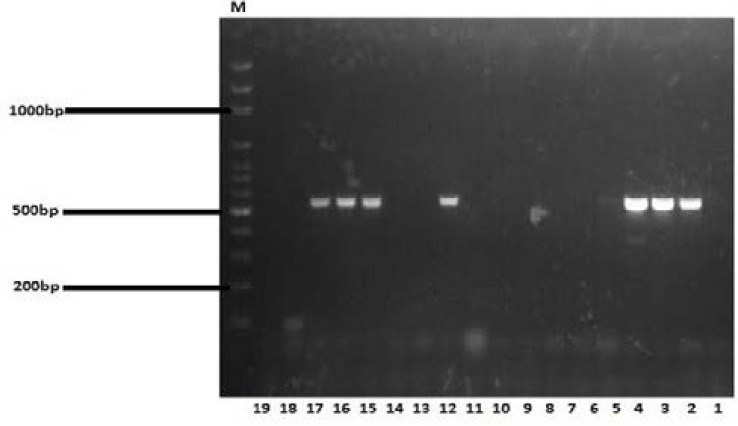
PCR was carried out on all staphylococcal isolates to detect mecA gene, a gene that confers resistance to methicillin and most β-lactam antibiotics. Figure 1 shows the Agarose gel electrophoresis of PCR product amplified from mecA genes for S. aureus and CoNS.
Figure 1.
Agarose gel electrophoresis of PCR product amplified from mecAgenes. M = DNA marker fragments. Lane 1 was the negative control, Lane 2 was the MRSA positive control; S. aureus ATCC 43300, Lane 3, 4, 12, 15, 16, and 17 indicate the mecA positive samples.. The DNA fragments of 533 bp were amplified from mecA gene.
On the whole, PCR analysis showed that 39.6% of Staphylococcus species recovered was mecA gene positive with 38.0% of S. aureus and 41.5% of the CoNS having mecA gene respectively. mecA gene was markedly distributed in wound swabs (30.5%) and was absent in high vaginal swabs and catheter tips (Table 4). However, there was no significant association between isolation site and the distribution of mecA gene among S. aureus (X2 = 0.7735, P > 0.05), and CoNS (X2 = 0.4259, P > 0.05). Seventeen (17) mecA+ S. aureus were oxacillin resistant (Sensitivity 89.5%) while 16 mecA+ CoNS were oxacilin resistant (Sensitivity 94.1%). Twenty-eight (28) of the 31 mecA- S. aureus strains were oxacilin sensitive (Specificity 90.3%) while Nineteen (19) of the 24 mecA- CoNS were oxacillin sensitive (specificity 79.1%).
Table 4.
Distribution of mecA gene among Staphylococcus species from clinical samples
| Clinical Sample |
N |
S. aureus mecA+ |
N | CoNS mecA+ |
| Wound swab | 13 | 4 (30.7) | 12 | 7 (58.3) |
| Endocervical swab |
1 | 0 | 3 | 1 (33.3) |
| High vaginal swab |
2 | 0 | 1 | 0 |
| Eye swab | 2 | 1 (50.0) | 2 | 0 |
| Ear swab | 6 | 3 (50.0) | 3 | 2 (66.6) |
| Urethral swab | 2 | 1 (50.0) | 1 | 1 (100) |
| Seminal fluid | 9 | 5 (55.5) | 1 | 0 |
| Blood | 7 | 2 (28.5) | 10 | 4 (40.0) |
| Urine | 6 | 2 (33.3) | 6 | 1 (16.6) |
| Aspirates | 1 | 1 (100) | 1 | 1 (100) |
| Catheter tips | 1 | 0 | 1 | 0 |
| Total | 50 | 19 (38.0) | 41 | 17 (41.5) |
CoNS- Coagulase Negative Staphylococci, N = Number of isolates, value in bracket = number in percentages.
