Abstract
MicroRNAs have implicated in the relapse and metastasis of cervical cancer, which is the leading cause of cervical cancer-related mortality. However, the underlying molecular mechanisms need further elucidation. Our present study revealed that miR-221-3p is transcriptionally promoted in metastatic cervical cancer tissues compared with non-metastatic cervical cancer tissues. Forced overexpression of miR-221-3p facilitated EMT and promoted cell migration and invasion in vitro and lymphatic metastasis in vivo. Twist homolog 2 (TWIST2) was found to be a key transcription factor binding to the promoter of miR-221-3p. Inhibitors of miR-221-3p drastically reduced the induction of EMT and decreased cell migration and invasion mediated by TWIST2. By combined computational and experimental approaches, THBS2 was recognized to be an important downstream target gene of miR-221-3p. In cervical cancer tissues, especially with lymphatic metastasis, miR-221-3p and TWIST2 were increased and THBS2 was decreased, suggesting that TWIST2 induces miR-221-3p expression and consequently suppresses its direct target THBS2 in lymphatic metastasis CC. Our findings uncover a mechanistic role for miR-221-3p in lymph node metastasis, suggesting that miR-221-3p is upregulated by the transcription factor TWIST2 and downregulates its target THBS2, which may potentially promote lymph node metastasis in cervical cancer.
Introduction
Cervical cancer (CC) is one of the most prevalent malignancies in women worldwide and is the leading cause of cancer death for women in developing countries1. Although widespread vaccination against human papilloma virus, periodic cancer screening and prompt surgical treatment have resulted in a significant decrease in the incidence of CC2,3, it remains one of the most common diseases causing mortality in women4,5. Squamous cell carcinoma, the most common histological subtype of CC, spreads principally by migrating into the lymphatics or by invading adjacent soft tissue6. Pelvic lymph node metastasis and local invasion of CC indicate a poor prognosis7. Therefore, it is important to develop effective treatments for invasive extension of this disease; to this end, it is essential to further elucidate the molecular mechanism of lymph node metastasis.
Cancer metastasis is a complex and multi-step process8. The initial stage of metastatic progression is essentially dependent on a prominent biological event referred to as epithelial-to-mesenchymal transition (EMT), which is characterized by specific morphogenetic changes, loss-of cell–cell adhesion, and increased cell motility9–12. MicroRNAs (miRNAs) are small, non-coding RNAs (18–23 nucleotides in size) that regulate gene expression by complementary base pairing with the 3′-untranslated region (UTR) of messenger RNA (mRNA) and trigger translation repression or RNA degradation13. Thus, miRNAs are considered to be master regulators of many important physiological processes, including cell proliferation, differentiation, development, and apoptosis14–16. Recently, genome-wide analyses indicated that 50% of miRNA genes are located in cancer-associated genomic regions or in fragile sites17. Moreover, miRNAs have been identified that function as oncogenes or tumor suppressor genes, and some act in the late stages of tumour progression. For example, the miR-200 family, especially miR-200c, is implicated in the metastasis and invasion of ovarian carcinoma18. Additionally, miR-29b represses SNAIL1 expression; hence, increased miR-29b expression can reverse EMT and decrease cell invasion19. Despite these studies, the role of miRNAs in the lymph node metastasis of CC has remained unclear. Therefore, understanding how miRNA molecules serve as master regulators in these important networks involved in cancer initiation, progression, metastasis and EMT open up significant innovative areas for therapy and diagnosis that have been sadly lacking for deadly CC20.
Considering the emerging roles of miRNAs in cancer biology, we set out to identify miRNAs that promote CC metastasis and addressed its underlying molecular mechanisms.
Methods and materials
Cell and transfection
Human CC cell lines SiHa and HeLa, and human embryonic kidney 293T cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). SiHa and 293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS). HeLa cells were cultured in MEM (Gibco) supplemented with 10% heat-inactivated FBS.
The miR-221-3p-overexpressing SiHa and HeLa cell lines were established using CMV-Fluc-IRES-RFP lentiviral particles (GeneChem, Shanghai, China). One milliliter of viral supernatant containing 4 Attogram (Ag) of polybrene was added to SiHa and HeLa cell lines for stable transduction. After 14 days, puromycin-resistant cell pools were established. Then, transfected cells were designated SiHa-luc-RFP-221-3p, SiHa-luc-RFP-NC, HeLa-luc-RFP-221-3p, and HeLa-luc-RFP-NC.
