Abstract
Introduction:
A number of studies indicated racial differences in cervical cancer outcomes and several factors are associated with it such as stage, comorbidities, treatment pattern, and socioeconomic status. However, the associations of tumor genomic patterns such as phosphatidylinositol 3-kinase catalytic subunit alpha (PIK3CA) gene mutations and amplifications with cervical cancer racial disparities are largely unexplored.
Objectives:
Therefore, the present investigation aimed to identify genetic alterations (mutations and copy number variations) in cervical cancer and determine whether the PIK3CA gene mutations and amplifications in cervical cancer differ across racial/ethnic groups in the United States.
Methods:
This study made use of The Cancer Genome Atlas (TCGA) database. TCGA database is a publicly available database that was created by a joint effort between the National Cancer Institute and the National Human Genome Research Institute. The two-tailed Fisher’s exact test was used to test for associations between the categorical variables, race, and PIK3CA gene mutation as well as PIK3CA gene amplification using the “Fisher test” function in R.
Results:
There were 309 cervical cancer samples, and of these, 194 samples had gene mutations and 295 samples had copy number alteration data. The top five mutated genes in TCGA dataset were PIK3CA, MUC4, KMT2C, SYNE1, and KMT2D. The top five amplified genes in TCGA dataset were MECOM, TP63, PRKCI, PIK3CA, and TRFC. The PIK3CA gene had the highest number of mutations with 53 mutation counts. The mutation rates were 62.5%, 31.3%, 25.4%, and 21.1% for American Indian, African American, White, and Asian, respectively. The amplification rates were 28.6%, 21.1%, 18.9%, and 12.5% for African American, Asian, White, and American Indian.
Conclusions:
There are many significantly mutated or amplified genes implicated in cervical cancer. Some of them are not grouped with the already known genes in relation to cervical cancer. For example, the KMT2C, SYNE1, KMT2D, EP300, RYR2, FLG, DMD, FBXW, MECOM, TRFC, RPL35A, LPP, TBL, FGF12, and SOX2 genes. They can be explored as therapeutic targets to improve cervical cancer treatment.
Keywords: Amplification, cervical cancer, ethnicity, MECOM, mutation, PIK3CA, race
Introduction
Despite the fact that cancer of the cervix is known to be a preventable cancer, it remains one of the major causes of cancer-related deaths in females under the age of 60.[1-3] Cervical cancer is the third most commonly diagnosed cancer and the fourth leading cause of cancer-related death in females worldwide with over 527,000 new cases,[1,2] and perhaps the second most common cancer among women in the third world countries.[3-5] In the United States, over 12,000 women were diagnosed with cervical cancer in 2015.[6]
The PIK3CA gene is the most commonly implicated gene in human cancer.[7-12] The PIK3CA gene is located on the long (q) arm of chromosome 3 at position 26.3 (3q26.3).[13] The PIK3CA gene provides information for making the p110 alpha (p110α) protein, which is the catalytic subunit of the phosphatidylinositol 3-kinase (PI3K) enzyme.[13] This is an important enzyme in the PI3K pathway. The PI3K pathway is essential for several cellular activities such as cell metabolism, cell survival, cell growth, and proliferation.[13]
The PI3K enzyme phosphorylates phosphatidylinositol -4,5-bisphosphate (PIP2) to phosphatidylinositol -3,4,5- triphosphate (PIP3).[13] The PIP3 induces the activation of kinase enzyme, protein kinase B (PKB), or AKT. The activated PKB, in turn, regulates the function of enzymes involved in cell growth and division such as mammalian target of rapamycin complex 1 (mTORC1).[14] The mTORC1 serves as the main regulator of cell metabolism, growth, and survival. The mTORC1 regulates cell growth and division by encouraging various anabolic actions such as synthesis of proteins, organelles, and lipids and by preventing catabolic processes.[14] PIK3CA gene amplification and mutations are two common causes of excessive activation of this pathway in cancer.[13] PIK3CA pathway over activation has been associated both with higher rates of local recurrence after radiotherapy and with decreased survival.[7,8,12] In addition, PIK3CA gene mutations have been associated with decreased survival after radical chemoradiotherapy.[7]
A number of studies indicated racial differences in cervical cancer outcomes and several factors are associated with it such as stage,[15-23] comorbidities,[24] treatment differences,[15,16,18,20,24,25] and socioeconomic status.[16,17,24,26-32] However, the associations of tumor genomic patterns such as PIK3CA gene mutation as well as PIK3CA gene amplification with cervical cancer racial disparities are unknown. Thus, the central purposes of this study were to identify genetic mutation as well as copy number variations in The Cancer Genome Atlas (TCGA) cervical cancer data and to examine racial differences in both PIK3CA gene mutation and PIK3CA gene amplification. Identifying tumor genomic patterns across different racial groups can lead to the discovery of therapeutic targets and improved cervical cancer treatments that will contribute to alleviating cervical cancer outcomes.[33]
Methods
This study made use of TCGA database. TCGA database is a publicly available database that was created by a joint effort between the National Cancer Institute and the National Human Genome Research Institute. TCGA database has comprehensive key genomic changes in 33 types of malignancy, including cervical cancer.[34] The two CESC genomic profiles used were mutation data from whole exome sequencing and putative copy-number alteration data from GISTIC 2.0. Clinical profiles including race were also accessed. The CESC clinical and genomic profiles were submitted to TCGA between 2011 and 2014. These were accessed through the cBioPortal for Cancer Genomics analytic tool. The cBioPortal is an open-access analytic tool that allows visualization, downloading, and analyzing of TCGA datasets.[35,36] The R statistical software was also utilized for the analysis. The two-tailed Fisher’s exact test was used to test for associations between the categorical variables, race, PIK3CA gene mutation, and PIK3CA gene amplification using the “Fisher test” function in R. The two-tailed Fisher’s exact test was selected considering the small sample size. Statistical significance was defined as P < 0.05. The analysis excluded variables that have no available values or sample values < 5.
