Abstract
Introduction:
Systemic lupus erythematosus (SLE) is an autoimmune disease with various presentations. This variation is due to the interaction of hormonal, environmental, and genetic factors. Associations between human leukocyte antigens and SLE have long been recognized in different ethnic populations and have been suggested to represent the most important association.
Objectives:
The objectives of this paper were to determine susceptibility and protection human leukocyte antigens (HLA) Class II markers for SLE and to highlight, for the first time, associations between HLA alleles and clinical and serological features in South Tunisia.
Methods:
We conducted a case–control study on 75 SLE patients and 123 healthy controls. The HLA Class II DRB1/DQB1 of all patients and controls was genotyped using polymerase chain reaction-sequence specific primer technique. Statistical analysis was performed using SPSS software.
Results:
HLA-DRB1*03 was the principal Class II allele associated with the genetic susceptibility to SLE (pc = 0.02; OR = 2.57; CI = [1.39–4.75]; this allele was also associated with anti-SSB production (P = 0.016; OR = 4.00; CI = [1.24–12.96]). HLA-DRB1*01 was significantly more expressed in SLE patients with neurologic disorders (P = 0.013; OR = 20.25; CI = [1.87–219.21]). No allele was found to be protective against SLE in our study group.
Conclusion:
Our results show that in South Tunisia SLE is associated with HLA-DRB1*03 and that some clinical features of SLE may be influenced by specific DRB1 and DQB1 alleles.
Keywords: Association, disease clinical expression, human leukocyte antigens, systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) is the prototype of autoimmune diseases with various presentations from mild to life-threatening multiple organ. The autoimmune process results in the production of autoantibodies directed against components of the cell nucleus such as double-stranded DNA, nucleosome, and Sm protein… After binding to autoantigens, these autoantibodies may settle on organs such as joints, skin, kidneys, heart, lungs, central nervous system, and hematopoietic system. Patients exhibit different combinations of symptoms and laboratory features. The variation of the SLE presentation is due to the interaction of hormonal, environmental, and genetic factors.[1]
In SLE, the search for susceptibility genes has used the candidate gene strategy through case–control association studies and recently genome wide association studies. These studies involved genes implicated either in SLE physiopathology or in the autoimmune response. In this regard, associations between human leukocyte antigens (HLA) and SLE have long been recognized in different ethnic populations and have been suggested to represent the most important association with this autoimmune disease, especially HLA-DRB1 and HLA-DQB1.[2,3]
In Tunisia, one study was conducted on the association between HLA genes and SLE in northern region.[4] As the status of HLA in southern population has previously been demonstrated to be different from the northern one,[5,6] we aimed in this study to search the HLA Class II markers of susceptibility and protection from SLE and to determine the association between HLA subtypes and clinical and serological features in our South Tunisian population.
Material and Methods
Patients and controls
This case–control study was approved by the ethics committee of Habib Bourguiba University Hospital of Sfax, Tunisia. We recruited 75 patients with SLE and 123 healthy controls, from the south of Tunisia. Patients and controls gave written consent to participate.
Patients included in this study fulfilled the American College of Rheumatology criteria for the diagnosis of SLE. An exhaustive information sheet containing clinical and serological features was filled for each patient. We defined a group of patients with severe form (lupus nephritis, pericarditis, pleurisy, and neurologic manifestations).
Serological study
Patients’ sera were analyzed by indirect immunofluorescence technique using Hep2 cells (Biosystem®, Spain) to detect antinuclear antibodies (ANA). Samples with positive fluorescence were then tested for anti-dsDNA on Crithidia luciliae substrate slides (Biosystem®, Spain). The specificities of ANA were determined by immunodot (Euroimmun®, Germany).
Each patient was also assayed for rheumatoid factor; C3 and C4 fractions of complement by nephelemetry, anti-cardiolipin, and anti-β2 glycoprotein I (β2gpI) antibodies by enzyme-linked immunosorbent assay (Orgentec®, Germany).
HLA Class II genotyping methods
We extracted genomic DNA from ethylenediaminetetraacetic acid peripheral blood using a phenol/chloroform technique. HLA Class II genes were typed using a polymerase chain reaction-sequence specific primer (PCR) kits (One Lambda Inc.®, CA, U.S.A.). This kit permit to amplify HLA-DRB1, DRB3, DRB4, DRB5, and DQB1 fragments. The PCR products were detected in ethidium bromide stained agarose gel and then visualized under ultraviolet illumination. The reaction results were analyzed using the kit instructions.
