Abstract
Summary
Optical mapping is a molecular technique capturing specific patterns of fluorescent labels along DNA molecules. It has been widely applied in assisted-scaffolding in sequence assemblies, microbial strain typing and detection of structural variations. Various computational methods have been developed to analyze optical mapping data. However, existing tools for processing and visualizing optical map data still have many shortcomings. Here, we present OMTools, an efficient and intuitive data processing and visualization suite to handle and explore large-scale optical mapping profiles. OMTools includes modules for visualization (OMView), data processing and simulation. These modules together form an accessible and convenient pipeline for optical mapping analyses.
Availability and implementation
OMTools is implemented in Java 1.8 and released under a GPL license. OMTools can be downloaded from https://github.com/aldenleung/OMTools and run on any standard desktop computer equipped with a Java virtual machine.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Optical mapping is a technique for imaging DNA molecules along which specific labels are captured. These labels form distinct patterns along DNA molecules based on their nucleotide sequences. Compared with the short read length in next-generation sequencing (∼150 bp), DNA molecules in optical mapping data are several orders longer, at ∼100–1000 kb (Lam et al., 2012). Because of the much greater read length, optical mapping has been used for assisted scaffolding, (Dong et al., 2013), structural variations detection (Cao et al., 2014, Mak et al., 2016) and microbial strain typing (Schwan et al., 2010). There are currently two instrument platforms available for generating optical mapping data: OpGen Inc. and BioNano Genomics Inc. Emerging computational methods have been developed for data analysis. To gain knowledge and insights from the optical mapping data, human analysis and interpretation is required and usually involves data visualization. At present, three visualization tools for optical mapping are available: BioNumerics v7 (Applied Maths NV), IrysView (Shelton et al., 2015) and JBrowse (Skinner et al., 2009). However, none of them could provide multiple visualization styles in accordance to the types of application. Here, we present the software package OMTools, which comprises modules for visualization (OMView), processing optical mapping data and alignment results, and simulation. The package has fast loading time, easy installation and support to multiple data formats (Supplementary Tables S1 and S2).
2 OMView
OMView is a multipurpose visualization module for optical mapping data analysis. Five types of visualizations (Supplementary Table S3) are implemented in OMView for different objectives: regional view, anchor view, alignment view, multiple alignments view and molecule view (Fig. 1A–E). In all types of visualizations, a rectangular block represents the DNA backbone, while vertical bars on the blocks represent signals on the DNA molecule. We explain the purpose and a related example for each type of visualization below.
Fig. 1.
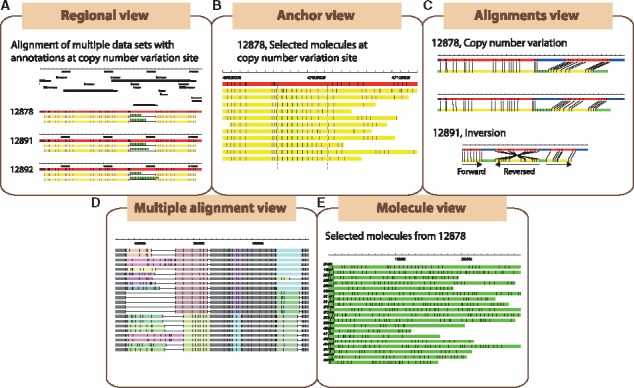
Major visualization modes in OMView. Five types of views are available to visualize optical mapping data for different purposes: (A) Regional view, (B) Anchor view, (C) Alignment view, (D) Multiple alignments view and (E) Molecule view
The regional view displays all alignments as an overview at a selected region. In each panel, the reference DNA is represented by a thick red line, while aligned and unaligned portions of a molecule are represented by yellow and green lines, respectively. Molecule signals matching the reference signals are in pink while the unaligned ones are in black. Contigs alignment along the reference is also displayed in a similar manner. Each aligned molecule is stretched accordingly so that the first and last matching signals are located at the respective horizontal positions of the reference signals. For a molecule that has individual portions separately aligned to different reference regions, OMView depicts the relationships between consecutive aligned portions (insertion, deletion, inversion and translocation). Multiple panels could be created to display alignment results from different datasets for comparison at the same region, as exemplified with a case of copy number variations on chromosome 18 in the 1000 Genomes trio dataset NA12878, NA12891 and NA12892 in Figure 1A (Mak et al., 2016). Additional panels could be loaded to visualize annotations on the reference, such as gene annotations or gaps depicted as black rectangles above the alignments.