Results
There were 309 cervical cancer samples, and of these, 194 samples had gene mutations and 115 samples did not have gene mutations. There were 295 samples with gene copy number variations and 14 samples without gene copy number variations. There were many significantly mutated genes. The top 10 mutated genes in TCGA dataset were PIK3CA, MUC4, KMT2C, SYNE1, KMT2D, EP300, RYR2, FLG, DMD, and FBXW7.
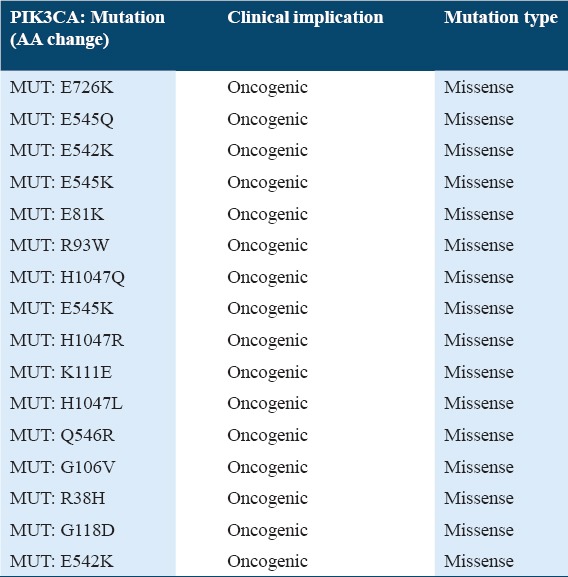
Table 1 shows the top 10 mutated genes in 194 profiled samples. The PIK3CA gene has the highest number of mutations with 53 mutation counts and 23.7%. The FBXW7 gene, the tenth frequent mutated gene has 19 mutation counts and 9.8%. The 53 PIK3CA gene mutations were all missense point mutations. Table 2 shows the common PIK3CA gene mutations in the dataset. Out of the 53 PIK3CA missense mutations, 48 mutations were known to be oncogenic (i.e., the mutated PIK3CA proteins have augmented catalytic action, leading to increased downstream signaling and oncogenic alteration). Thus, the percentage of PIK3CA mutations with oncogenic effect was about 91%.
Table 1.
Ten 10 mutated genes in 194 profiled samples
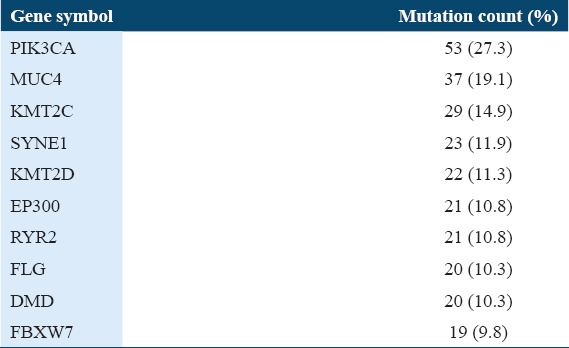
Table 2.