There was no association between gender and the distribution of mecA gene among the Staphylococci (S. aureus; X2 = 0.3253, CoNS; X2 = 0.4205; P > 0.05). Similarly, mecA gene did not show significant association for the any one of the Staphylococci (X2 = 0.9604; P >0.05)
Discussion
In this study, 28.8% of Staphylococci isolated were from wound swabs. In a study which had Staphylococcal isolates recovered from eight hospitals which cut across South-Western, North-central and North-Eastern Nigeria, more than 80% of the total number of S. aureus isolates recovered was from infected wounds13. It has been observed that the skin of 80–90% of people is colonized with S. epidermidis and that most CoNS infections are acquired from patients own flora26. The CoNS have also shown a higher frequency of isolation from wound swabs when compared to other samples18. In Nigeria, CoNS is one of the common causes of infections of open fractures in wounds and delay in wound debridement has been reported to be a major predisposing factor to wound infection28. Similarly, S. aureus is found on the skin, axilla, anterior nares and groins as normal flora27. It is therefore not hard to imagine that it would be implicated in wound infection as there is a breach in the structural integrity of the skin.
Resistance was observed to oxacillin in this study, 44.0% and 46.3% for S. aureus and the CoNS respectively. This is in contrast to an Iranian study, where resistance was 88% and 60.3% for S. aureus and CoNS respectively29. Oxacillin and methicilin are hardly used in our setting as compared with the Iranian study which noted the rampant use of oxacillin. Our report is however similar to observations by Olowe et al in Osogbo, Nigeria, where 40.4% of S. aureus clinical isolates were resistant to oxacillin. Similarly, demonstrable levels of resistance was observed for meropenem (S. aureus- 26%, CoNS- 46.3%), though the drug is expensive and one of last resort in our locality30.
The study also shows that majority of staphylococcal isolates were multidrug resistant, with MAR indices > 0.2 (S. aureus — 60%, CoNS — 80.5%). Worthy of note is the high prevalence of multidrug resistant CoNS. The finding is similar to an observation by Akinjogunla and Enabulele (2010). However, in their study on Staphylococcal isolates from ear swabs of patients with otitis media, they found that 19.2% of S. aureus and 9.2% of CoNS had MAR indices above 0.831. An MAR index higher than 0.2 has been said to be indicative of isolates originating from an environment where antibiotics were often used32, the high indexes from this study therefore give credence to the observation of many researchers in Nigeria on the prevailing practice of indiscriminate use of antibiotics and lack of Laboratory guidance before institution of antimicrobial therapy16,17, as they may have induced resistance mechanisms over time in these versatile opportunists.
Using PCR method, 38.0% of S. aureus isolates and 41.5% of the CoNS had mecA gene respectively. Using phenotypic method (with oxacillin antibiotic disc) however, the distribution of MRSA and MRCoNS was 44.0% and 46.3% respectively. Differing rates in susceptibility pattern has been observed by researchers. Olowe et al in drawing comparisons between methicillin, oxacillin, cefoxitin and the gold standard PCR, observed that all the antibiotics mentioned slightly overestimated methicillin resistance in S. aureus30. Hyperproduction of β-lactamase enzyme has been observed among mecA negative MRSAs in Nigeria35. Besides the challenge of heterogeneity of the strains, this could be a resistance mechanism for oxacillin-resistant mecA gene negative Staphylococci as observed in this study, more research is therefore needed. There was no significant association between the distribution of Staphylococci and the distribution of mecA gene among clinical samples in this study (P > 0.05), as all samples showed some percentage distribution of the gene with the exception of catheter tips and high vaginal swabs. mecA gene was markedly distributed among the staphylococci recovered from wound swabs in this study (S. aureus-30.7%, CoNS-58.3%). This is comparable to a study in Osogbo (S. aureus- 52.2%)30, and two multi-centre studies evaluating the prevalence of mecA gene among the staphylococci from clinical samples13,17.