Statistical analysis
We used the SPSS software (version 20.0, Chicago, USA) to analyze our results. We first compared DRB1, DRB3, DRB4, DRB5, and DQB1 alleles’ distribution in SLE patients and healthy controls. We further examined differences in alleles’ distribution according to sex and to age. We finally tested the impact of each allele on the different disease clinical and serological features. We searched significant correlations using χ2 test. P values were considered as significant if <0.05. P values were corrected (pc) for the number of comparisons of DRB1* and DQB1* alleles with a frequency above 5% in either patients or controls (Bonferroni correction). We used Student’s t-test to assess correlations between DRB1, DRB3, DRB4, DRB5, and DQB1 alleles and ECLAM score and age.
Haplotype frequencies were estimated for controls and patients separately with the full precise iteration algorithm implemented in the SHE sis software http://analysis.bio-x.cn/myAnalysis.php. Haplotype frequency < 3% in both controls and cases has been dropped. Association between haplotypes and SLE was assessed by the χ2 test or the exact test as implemented in the program SHEesis.[7,8]
Post hoc power analysis was determined using the computer G power software with an error rate alpha of 0.05.
Results
The population studied was formed by 75 patients (65 women and 10 men) with a mean age = 32 ± 14 years and 123 controls (72 women and 51 men) with a mean age = 32 ± 9.28 years.
Clinical and immunological characteristics
Clinical symptoms consisted essentially on anemia, polyarthralgia, malar rush, and lupus nephritis. ECLAM score available for 68 patients ranged between 3 and 9 (mean = 5.72 ± 1.54) [Table 1].
Table 1.
Clinical and immunological manifestations with a frequency ≥5% and their association with HLA Class II
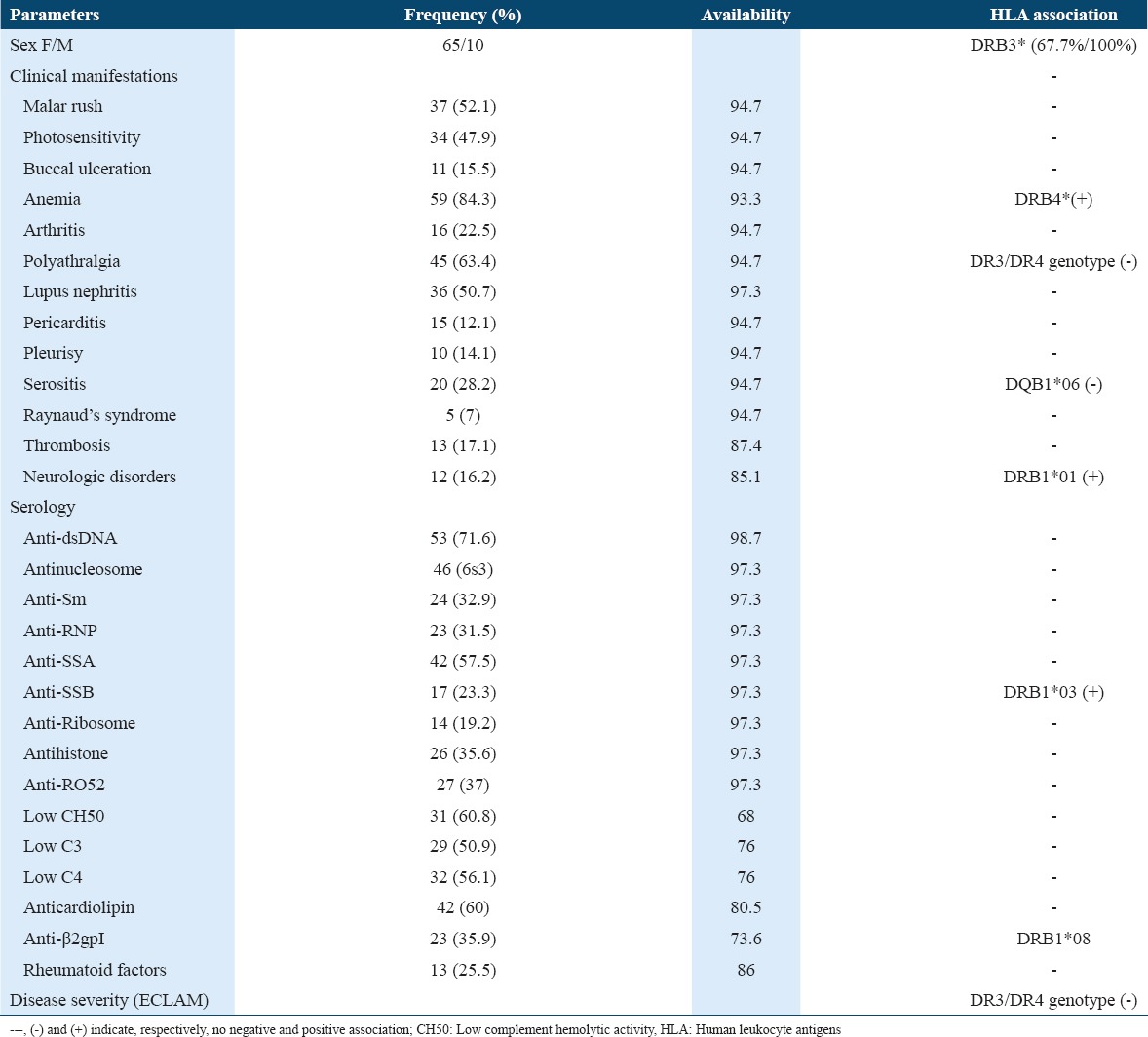
HLA-DR/DQ alleles
Our results showed significant differences between patients and controls concerning the distribution of HLA alleles essentially DRB1*03, DRB1*04, DRB1*15, and DQB1*05 [Table 2].