A specifically enhanced metastatic subline of SiHa cell line named SiHa-twist2 (SiHa-tw2) and a weakly metastatic subline of SiHa cells named SiHa-shtwist2 (SiHa-shtw2) have been established by repeated selection in our previous studies21,22. SiHa-tw2 cells overexpressed twist homolog 2 (TWIST2) and SiHa-shtw2 cells downexpressed TWIST2 in SiHa cells, which were cultured in DMEM (Gibco) supplemented with 10% heat-inactivated FBS.
miRNAs microarray
Tissues (six CC tissues derived from stage I and II, respectively) and cells (SiHa, SiHa-shtw2, SiHa-tw2 cells) were collected and the expression profiles of miRNAs were determined using Agilent miRNA microarray 21.0. In brief, total RNA was extracted from cells or tissues samples using the miRVana miRNA Isolation Kit (Ambion Inc., TX, USA). The quality and quantity of RNA samples were assessed by a 2100 Bioanalyzer using the RNA 6000 Pico LabChip kit (Agilent Technologies, Santa Clara, CA). The miRNA microarray was carried out at Shanghai Biotechnology Corporation (Shanghai, China). Data analysis was performed using GeneSpringGX software 11.0 (Agilent Technologies).
qRT-PCR analysis of mRNA and miRNA expression
Total RNA was extracted from cells and tissues with TRIzol reagent (Invitrogen). The relative quantity of mRNA and miRNA were determined by real-time polymerase chain reaction (RT-PCR). GAPDH was chosen as the reference gene. U6 was chosen as an internal control for miRNAs. The primer sequences are shown in Table S3. Quantitative mRNA and miRNA expression was measured with ABI Prism 7500 Software v2.0.6 and calculated based on the comparative CT method. The expression level of each mRNA or miRNA was normalized to that of GAPDH or U6, and expression as n-fold difference relative to the control.
Western blotting
Cell lysates were prepared as described previously22. A total of 50 μg of protein was separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and subsequently transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% bovine serum albumin for 1 h before being incubated overnight at 4 °C with the primary antibody, including anti-E-cadherin (Epitomics Biotechnology, 1:1000), anti-N-cadherin (Epitomics Biotechnology, 1:500), anti-Vimentin (Epitomics Biotechnology, 1:400), anti-TWIST2 (Abcam, 1:400), and anti-THBS2 (Novus Biotechnology, 1:800). β-actin (Abcam, 1:1000) was used as an internal control for protein loading and analysis. The membranes were washed three times with PBST (PBS buffer containing 0.1% Tween-20) and then incubated with HRP-conjugated secondary antibodies for 1 h. After the final wash with PBST, the antibody staining intensities were detected using ECL chemiluminescence reagents (Pierce).
Boyden chamber assay
The Boyden invasion chambers were rehydrated with DMEM (serum-free) for 2 h at 37 °C. Medium containing 10% serum was used as a chemoattractant in the lower chambers. Then, 1 × 105 tumour cells in serum-free DMEM were added to the upper compartment of the chamber. After incubation for 48 h, the noninvasive cells were removed with a cotton swab. Invading cells on the undersides of membranes were stained with haematoxylin and counted under a light microscope in five random visual fields (×200). Each experiment was repeated three times.
Wound-healing assay
Cells in the exponential phase of growth were harvested and seeded in a six-well plate at a density of 5 × 105 cells/well. A scratch wound was generated using sterile 10-µl pipet tip, and floating cells were removed by washing with PBS. Images of the scratches were taken using an inverted microscope at ×100 magnification at 0 h and 48 h of incubation. The percentage of healed wound area was measured as a ratio of occupied area to the total area using Image Olympus IX71 (Olympus Inc).
Popliteal lymph node metastasis model
Female athymic nude (nu/nu) mice (4-week old) were purchased from Beijing HFK Bio-Technology Co, LTD., (Beijing, China) for studies approved by the Committee on the Ethics of Animal Experiments of Southern Medical University. The mice were maintained in the accredited animal facility of Southern Medical University. First, 1 × 107 SiHa-luc-RFP-221-3p/SiHa-luc-RFP-NC cells or HeLa-luc-RFP-221-3p/ HeLa-luc-RFP-NC cells in 50 μl serum-free DMEM/Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) (9:1) were injected subcutaneously into their claw pads, respectively. Tumour size (mm3) was measured and calculated by the formula: Volume = (width)2+ × length/2. Metastases were detected when primary tumors reached the size of ~150 mm3. The quantity of metastases was tracked in living mice by optical imaging of luciferase activity using the IVIS Spectrum system (Caliper, Xenogen, USA). After anaesthetization with 4% pentobarbital sodium, mice were imaged 10 min after intraperitoneal injection of 100 mg/kg d-luciferin. When the mice were euthanized, their popliteal lymph nodes were excised. The metastases of tumour cells in the lymph nodes were confirmed by detecting expressed-expressed RFP under an Olympus SZX16 (Olympus Inc.) dissecting microscope. The percentage of metastasis-positive lymph nodes in each mouse was calculated.