Noteworthy in this study is the rising prevalence of MRSA and MRCoNS from blood of patients showing signs of septicaemia, as 28.5% of S. aureus and 40.0% of the CoNS were mecA positive. This is in sharp contrast to some studies in Nigeria in which all S. aureus isolated from the blood of patients showing similar symptoms were mecA gene negative17,30, however, the gene was detected among CoNS notably S. haemolyticusca having septicemia in Lagos, Nigeria13. Some researchers have observed the rising prevalence of MRCoNS from patients with septicaemia in India; 12% and 54% respectively33, 34.
In our country, little attention has been accorded the CoNS to the extent of screening for mecA gene among them. Literature is however growing on its pathogenicity and increased isolation from clinical infections18,19. The marked distribution of mecA gene among these isolates (41.5%), as well as the insignificant difference in their isolation rate and distribution of mecA gene when compared with S. aureus recovered from clinical samples is a pointer to the fact that they should be accorded equal status as opportunistic pathogens (P > 0.05).
Conclusion
Summarily, this study notes a rising percentage in the distribution of mecA gene among the Staphylococci recovered from clinical samples in Benin when compared with previous studies in our country, it reports increasing level of multidrug resistant staphylococci with high MAR index, it reiterates the continued practice of indiscriminate use of antibiotics and emphasizes laboratory guidance before institution of anti-microbial therapy. Efforts should also be made to enact regulations on antibiotic usage. Periodic review of susceptibility pattern, molecular epidemiological surveys and surveillance is equally imperative. These when implemented promises to stem the tide of anti-mi crobial resistance.
Table 3.
Multiple antibiotic resistance (MAR) index of Staphylococcus species recovered from clinical samples.
| MAR Index | S. aureus | CoNS |
| 0.2 | 5 (16.0) | 1(2.8) |
| 0.3 | 2 (4.0) | 3 (8.3) |
| 0.4 | 1 (2.0) | 1 (2.8) |
| 0.5 | 6 (12.0) | 5 (13.8) |
| 0.6 | 1 (2.0) | 1(2.8) |
| 0.7 | 2 (4.0) | 4 (11.1) |
| 0.8 | 5 (10.0) | 3 (8.3) |
| 0.9 | 8 (16.0) | 7 (19.4) |
| 1.0 | 7 (14.0) | 8 (22.2) |
Number in parentheses = value in percentage.
CoNS- Coagulase negative Staphylococci
Conflict of interest
None declared.
References
- 1.Projan SJ, Novick RP. The molecular basis of pathogenicity. In: Crossley KB, Archer GL, editors. The Staphylococci in human diseases. New York: Churchill Livingstone; 1997. pp. 55–81. [Google Scholar]
- 2.Roth RR, James WD. Microbial ecology of the skin. Annual Revision Microbiology. 1988;42:441. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- 3.Longauerova A. Review: Coagulase negative Staphylococci and their participation in pathogenesis of human infections. Bratislavia Medical Journal. 2006;107:448–452. [PubMed] [Google Scholar]
- 4.Barber M. Methicillin-resistant staphylococci. Journal of Clinical Pathology. 1961;14:385–393. doi: 10.1136/jcp.14.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jevons M. Celbenin resistant Staphylococci. British Medical Journal. 1961;1:124–125. [Google Scholar]
- 6.Parker MT, Hewitt MJH. Methicillin resistance in Staphylococcus aureus. Lancet. 1970;1:800–804. doi: 10.1016/s0140-6736(70)92408-6. [DOI] [PubMed] [Google Scholar]
- 7.Haley RW, Hightower AW, Khabbaz RF. The emergence of methicillin resistant Staphylococcus aureus in United States hospitals. Annals of Internal Medicine. 1982;97:297–308. doi: 10.7326/0003-4819-97-3-297. [DOI] [PubMed] [Google Scholar]