Table 2.
Frequencies of HLA-DRB1 and DQB1 alleles in patients and controls
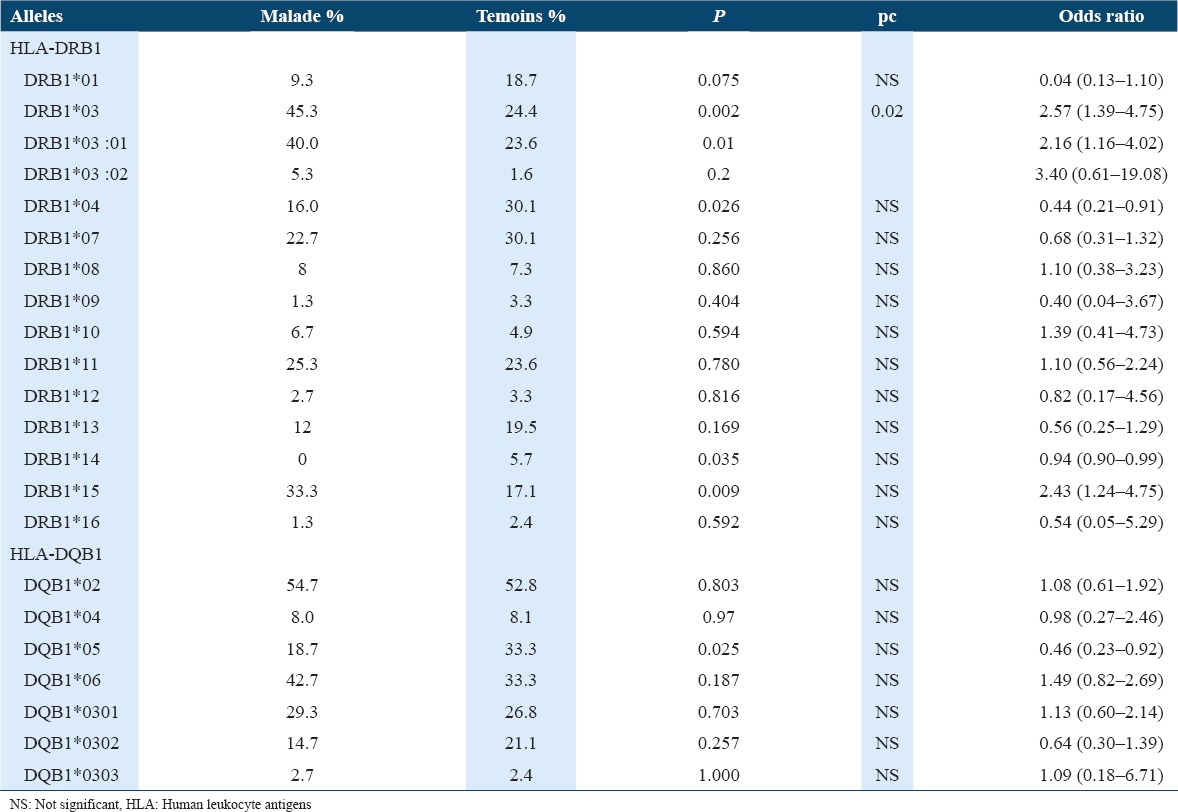
HLA-DRB1*03 and DRB1*15 showed a positive association with a post hoc power of 54% and 38%, respectively. However, HLA-DRB1*04 and HLA-DQB1*05 showed a negative association. After Bonferroni correction (pc < 0.05), only HLA-DRB1*03 preserved as a positive association (pc = 0.02).