Types of mutation in PIK3CA gene
The white race has the highest number of samples (211), followed by Black or African American (31), Asian (20), American Indian or Alaska Native (8) while native Hawaiian or other Pacific Islander close off with having the lowest number of samples. Table 3 shows the comparison of PIK3CA gene mutation status by race. The racial group with the highest mutation rate was the American Indian or Alaska Native (62.5%), followed by Black or African American (31.3%), white (25.4%), and Asian (21.1%). The Fisher’s exact result was 0.0792 for the PIK3CA gene mutation status by race.
Table 3.
PIK3CA gene mutation status by race
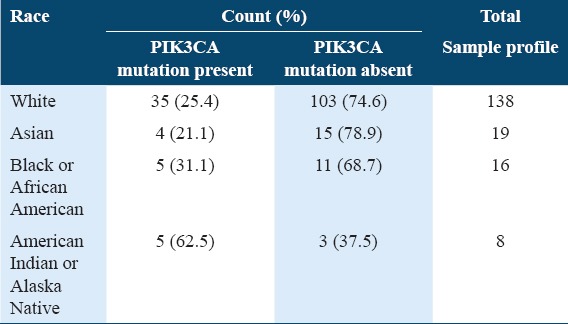
Table 4 shows the top 10 amplified genes in 295 profiled samples. The top 10 amplified genes in TCGA dataset were MECOM, TP63, PRKCI, PIK3CA, TRFC, RPL35A, LPP, TBL, FGF12, and SOX2. The MECOM gene has the highest numbers of amplification with 62 counts and 20.3%. The PIK3CA gene, the fourth frequent amplified gene has 60 counts and 20.3%. The 60 PIK3CA copy number variations were all due to the amplification of PIK3CA gene. There was no PIK3CA copy number variation that was due to deletion.
Table 4.
Ten 10 CNA genes in 295 profiled samples
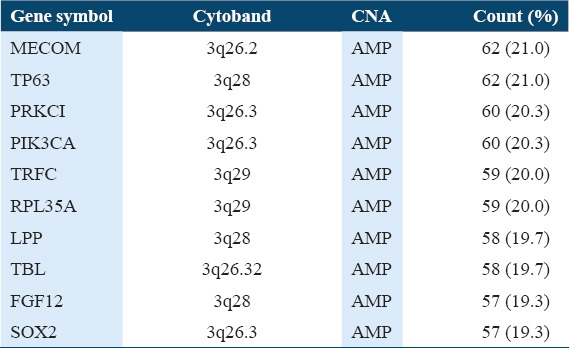
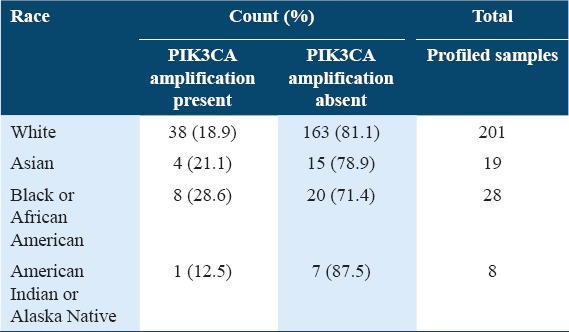
Table 5 shows the comparison of PIK3CA gene amplification status by race. The racial group with the highest rates of PIK3CA amplification was the Black or African American (28.6%), followed by the Asian (21.1%), white (18.9%), and American Indian or Alaska Native (12.5%). The Fisher’s exact result was 0.234 for the PIK3CA gene amplification status by race.
Table 5.
PIK3CA amplification status by race
Discussion
There are many significantly mutated or amplified genes implicated in cervical cancer. Some of them are not well-known genes in relation to cervical cancer. For example, the KMT2C, SYNE1, KMT2D, EP300, RYR2, FLG, DMD, and FBXW genes are not commonly implicated gene mutations in cervical cancer, but they are significantly mutated in TCGA cervical cancer dataset. Similarly, MECOM, TRFC, RPL35A, LPP, TBL, FGF12, and SOX2, are not already well-established gene amplifications implicated in cervical cancer, but they are significantly amplified in TCGA dataset.
The Fisher’s exact test showed that there were no significant differences in PIK3CA gene mutation status and amplification status by race. This study has helped us to understand that PIK3CA gene mutation and PIK3CA gene amplification do not differ across racial groups with cervical cancer, unlike with breast cancer in which racial differences in certain genes contribute to racial disparities in mortality and survival.[37] However, there are many other significantly mutated/amplified genes such as KMT2C, SYNE1, KMT2D, EP300, RYR2, FLG, DMD, and FBXW that can be explored as therapeutic targets to improve cervical cancer treatment. The finding that PIK3CA is the most frequently mutated gene in this study is similar to the result of the study conducted by Wright et al.;[12] however, in addition to the PIK3CA gene, they also found the KRAS and EGFR genes.[12] Similarly, Xiang et al. and Lou et al. found the PIK3CA gene as the highest mutated gene in cervical cancer.[11,38]
Limitations of the study and future perspectives
As with any study, this investigation has limitations. The available TCGA dataset for cervical cancer was small; thus, the generalization of this study must be done with caution. Further evaluation with a larger dataset will be required to validate these findings. Focusing on different gene mutations and amplifications that may be implicated in racial differences are highly encouraged.