Transcription factor-binding site analysis
Jaspar (http://jaspar.genereg.net), an open-access database of transcription factor-binding preferences in multiple species23, was used to predict potential transcription factor-binding sites.
Specimens
A total of 55 archival, formalin-fixed, paraffin-embedded cervical specimens were obtained from the department of pathology at Nanfang Hospital from 2008 to 2010. The specimens included: 10 cases of normal cervical tissues, 23 cases of LNM-negative CC and 22 cases of LNM-positive CC. Fresh CC tissues were collected from 28 patients who underwent abdominal radical hysterectomy without prior radiotherapy and chemotherapy at Nanfang Hospital from 2013 to 2015. All 28 cases were submitted for pathological examination after operation; 14 were confirmed to contain lymph node metastasis, and 14 lacked lymph node metastasis (Table S2). These samples were collected immediately after resection, snap-frozen in liquid nitrogen, and stored at −80 °C until needed. Informed consent was obtained from each patient before using the samples.
Immunohistochemistry
Tissue sections were subjected to immunohistochemical analysis as described previously24 using the Avidin-Biotin Complex Vectastain Kit (Zsgb-Bio, Beijing, China). Rabbit anti-human TWIST2 antibody (Abcam Biotechnology) and rabbit anti-human THBS2 antibody (Novus Biotechnology) were used as primary antibodies. The stained tissues were reviewed and independently scored by two pathologists blinded to the clinical parameters. TWIST2 staining was observed primarily in the cytoplasm and nucleus, and THBS2 staining was observed in the cytoplasm. The expression status of TWIST2 and THBS2 of the entire area was determined as the product of the score for the average extent and intensity of positively stained cells24. For semiquantitative evaluation of the expression level in tissue, an immunoreactivity scoring system HSCORE was used25,26. The HSCORE was calculated using the following formula: HSCORE = ∑Pi(i + 1), where i is the staining intensity of cells and Pi is the percentage of cells at each level of intensity. HSCORE ⩽ 2 was classified as a low-level expression, and HSCORE > 2 was classified as a high-level expression. In 95% of the samples, the evaluations of the two observers were identical. The remaining slides were re-evaluated, and consensus decisions were made. The scoring system has been described previously.
In situ hybridization
Tissue sections were obtained from human cervical tissues as we used previously, and spontaneous primary CC tumors and matched lymph node metastases were harvested from mice. In brief, after deparaffinization of the slides in xylene and ethanol, the slides were incubated with 3% H2O2 for 10 min at room temperature and then digested with pepsin for 10 min at 37 °C. After rinsing in water, the slides were fixed with 1% PFA in DEPC (Generay, China) for 5 min. Slides were prehybridized in hybridization buffer at 42 °C with miR-221-3p or U6 LNA probe (Exiqon, Woburn, MA, USA). Following stringency washes at 60 °C and blocking for 30 min at room temperature, slides were then incubated with streptavidin-biotin complex. After washing three times with PBS, the slides were incubated with horseradish peroxidase polymer conjugate for an additional 30 min at room temperature. Subsequently, they were stained with 3,3-diaminobenzidine and counterstained with haematoxylin (Sigma-Aldrich, USA). The slides were then independently scored by two pathologists. The HSCORE scoring system has been described previously.
miRNA target prediction
The analysis of miR-221-3p predicted targets was determined using TargetScan (http://www.targetscan.org/), PicTar (http://pictar.bio.nyu.edu/), and miRwalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/). To identify the genes commonly predicted by the three different algorithms, miRGen (http://diana.pcbi.upenn.edu/miRGen/v3/miRGen.html) was used.
Incomplete complementation of the base of miR-221-3p to the 3′-UTR region of THBS2 was predicted by TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do).