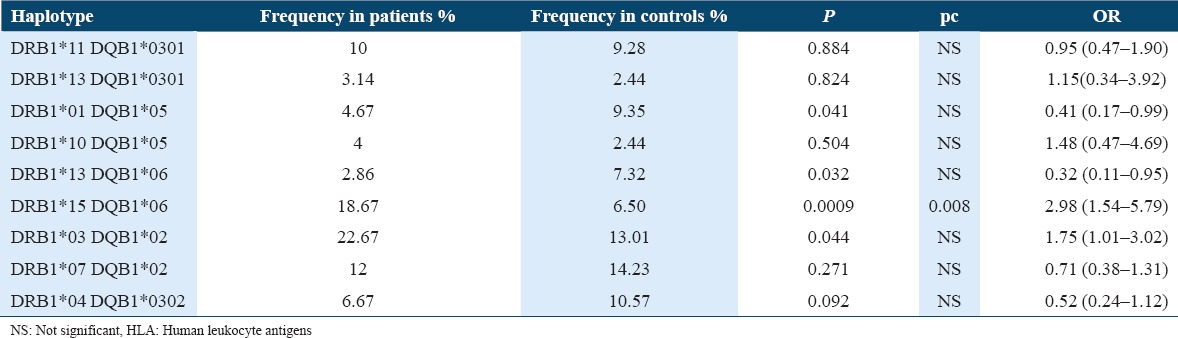
Haplotype analysis
Significant differences were found in haplotypes when we compared SLE patients against healthy controls (P = 0.0017, pc = 0.015). The most evident difference was noted for the DRB1*15-DQB1*06 haplotype (P = 0.0009; pc = 0.008; OR = 2.98 [1.54–5.79]) [Table 3].
Table 3.
HLA-DR/DQ haplotype association analysis
Clinical and serological correlations with HLA alleles
With respect to clinical manifestations, negative and positive associations were noted [Table 1].
There was no association between HLA alleles and the severe form of SLE. The heterozygote genotype HLA-DRB1*03/DRB1*04 was protective from polyarthralgia (P = 0.005; OR = 0.081; CI = [0.067–0.97]); this genotype was also associated with a lower value of ECLAM score (4.25/5.81; P = 0.001). A negative association was also noted between HLA-DQB1*06 and serositis (pericarditis and pleurisy)(P = 0.047; OR = 0.32; CI = [0.10–1.01]).
There was a positive association between HLA-DRB1*01 and neurologic disorders (P = 0.013; OR = 20.25; CI = [1.87–219.21]). HLA-DRB1*03 predisposed to the anti-SSB antibodies production (12 patients/17) (P = 0.016; OR = 4.00; CI = [1.24–12.96] and HLA-DRB1*08 to the anti-β2gpI antibodies production (P = 0.036; OR = 10.14; CI = [1.05–98.217]).
A positive association was found between DRB4* and anemia (P = 0.041; OR = 7.88; CI = [0.95–65.57]). All men were DRB3*; this positive association did not reach the cutoff of significance (P = 0.053; OR = 1.48; CI = [1.25–1.75]).
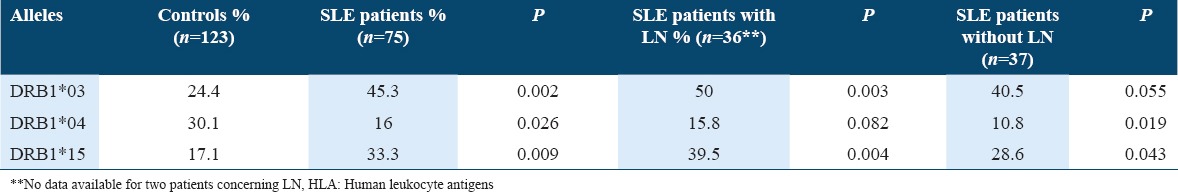
When we subdivided our patients in two groups (with and without lupus nephritis), the predisposal character of DRB1*03 and DRB1*15 persisted in the group with lupus nephritis (P = 0.003 and 0.004, respectively) but disappeared in the other group (P = 0.055 and 0.043, respectively) [Table 4].
Table 4.