Conclusions
There are many significantly mutated or amplified genes implicated in cervical cancer. Some of them are not grouped with the already known genes in relation to cervical cancer. They can be explored as therapeutic targets to improve cervical cancer treatment. PIK3CA gene mutation and amplification in cervical cancer do not differ significantly across the various racial groups. The findings of this study are important as they will allow scientists to focus on different gene mutations/amplifications that may be implicated in racial differences. Furthermore, this can push for the creation of targeted prevention and treatment programs that will address other attributed causes, other than those related to genomics.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Galani E, Christodoulou C. Human papilloma viruses and cancer in the post-vaccine era. Clin Microbiol Infect. 2009;15:977–81. doi: 10.1111/j.1469-0691.2009.03032.x. [DOI] [PubMed] [Google Scholar]
- 3.Odekunle FF. Contributions of epidemiological and laboratory methods to the establishment of the causal association of human papillomavirus with cancer of the cervix. Int J Recent Adv Multidiscip Res. 2017;4:2166–70. [Google Scholar]
- 4.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 6.Downs LS, Smith JS, Scarinci I, Flowers L, Parham G. The disparity of cervical cancer in diverse populations. Gynecol Oncol. 2008;109(2 Suppl):S22–30. doi: 10.1016/j.ygyno.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 7.McIntyre JB, Wu JS, Craighead PS, Phan T, Köbel M, Lees-Miller SP, et al. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol Oncol. 2013;128:409–14. doi: 10.1016/j.ygyno.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Kim TJ, Lee JW, Song SY, Choi JJ, Choi CH, Kim BG, et al. Increased expression of pAKT is associated with radiation resistance in cervical cancer. Br J Cancer. 2006;94:1678–82. doi: 10.1038/sj.bjc.6603180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC, Whang-Peng J, et al. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–44. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 10.Lee JE, Huh SJ, Park W, Lim DH, Ahn YC, Park CS, et al. Radical radiotherapy for locally advanced cancer of uterine cervix. Cancer Res Treat. 2004;36:222–7. doi: 10.4143/crt.2004.36.4.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang L, Jiang W, Li J, Shen X, Yang W, Yang G, et al. PIK3CA mutation analysis in Chinese patients with surgically resected cervical cancer. Sci Rep. 2015;5:14035. doi: 10.1038/srep14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright AA, Howitt BE, Myers AP, Dahlberg SE, Palescandolo E, Van Hummelen P, et al. Oncogenic mutations in cervical cancer: Genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer. 2013;119:3776–83. doi: 10.1002/cncr.28288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genetics Home Reference Your Guide to Understanding Genetic Conditions. [Last accessed on 2016 Aug 1]. Available from: https://www.ghr.nlm.nih.gov/gene/PIK3CA .
- 14.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–94. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leath CA, 3rd, Straughn JM, Jr, Kirby TO, Huggins A, Partridge EE, Parham GP. Predictors of outcomes for women with cervical carcinoma. Gynecol Oncol. 2005;99:432–6. doi: 10.1016/j.ygyno.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 16.Brooks SE, Baquet CR, Gardner JF, Moses G, Ghosh A. Cervical cancer-the impact of clinical presentation, health and race on survival. J Assoc Acad Minor Phys. 2000;11:55–9. [PubMed] [Google Scholar]
- 17.Morgan MA, Behbakht K, Benjamin I, Berlin M, King SA, Rubin SC. Racial differences in survival from gynecologic cancer. Obstet Gynecol. 1996;88:914–8. doi: 10.1016/s0029-7844(96)00342-0. [DOI] [PubMed] [Google Scholar]
- 18.Eggleston KS, Coker AL, Williams M, Tortolero-Luna G, Martin JB, Tortolero SR. Cervical cancer survival by socioeconomic status, race/ethnicity, and place of residence in Texas 1995-2001. J Womens Health (Larchmt) 2006;15:941–51. doi: 10.1089/jwh.2006.15.941. [DOI] [PubMed] [Google Scholar]