- 8.Taiwo SS. Methicillin Resistance in Staphylococcus aureus: A Review of the Molecular Epidemiology, Clinical Significance and Laboratory Detection Methods. West African Journal of Medicine. 2009;28(5):281–290. doi: 10.4314/wajm.v28i5.54998. [DOI] [PubMed] [Google Scholar]
- 9.Diekema DJ, Pfaller MA, Schmitz FJ, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clinical Infectious Disease. 2001;32(2):114. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 10.Lowy FD. Staphylococcus aureus infections. New England Journal of Medicine. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 11.Cosgrove S, Sakoulas G, Perencevich E, Schwaber M, Karchmer A, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clinical Infectious Disease. 2003;36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 12.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrobial Agents Chemotheraphy. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shittu A, Oyedara O, Abegunrin F, Okon K, Raji A, Taiwo S, Ogunsola F, Onyedibe K, Elisha G. Characterization of methicillin-susceptible and -resistant staphylococci in the clinical setting: a multicentre study in Nigeria. BMC Infectious Diseases. 2012;12:286. doi: 10.1186/1471-2334-12-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotimi VO, Orebanjo O, Banjo TO, Nwobu R, Onyenefa PI. Occurrence and antibiotic susceptibility profile of methicillin-resistant Staphylococcus aureus in LUTH Lagos. Central African Journal of Medicine. 1987;33:95–98. [PubMed] [Google Scholar]
- 15.Onemu OS, Ophori EA. Prevalence Of Multi-Drug Resistant Staphylococcus aureus In Clinical Specimens Obtained From Patients Attending The University Of Benin Teaching Hospital, Benin City, Nigeria. Journal of Natural Sciences Research. 2013;5:154–159. [Google Scholar]
- 16.Olayinka BO, Olayinka AT, Onaolapo JA, Olurinola PF. Pattern of resistance to vancomycin and other antimicrobial agents in staphylococcal isolates in a University Teaching Hospital. African Journal of Clinical and Experimental Microbiology. 2005;6:21–27. [Google Scholar]
- 17.Alli OAT, Ogbolu DO, Akorede E, Onemu OM, Okanlawon BM. Distribution of mecA gene amongst Staphylococcus aureus isolates from SouthWestern Nigeria. African Journal of Biomedical Research. 2011;14:9–16. [Google Scholar]
- 18.Azih A, Enabulele I. Species Distribution and Virulence Factors of Coagulase Negative Staphylococci Isolated From Clinical Samples From the University of Benin Teaching Hospital, Edo State, Nigeria. Journal of Natural Sciences Research. 2013;3(9):38–43. [Google Scholar]
- 19.Ogbolu DO, Daini OA, Alli OT, Adesina OA, Odekanmi AA, Okanlawon BM, Olusoga-Ogbolu FF, Oni AA. Coagulase Negative Staphylococci distribution in clinical samples in a tertiary hospital in Ibadan, Nigeria. Nigerian Journal of Health and Biomedical Sciences. 2009;8:1. [Google Scholar]
- 20.Abdalla AM, Silma LI, Masri MAR. Molecular detection of methicillin resistant Staphylococcus aureus strains (MRSA) Isolated from wound infections. American Journal of Research Communication. 2014;2(9):69–81. [Google Scholar]
- 21.Cheesbrough M. District Laboratory Practice in Tropical Countries Part 2. Cambridge: Cambridge University Press; 2009. [Google Scholar]
- 22.Clinical Laboratory Standards Institute (CLSI), author Performance standards for anti-microbial susceptibility testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2007. M100-S17. [Google Scholar]