HLA alleles of susceptibility and protection in patients with and without lupus nephritis
Discussion
HLA Class II molecules were reported to be implicated in the physiopathology of different autoimmune diseases.[9] Different studies showed the association of HLA alleles with SLE and with the clinical and the serological features of this autoimmune disease in different ethnic groups [Table 5].
Table 5.
HLA-DRB1*/DQB1* associations with SLE and clinical an serological features in different populations
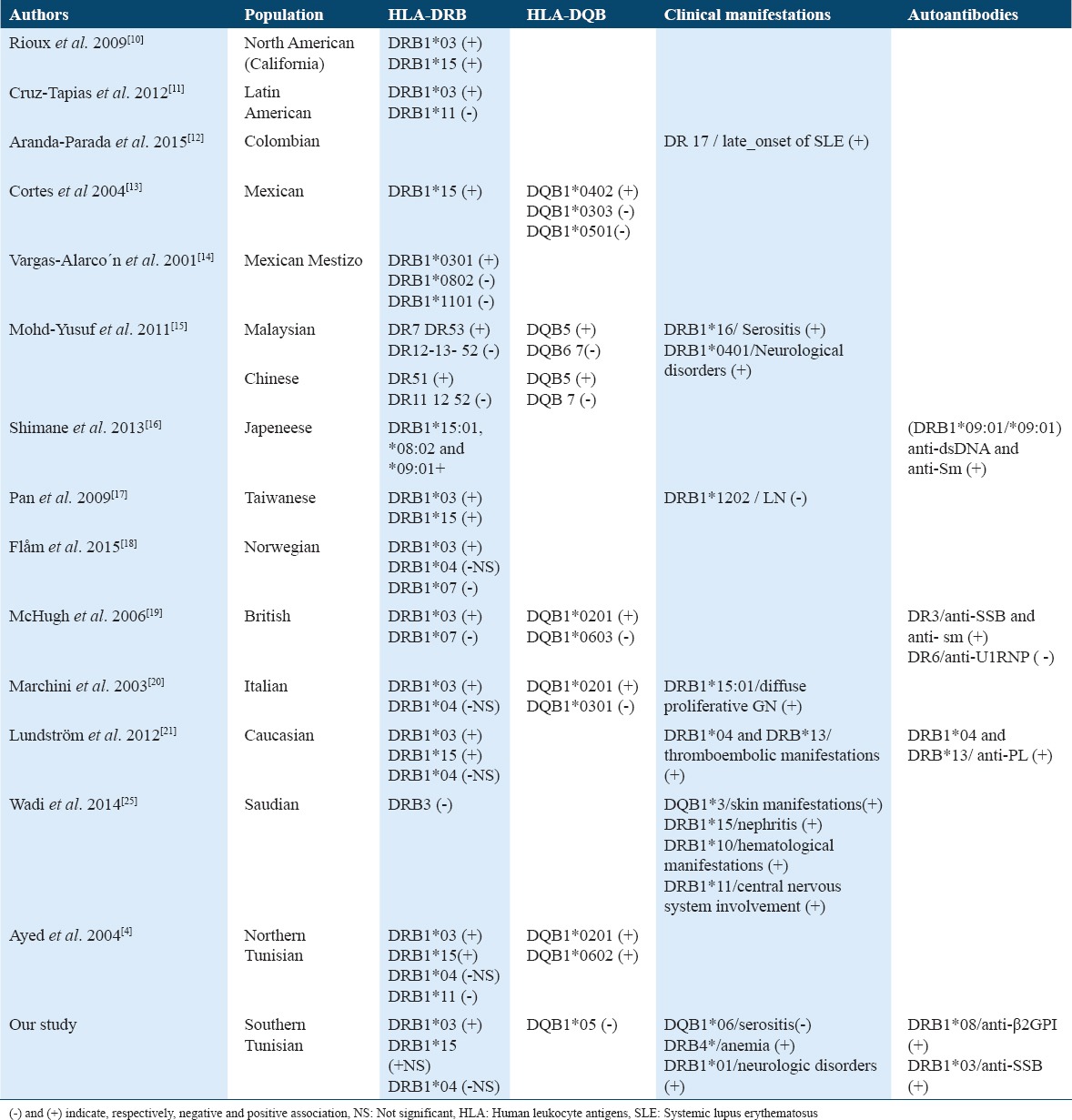
In Tunisia, Ayed et al. explored HLA in northern SLE patients, they reported the predisposal role of HLA-DRB1*03:01, HLA-DRB1*15:01, HLA-DQB1*02:01, and HLA-DQB1*06:02; while HLA-DRB1*11 was reported to be protective.[4] In previous studies, we reported important differences in HLA alleles associations with several autoimmune diseases between northern and southern population in Tunisia.[5,6] These findings motivated us to study HLA in southern SLE patients. We demonstrated once again that HLA-DRB1*03 and HLA-DRB1*15 were positively associated with SLE. These alleles were also reported to be associated with lupus in Asian, Caucasian, and Afro-American SLE patients.[2-24] However, we found that HLA-DRB1*04 was protective from SLE in southern Tunisian population. In the previous published studies, the protective alleles reported were HLA-DRB1*07 and HLA-DRB1*11.[2-4] Lundström et al. reported a lower frequency of HLA-DRB1*04 in SLE Caucasian patients, they also demonstrated that SLE patients carrying HLA-DRB1*04 alleles had an increased risk for vascular events.[21] Wadi et al. reported a positive association of DRB1*11 with neurologic disorders in lupus patients.[25] For our patients, these disorders were associated with HLA-DRB1*01. The same allele has been reported to be associated with symptomatic acute parvovirus B19 (PVB19) infection, particularly with meningoencephalitis caused by this virus.[26,27] On the other hand, PVB19 has been suggested as an environmental trigger of SLE.[28]
Our results showed a positive association between HLA-DRB4 and hematological disorders. Machulla et al. reported an increasing of HLA-DRB4 frequency among patients with chronic lymphocytic leukemia (P < 0.0025).[29]
To explain the implication of HLA polymorphism in SLE, functional analysis of susceptibility and protective molecules has been conducted. These studies revealed physicochemical differences of critical amino acids residues shaping the peptide-binding groove in the DRβ chain. Protective alleles such as HLA-DRB1*07 and HLA-DRB1*11 encode an arginine residue which is the strongest basic residue. Risk alleles (HLA-DRB1*03 and HLA-DRB1*15) determine the presence of a neutral (alanine) or less basic (lysine) residue at position β71.[2]