The anchor view is mainly designed for validating structural variations. It displays alignments of which molecule signals could match two selected signals on the reference. Under this view, molecules are shown at the original measured lengths such that structural variations can be easily seen. After the automatic sorting of alignments according to the distance between the two signals on molecules by a sub-module in OMView, the presence as well as the zygosity of insertions/deletions could be easily determined. A similar example of copy number variations described before in anchor view is shown in Figure 1B.
The alignment view illustrates the alignment of one single molecule against the reference with more details. This is especially important in visualizing partial alignments with complicated relationships. The alignment view employs the aforementioned coloring scheme, where an extended blue block is added to represent unaligned portion of the reference. Multiple panels of alignments are displayed if there are individual portions separately aligned to different reference regions. Below each alignment shows detailed information including aligned signals, alignment score and CIGAR (Concise Idiosyncratic Gapped Alignment Report) string. Certain alignments picked from the previous copy number variations example and an alignment demonstrating an inversion is shown in Figure 1C (Mak et al., 2016).
The multiple alignments view, designed for whole-genome comparisons, depicts multiple optical map alignment of a dataset. Here, matching patterns are represented by a list of collinear-aligned-blocks. Each row of rectangular blocks represents one optical map genome, while each collinear-aligned-block contains a column of rectangular blocks with the same color that represents a matching pattern across different genomes. Down the same column position, a solid line represents a gap and a different colored block represents different matching patterns. Users can therefore easily visualize the structural difference within variable regions across multiple samples. Figure 1D offers an example of multiple alignments. The user-interface also allows users to manually modify the multiple alignment results and produce statistics of collinear blocks.
Finally, the molecule view is a module for inspection of molecules to offer a general impression on the dataset as shown in Figure 1E. One could sort the molecules by name, size or number of signals, and a constant number of molecules can be viewed page-by-page.
3 Additional features
OMTools contains some useful modules that can be executed within the same software framework for processing of optical mapping data (Supplementary Table S4). Data processing is important for any downstream analysis. OMTools provides filtering tools on optical mapping data such as filtering by size or number of signals. Since molecules with high density or low complexity impede alignment and assembly, OMTools also offers a signal density and complexity filter. A separate module could be applied to detect data duplication errors. Similarly, OMTools provides filtering tools on alignment results. A partial alignment joining module separated from OMBlast (Leung et al., 2016) could be employed to treat alignments from other alignment methods as partial alignments to connect them into final alignments. OMTools can also merge and generate statistics for results from various alignment methods.
A set of simulation modules enables data simulation with a variety of modeling parameters. Various categories of errors including missing and extra signals, scaling, measurement and resolution error are modeled, with optional structural variations added on reference or data to test software related to structural variation detection.
4 Conclusions
OMTools offers a fundamental toolbox for optical mapping data processing. OMView serves as a powerful visualization tool for data analysis and illustration. On top of the existing modules and methods included in the OMTools Java library, researchers could build additional analysis modules with minimal effort.
Supplementary Material
Funding
This work was partially supported by the Health and Medical Research Fund 12110542, RGC Collaborative Research Fund [CUHK3/CRF/11G and C4042-14G], Theme-based research schemes T12-403/11, T12-401/13R and T12-402/13-N of the Hong Kong Government, and the Lo Kwee-Seong Biomedical Research Fund and Lee Hysan Foundation.
References
- Cao H. et al. (2014) Rapid detection of structural variation in a human genome using nanochannel-based genome mapping technology. Gigascience, 3, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y. et al. (2013) Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol., 31, 135–141. [DOI] [PubMed] [Google Scholar]
- Lam E.T. et al. (2012) Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol., 30, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K.-Y. et al. (2016) OMBlast: alignment tool for optical mapping using a seed-and-extend approach. Bioinformatics, 33, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A.C.Y. et al. (2016) Genome-wide structural variation detection by genome mapping on nanochannel arrays. Genetics, 202, 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan W.R. et al. (2010) Use of optical mapping to sort uropathogenic Escherichia coli strains into distinct subgroups. Microbiology, 156, 2124–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton J.M. et al. (2015) Tools and pipelines for BioNano data: molecule assembly pipeline and FASTA super scaffolding tool. BMC Genomics, 16, 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M.E. et al. (2009) JBrowse: a next-generation genome browser. Genome Res., 19, 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