- 23.Clinical and Laboratory Standard Institute (CLSI), author Performance standards for anti-microbial disk susceptibility tests. Approved standard N2A8. 25. Vol. 15. Wayne, PA: Clinical and Laboratory Standard Institute; 2005. p. 1. PubMed. [Google Scholar]
- 24.Smith SI, Fowora MA, Otegbayo JA, Abdulkareem FB, Omonigbehin EA, Adegboyega A, Contreras M, Haas Comparison of PCR with other diagnostic techniques for the detection of H. pylori infection in patients presenting with gastroduodenal symptons in Nigeria. International Journal of Molecular Epidemiology and Genetics. 2011;15(2):178–184. [PMC free article] [PubMed] [Google Scholar]
- 25.Del Vecchio VG, Petroziello JM, Gress MJ, McCleskey FK, Melcher GP, Crouch HK, Lupski JR. Molecular Genotyping of Methicillin-Resistant Staphylococcus aureus via Fluorophore-Enhanced Repetitive-Sequence PCR. Journal of Clinical Microbiology. 1995;33:2141–2144. doi: 10.1128/jcm.33.8.2141-2144.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lark RI, VanderHyde K, Deeb GM, Dietrich S, Massay JP, Chenoweth C. An outbreak of coagulose negative Staphylococcal surgical site infections following Aortic valve replacement. Infection Control and Hospital Epidemiology. 2001;22(10):618–623. doi: 10.1086/501832. [DOI] [PubMed] [Google Scholar]
- 27.Ikem IC, Oginni LM, Bamgboye EA, Ako-Nai AK, Onipede AO. The bacteriology of Open fractures in Ile-Ife, Nigeria. Nigerian Journal of Medicine. 2004;13(4):359–365. [PubMed] [Google Scholar]
- 28.Davies CP. Normal flora. In: Baron S, editor. Medical Microbiology. 4th Edition. Galveston, Texas: The University of Texas Medical Branch; 1996. pp. 113–119. PubMed. [Google Scholar]
- 29.Rahimi F, Bouzari M, Maleki Z, Rahimi F. Antibiotic susceptibility pattern among Staphylococcus spp. with emphasis on detection of mecA gene in methicillin resistant Staphylococcus aureus isolates. Iranian Journal of Clinical Infectious Diseases. 2009;4(3):143–150. [Google Scholar]
- 30.Olowe OA, Kukoyi OO, Taiwo SS, Ojurongbe O, Opaleye OO, Bolaji OO, Adegoke AA, Makanjuola OB, Ogbolu DO, Alli OA. Phenotypic and molecular characteristics of methicillin-resistant Staphylococcus aureus isolates from Ekiti state, Nigeria. Infection and Drug Resistance. 2003;6:87–92. doi: 10.2147/IDR.S48809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akinjogunla OJ, Enabulele IO. Virulence Factors, Plasmid Profiling and Curing analysis of Multidrug Resistant Staphylococcus aureus and Coagulase negative Staphylococcus species isolated from Patients with Acute Otitis Media. Journal of American Science. 2010;6(11):1022–1033. [Google Scholar]
- 32.Paul S, Bezbarauh RL, Roy MK, Ghosh AC. Multiple antibiotic resistance (MAR) index and its reversion in Pseudomonas aeruginosa. Letters in Applied Microbiology. 1997;24:169–171. doi: 10.1046/j.1472-765x.1997.00364.x. [DOI] [PubMed] [Google Scholar]
- 33.Baumgarth S, Susan EH, Joseph MC, Richard AP. Sepsis with coagulase negative staphylococci in critically ill newborns. American Journal of Diseases in Children. 1983;137:461–463. doi: 10.1001/archpedi.1983.02140310043012. [DOI] [PubMed] [Google Scholar]
- 34.Shubhra S, Gopa B, Agarwa SK, Anuradha R, Piyush T, Mala K, Shraddha S, Singh RK. Prevalence of mecA gene positive coagulase negative Staphylococci in NICU of a tertiary care hospital. Biomedical Research. 2009;20(2):94–98. [Google Scholar]
- 35.Olayinka BO, Olayinka AT, Obajuluwa AF, Onaolapo JA, Olurinola PF. Absence of mecA gene in methicillin-resistant Staphylococcus aureus isolates. African Journal of Infectious Disease. 2009;3(2):49–56.. [Google Scholar]