It has been suggested that HLA Class II alleles are more related to autoantibody generation than to the disease itself.[2] The association between the MHC Class II region and specific autoantibody generation is consistent with the concept of an antigen driven process involving T-helper cell recognition.[19] SLE autoantibody response can be initiated by multiple microbial T-epitope mimics in a DR restricted manner. This supports the hypothesis that autoimmune response to SLE-related antigens is initiated by multiple environmental T-epitope mimics that are cross-reactive with the autoantigen of interest.[30]
The association of HLA-DR3 and anti-SSB is well supported from previous studies of patients with SLE and Sjogren’s syndrome.[31] In our SLE patients, 12 of 17 SSB positive patients were HLA-DR3 positive. McHugh and al in 2006 had reported a strong association between anti-La antibodies and HLA-DR3 positive; they suggested that the presence of a HLA-DR3 containing haplotype greatly facilitates anti-SSB autoantibody generation and that there may be a strict MHC Class II requirement for processing of SSB peptides.[19] The association between HLA and anti-SSB autoantibodies was not established in the study of Ayed et al.[4] The differences we found with these authors provide an additional argument supporting the view that southern and northern populations in Tunisia exhibit different epidemiological features and a particular genetic background.
Finally, concerning HLA haplotype, the most evident association we found was with the haplotype DRB1*15-DQB1*06, association already described by others.[4-19]
In conclusion, the present findings confirm once again the implication of HLA-DR3 in SLE susceptibility and in anti-SSB production while no protective allele was found. This differs from reports of studies from north Tunisia in which no correlation with autoantibodies production was described, whereas HLA-DR11 was protective. These differences may be due to the disparity in allele’s frequencies that could be related to admixture with different populations.
Acknowledgments
We thank Mrs. Gaddour Lilia and Hakim Feiza in the Immunology Department, Hedi Chaker Hospital, Sfax, Tunisia, for their technical assistance. This work was supported by grants from the “Direction Generale de la Recherche Scientifique et Technique” (DGRST) of Tunisia.