- 19.del Carmen MG, Montz FJ, Bristow RE, Bovicelli A, Cornelison T, Trimble E. Ethnic differences in patterns of care of stage 1A(1) and stage 1A(2) cervical cancer: A SEER database study. Gynecol Oncol. 1999;75:113–7. doi: 10.1006/gyno.1999.5543. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell JB, McCormack LA. Time trends in late-stage diagnosis of cervical cancer. Differences by race/ethnicity and income. Med Care. 1997;35:1220–4. doi: 10.1097/00005650-199712000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Mandelblatt J, Andrews H, Kerner J, Zauber A, Burnett W. Determinants of late stage diagnosis of breast and cervical cancer: The impact of age, race, social class, and hospital type. Am J Public Health. 1991;81:646–9. doi: 10.2105/ajph.81.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones WB, Shingleton HM, Russell A, Chmiel JS, Fremgen AM, Clive RE, et al. Patterns of care for invasive cervical cancer. Results of a national survey of 1984 and 1990. Cancer. 1995;76(10 Suppl):1934–47. doi: 10.1002/1097-0142(19951115)76:10+<1934::aid-cncr2820761310>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Mundt AJ, Connell PP, Campbell T, Hwang JH, Rotmensch J, Waggoner S. Race and clinical outcome in patients with carcinoma of the uterine cervix treated with radiation therapy. Gynecol Oncol. 1998;71:151–8. doi: 10.1006/gyno.1998.5203. [DOI] [PubMed] [Google Scholar]
- 24.Katz A, Eifel PJ, Moughan J, Owen JB, Mahon I, Hanks GE. Socioeconomic characteristics of patients with squamous cell carcinoma of the uterine cervix treated with radiotherapy in the 1992 to 1994 patterns of care study. Int J Radiat Oncol Biol Phys. 2000;47:443–50. doi: 10.1016/s0360-3016(00)00417-x. [DOI] [PubMed] [Google Scholar]
- 25.Coker AL, Du XL, Fang S, Eggleston KS. Socioeconomic status and cervical cancer survival among older women: Findings from the SEER-Medicare linked data cohorts. Gynecol Oncol. 2006;102:278–84. doi: 10.1016/j.ygyno.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 26.Bradley CJ, Given CW, Roberts C. Health care disparities and cervical cancer. Am J Public Health. 2004;94:2098–103. doi: 10.2105/ajph.94.12.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenwald HP, Polissar NL, Dayal HH. Race, socioeconomic status and survival in three female cancers. Ethn Health. 1996;1:65–75. doi: 10.1080/13557858.1996.9961771. [DOI] [PubMed] [Google Scholar]
- 28.Murphy M, Goldblatt P, Thornton-Jones H, Silcocks P. Survival among women with cancer of the uterine cervix: Influence of marital status and social class. J Epidemiol Commun Health. 1990;44:293–6. doi: 10.1136/jech.44.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samelson EJ, Speers MA, Ferguson R, Bennett C. Racial differences in cervical cancer mortality in Chicago. Am J Public Health. 1994;84:1007–9. doi: 10.2105/ajph.84.6.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shelton D, Paturzo D, Flannery J, Gregorio D. Race, stage of disease, and survival with cervical cancer. Ethn Dis. 1992;2:47–54. [PubMed] [Google Scholar]
- 31.Singh GK, Miller BA, Hankey BF, Edwards BK. Persistent area socioeconomic disparities in U.S. incidence of cervical cancer, mortality, stage, and survival 1975-2000. Cancer. 2004;101:1051–7. doi: 10.1002/cncr.20467. [DOI] [PubMed] [Google Scholar]
- 32.Farley JH, Hines JF, Taylor RR, Carlson JW, Parker MF, Kost ER, et al. Equal care ensures equal survival for African-American women with cervical carcinoma. Cancer. 2001;91:869–73. [PubMed] [Google Scholar]
- 33.Odekunle FF. Racial and socioeconomic disparities in cervical cancer mortality. Int J Health Sci Res. 2017;7:232–6. [Google Scholar]
- 34.TCGA Research Network. [Last accessed on 2016 Sep 23]. Available from: http://www.cancergenome.nih.gov .
- 35.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keenan T, Moy B, Mroz EA, Ross K, Niemierko A, Rocco JW, et al. Comparison of the genomic landscape between primary breast cancer in African American versus white women and the association of racial differences with tumor recurrence. J Clin Oncol. 2015;33:3621–7. doi: 10.1200/JCO.2015.62.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou H, Villagran G, Boland JF, Im KM, Polo S, Zhou W, et al. Genome analysis of latin american cervical cancer: Frequent activation of the PIK3CA pathway. Clin Cancer Res. 2015;21:5360–70. doi: 10.1158/1078-0432.CCR-14-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]