Luciferase activity assay
For the binding of miR-221-3p to THBS2 3′UTR, the 3′UTR segment of the THBS2 gene was amplified by PCR and inserted into the vector. A mutant construct in the miR-221-3p-binding sites of THBS2 3′UTR region was also generated using Quick Change Site-Directed Mutagenesis Kit (Agilent, Roseville City, CA, USA). Co-transfection of THBS2 3′-UTR or mutTHBS2 3′-UTR plasmid with miR-221-3p mimic into the cells was accomplished using Lipofectamine 2000 (Invitrogen). For the binding of TWIST2 to the miR-221-3p promoter (which was purchased from Taihe Biotechnology CO., LTD, Beijing, China), the coding region of TWIST2 and the 2-kb region directly upstream of the miR-221-3p transcription-binding site were amplified by PCR and then inserted into the vectors, respectively. Luciferase activity was measured 48 h after transfection by the Dual-Luciferase Reporter Assay System (Promega). Each assay was repeated in three independent experiments.
Statistical analysis
SPSS (version 20.0) software package was used for statistical analysis. The results were expressed as the mean value ± SEM and interpreted by one-way ANOVA. Correlation analyses were done using Spearman rank test. The odds ratio of LN metastasis was analyzed by χ2-test where applicable. Differences were considered to be statistically significant when p < 0.05.
Results
Identification of miR-221-3p as a metastasis-promoting miRNA in CC
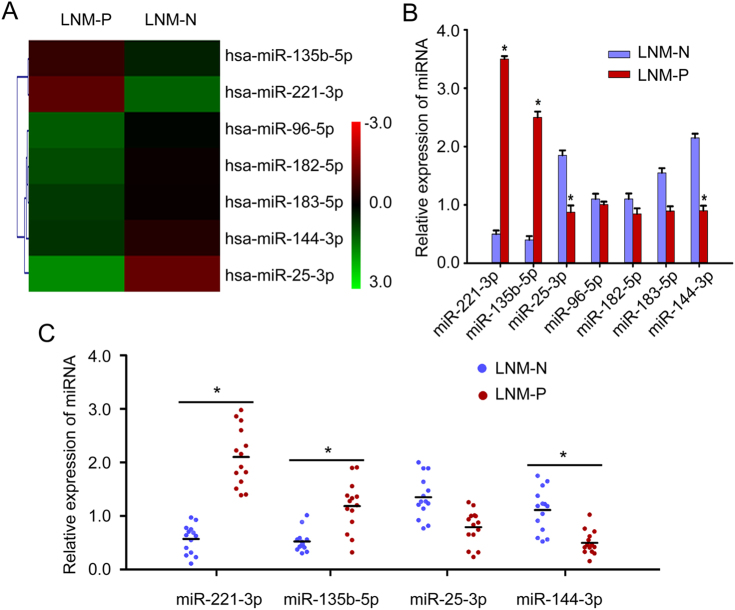
To investigate the differences in miRNA profiles specially related to lymph node metastasis in CC, six primary CC tissues derived from stage І-ІІ patients with (n = 3) or without (n = 3) lymph node metastasis were collected, and the miRNA expression profiles were determined by Agilent miRNA microarray 21.0 (Fig. 1a, Table S1) (Deep-sequencing data have been deposited in GEO with accession number: GSE102969). The differential expression of seven representative miRNAs (top seven miRNAs included: miR-135b-5p, miR-221-3p, miR-25-3p, miR-96-5p, miR-182-5p, miR-183-5p, and miR-144-3p) was verified using qRT-PCR in the same tissues used for microarray analysis. The result showed that a sevenfold higher level of miR-221-3p (p < 0.001), 6.1-fold higher level of miR-135b-5p (p < 0.001), 2.11-fold lower level of miR-25-3p expression (p < 0.05) and 2.39-fold lower level of miR-144-3p (p < 0.05) were observed in lymph node metastasis CC tissues compared with non-lymph node metastasis CC tissues, respectively (Fig. 1b). It is noticeable that miR-221-3p emerged as the most significantly different miRNA between CC tissues of the two groups.