References
- 1.Ahmadpoor P, Dalili N, Rostami M. An update on pathogenesis of systemic lupus erythematosus. Iran J Kidney Dis. 2014;8:171–84. [PubMed] [Google Scholar]
- 2.Castaño-Rodríguez N, Diaz-Gallo LM, Pineda-Tamayo R, Rojas-Villarraga A, Anaya JM. Meta-analysis of HLA-DRB1 and HLA-DQB1 polymorphisms in Latin American patients with systemic lupus erythematosus. Autoimmun Rev. 2008;7:322–30. doi: 10.1016/j.autrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Relle M, Weinmann-Menke J, Scorletti E, Cavagna L, Schwarting A. Genetics and novel aspects of therapies in systemic lupus erythematosus. Autoimmun Rev. 2015;14:1005–18. doi: 10.1016/j.autrev.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Ayed K, Gorgi Y, Ayed-Jendoubi S, Bardi R. The involvement of HLA -DRB1*, DQA1*, DQB1*and complement C4A loci in diagnosing systemic lupus erythematosus among Tunisians. Ann Saudi Med. 2004;24:31–5. doi: 10.5144/0256-4947.2004.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abida O, Zitouni M, Kallel-Sellami M, Mahfoudh N, Kammoun A. Tunisian endemic pemphigus foliaceus is associated with the HLA-DR3 gene: Anti-desmoglein 1 antibody-positive healthy subjects bear protective alleles. Br J Dermatol. 2009;161:522–52. doi: 10.1111/j.1365-2133.2009.09207.x. [DOI] [PubMed] [Google Scholar]
- 6.Kamoun A, Mahfoudh N, Frigui M, Bahloul Z, Makni H. Association of HLA class I antigens with behçet disease in South Tunisia. Pathol Biol (Paris) 2012;60:e59–64. doi: 10.1016/j.patbio.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–8. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. Apartition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: Update of the SHEsis ( http://analysis.bio-x.cn) Cell Res. 2009;19:519–23. doi: 10.1038/cr.2009.33. [DOI] [PubMed] [Google Scholar]
- 9.Miyadera H, Tokunaga K. Associations of human leukocyte antigens with autoimmune diseases: Challenges in identifying the mechanism. J Hum Genet. 2015;60:697–702. doi: 10.1038/jhg.2015.100. [DOI] [PubMed] [Google Scholar]
- 10.International MHC and Autoimmunity Genetics Network. Rioux JD, Goyette P, Vyse TJ, Hammarström L, Fernando MM, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A. 2009;106:18680–5. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz-Tapias P, P´erez-Fern´ez OM, Rojas-Villarraga A, Rodr´ıguez-Rodr´ıguez A, Arango MA, Anaya JM. . Shared HLA class II in six autoimmune diseases in Latin America: A meta-analysis. Autoimmune Dis. 2012;2012:569728. doi: 10.1155/2012/569728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peñaranda-Parada E, Quintana G, Yunis JJ, Mantilla R, Rojas W, Panqueva U, et al. Clinical, serologic, and immunogenetic characterization (HLA-DRB1) of late-onset lupus erythematosus in a Colombian population. Lupus. 2015;24:1293–9. doi: 10.1177/0961203315588576. [DOI] [PubMed] [Google Scholar]
- 13.Cortes LM, Baltazar LM, Lopez-Cardona MG, Olivares N, Ramos C, Salazar M, et al. HLA class II haplotypes in Mexican systemic lupus erythematosus patients. Hum Immunol. 2004;65:1469–76. doi: 10.1016/j.humimm.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Vargas-Alarcón G, Salgado N, Granados J, Gómez-Casado E, Martinez-Laso J, Alcocer-Varela J, et al. Class II allele and haplotype frequencies in Mexican systemic lupus erythematosus patients: The relevance of considering homologous chromosomes in determining susceptibility. Hum Immunol. 2001;62:814–20. doi: 10.1016/s0198-8859(01)00267-1. [DOI] [PubMed] [Google Scholar]
- 15.Mohd-Yusuf Y, Phipps ME, Chow SK, Yeap SS. HLA-A*11 and novel associations in Malays and Chinese with systemic lupus erythematosus. Immunol Lett. 2011;30(139):68–72. doi: 10.1016/j.imlet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Shimane K, Kochi Y, Suzuki A, Okada Y, Ishii T, Horita T, et al. An association analysis of HLA-DRB1 with systemic lupus erythematosus and rheumatoid arthritis in a Japanese population: Effects of *09: 01 allele on disease phenotypes. Rheumatology (Oxford) 2013;52:1172–82. doi: 10.1093/rheumatology/kes427. [DOI] [PubMed] [Google Scholar]
- 17.Pan CF, Wu CJ, Chen HH, Dang CW, Chang FM, Liu HF, et al. Molecular analysis of HLA-DRB1 allelic associations with systemic lupus erythematous and lupus nephritis in Taiwan. Lupus. 2009;18:698–704. doi: 10.1177/0961203308101955. [DOI] [PubMed] [Google Scholar]
- 18.Flåm ST, Gunnarsson R, Garen T Norwegian MCTD Study Group. Lie BA, Molberg Ø, editors. The HLA profiles of mixed connective tissue disease differ distinctly from the profiles of clinically related connective tissue diseases. Rheumatology (Oxford) 2015;54:528–35. doi: 10.1093/rheumatology/keu310. [DOI] [PubMed] [Google Scholar]
- 19.McHugh NJ, Owen P, Cox B, Dunphy J, Welsh K. MHC class II, tumour necrosis factor a, and lymphotoxin a gene haplotype associations with serological subsets of systemic lupus erythematosus. Ann Rheum Dis. 2006;65:488–494. doi: 10.1136/ard.2005.039842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchini M, Antonioli R, Lleò A, Barili M, Caronni M, Origgi L, et al. HLA class II antigens associated with lupus nephritis in Italian SLE patients. Hum Immunol. 2003;64:462–8. doi: 10.1016/s0198-8859(03)00017-x. [DOI] [PubMed] [Google Scholar]
- 21.Lundström E, Gustafsson JT, Jönsen A, Leonard D, Zickert A, Elvin K, et al. HLA-DRB1*04/*13 alleles are associated with vascular disease and ant phospholipid antibodies in systemic lupus erythematosus. Ann Rheum Dis. 2013;72:1018–25. doi: 10.1136/annrheumdis-2012-201760. [DOI] [PubMed] [Google Scholar]
- 22.Barcellos LF, May SL, Ramsay PP, Quach HL, Lane JA, Nititham J, et al. High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet. 2009;5:e1000696. doi: 10.1371/journal.pgen.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louthrenoo W, Kasitanon N, Wichainun R, Wangkaew S, Sukitawut W, Ohnogi Y, et al. The genetic contribution of HLA-DRB5*01: 01 to systemic lupus erythematosus in Thailand. Int J Immunogenet. 2013;40:126–30. doi: 10.1111/j.1744-313X.2012.01145.x. [DOI] [PubMed] [Google Scholar]
- 24.Furukawa H, Kawasaki A, Oka S, Ito I, Shimada K, Sugii S, et al. Human leukocyte antigens and systemic lupus erythematosus: A protective role for the HLA-DR6 alleles DRB1*13: 02 and *14: 03. PLoS One. 2014;9:e87792. doi: 10.1371/journal.pone.0087792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wadi W, Elhefny NE, Mahgoub EH, Almogren A, Hamam KD, Al-Hamed HA, et al. Relation between HLA typing and clinical presentations in systemic lupus erythematosus patients in Al-Qassim region, Saudi Arabia. Int J Health Sci (Qassim) 2014;8:159–65. doi: 10.12816/0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr JR, Mattey DL, Thomson W, Poulton KV, Ollier WE. Association of symptomatic acute human parvovirus B19 infection with human leukocyte antigen class I and II alleles. J Infect Dis. 2002;186:447–52. doi: 10.1086/341947. [DOI] [PubMed] [Google Scholar]
- 27.Kerr JR, Barah F, Chiswick ML, McDonnell GV, Smith J, Chapman MD, et al. Evidence for the role of demyelination, HLA-DR alleles, and cytokines in the pathogenesis of parvovirus B19 meningoencephalitis and its sequelae. J Neurol Neurosurg Psychiatry. 2002;73:739–46. doi: 10.1136/jnnp.73.6.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khlif S, Hachicha H, Frikha F, Feki S, Ben Ayed M, Bahloul Z, et al. Pediatric onset systemic lupus erythematosus: About a case. Pan Afr Med J. 2015;20:25. doi: 10.11604/pamj.2015.20.25.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machulla HK, Müller LP, Schaaf A, Kujat G, Schönermarck U, Langner J. Association of chronic lymphocytic leukemia with specific alleles of the HLA-DR4: DR53: DQ8 haplotype in German patients. Int J Cancer. 2001;92:203–7. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1167>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 30.Fu SM, Deshmukh US, Gaskin F. Pathogenesis of systemic lupus erythematosus revisited. 2011: End organ resistance to damage, autoantibody initiation and diversification, and HLA-DR. J Autoimmun. 2011;37:104–12. doi: 10.1016/j.jaut.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris DL, Fernando MM, Taylor KE, Chung SA, Nititham J, Alarcón-Riquelme ME, et al. MHC associations with clinical and autoantibody manifestations in European SLE. Genes Immun. 2014;15:210–7. doi: 10.1038/gene.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]