Fig. 1. Identification of miR-221-3p as a metastasis-promoting miRNA in cervical cancer.
a The certified result of microarray analysis. Hierarchical clustering of seven significantly dysregulated miRNA expression profiles in human primary cervical cancer tissues derived from cervical cancer patients with or without lymph node metastasis. b Validation of the selected miRNAs predicted to be dysregulated in cervical cancer with or without lymph node metastasis using qRT-PCR in the same tissues used for microarray analysis. Data are shown from three independent experiments and presented as fold expression normalized to U6 ± SD (standard deviation). c qRT-PCR analysis of the relative tissue expression of miR-221-3p, miR-135b-5p, miR-25-3p, and miR-144-3p in additional 28 (LNM-N = 14; LNM-P = 14) cases of human CC tissues. Each sample was analyzed in triplicate and normalized to U6. *p < 0.05
We further confirmed these data, particularly the differential expression of the four miRNAs (miR-221-3p, miR-135b-5p, miR-25-3p, and miR-144-3p) with statistical significance, in an additional 28 CC patients (14 CC patients with lymph node metastasis and 14 CC patients without lymph node metastasis, Table S2). MiR-221-3p displayed the most significant difference between lymph node metastasis CC tissues (2−ΔΔCT = 2.10 ± 0.43) and non-lymph node metastasis CC tissues (2−ΔΔCT = 0.60 ± 0.21) (p = 0.007, Fig. 1c). These results suggest the likelihood that miR-221-3p is associated with the potential for lymphatic metastasis in CC.
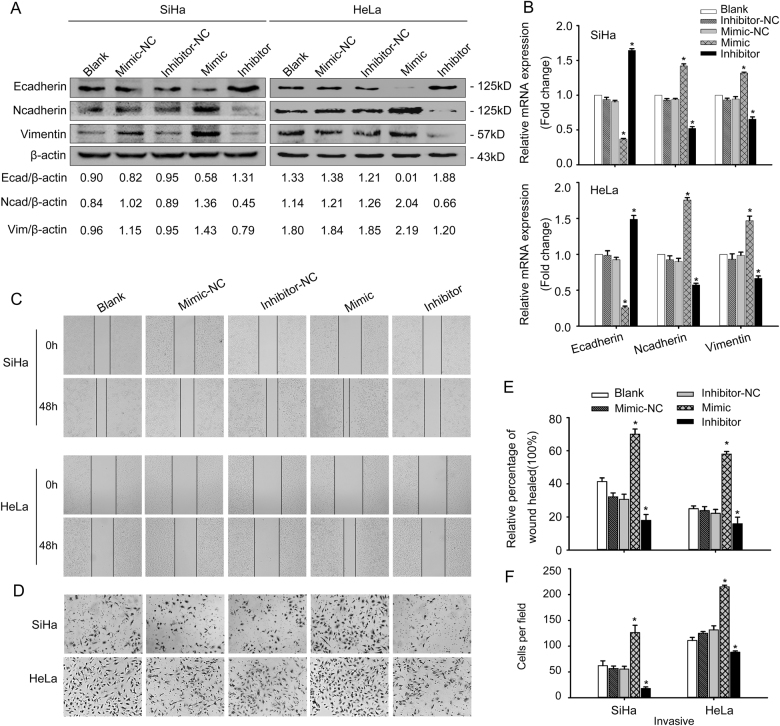
miR-221-3p promoted EMT of CC cells in vitro
Loss-of-function or gain-of-function assays were performed to ascertain whether miR-221-3p could promote CC cell EMT. Transfection of SiHa and HeLa cells with miR-221-3p mimic caused increased expression of Vimentin and N-cadherin protein and decreased expression of E-cadherin protein (p < 0.05, Fig. 2a). These changed expression profiles of EMT markers (E-cadherin, Vimentin, and N-cadherin) were also confirmed by qRT-PCR (*p < 0.05, Fig. 2b) and immunofluorescent results (Fig. S1). MiR-221-3p mimic-treated cells showed enhanced invasiveness and migration capacities compared with blank and NC groups by Boyden chamber and wound-healing assays (*p < 0.05, Fig. 2c–f). In contrast, this result was reversed by treatment with a miR-221-3p inhibitor (*p < 0.05, Fig. 2a, 2c–f). These data clearly substantiate that miR-221-3p contributes to enhance the transition from an epithelial to mesenchymal phenotype in CC cells.
Fig. 2. miR-221-3p promoted EMT in vitro.
a SiHa and HeLa cells were transfected with miR-221-3p mimic, miR-221-3p inhibitor, miR-221-3p mimic-nc, and miR-221-3p inhibitor-nc. Western blotting analysis of E-cadherin, N-cadherin, and Vimentin was performed of the five groups in SiHa and HeLa cells. β-actin was used as loading control. b qRT-PCR analysis of E-cadherin, N-cadherin, and Vimentin of the five groups in SiHa and HeLa cells. c Wound-healing assay of the five groups in SiHa and HeLa cells. d Boyden chamber assay of the five groups in SiHa and HeLa cells. e Cell migration was quantified as percentage of wound-healed area. f Average number of invading cells per field from three independent experiments. Data represent means ± SD of five randomly selected areas. *p < 0.05
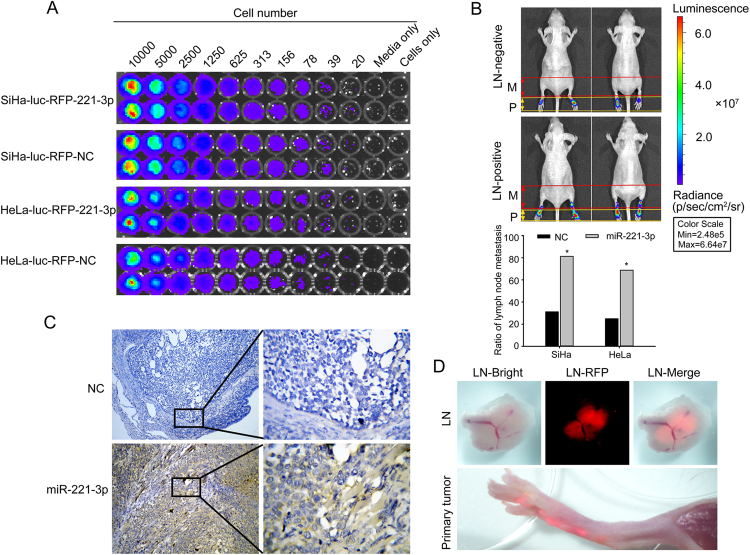
MiR-221-3p enhances lymphatic metastasis in vivo
The effect of miR-221-3p on lymph node metastasis in CC was investigated in vivo using a popliteal lymph node metastasis model. The SiHa-luc-RFP-221-3p, SiHa-luc-RFP-NC, HeLa-luc-RFP-221-3p and HeLa-luc-RFP-NC cells, which stably expressed firefly luciferase, were detected by using the IVIS Spectrum system (Fig. 3a). The results showed that the ratio of metastatic to total popliteal lymph nodes was markedly higher in the SiHa-luc-RFP-221-3p group (81.25%, 13/16) and HeLa-luc-RFP-221-3p group (68.75%, 11/16) than in the SiHa-luc-RFP-NC group (31.25%, 5/16) and HeLa-luc-RFP-NC group (25.00%, 4/16) (*p <0.05, Fig. 3b, Table 1). Detecting RFP-expressing primary tumors and popliteal lymph nodes under an Olympus SZX16 dissecting microscope also confirmed the above results (Fig. 3d). In situ hybridization with the miR-221-3p LNA probe further determined that tumors with increased miR-221-3p expression had higher rates of lymph nodes metastasis than did the decreased miR-221-3p expression group (Fig. 3c). These data provided strong evidence that the high expression of miR-221-3p was closely associated with lymph node metastasis.
Fig. 3. miR-221-3p enhances lymphatic metastasis in vivo.
a Correlation of bioluminescence intensity with cell quantity in SiHa-luc-RFP-221-3p, SiHa-luc-RFP-nc, HeLa-luc-RFP-221-3p, and HeLa-luc-RFP-NC cells in vitro. b In vivo bioluminescence images of lymphatic metastasis. Primary tumour (P) in claw pad of mice and metastases (M). Lymph nodes in popliteal and inguinal regions were measured by detecting bioluminescence signals. The ratio of metastasis-positive lymph nodes was calculated. *p < 0.05. c In situ hybridization of miR-221-3p expression in primary tumors of mice injected with SiHa-luc-RFP-221-3p, SiHa-luc-RFP-nc, HeLa-luc-RFP-221-3p or HeLa-luc-RFP-NC cells. d Primary tumour site and lymph nodes with tumour cells were identified by tumour cell-expressed RFP
Table 1.
Claw pad model and metastasis LN number
| Cell line | Experimental parameters | Metastases LN in vivo | ||||
|---|---|---|---|---|---|---|
| Mouse strain | n | End-point (weeks) | No. total LNs | No. metastasis LNs | Metastatic ratio (%) | |
| SiHa-luc-RFP-NC | BALB/c-nu | 8 | 7–9 | 16 | 5 | 31.25 |
| SiHa-luc-RFP-221-3p | BALB/c-nu | 8 | 7–9 | 16 | 13 | 81.25a |
| HeLa-luc-RFP-NC | BALB/c-nu | 8 | 7–9 | 16 | 4 | 25.00 |
| HeLa-luc-RFP-221-3p | BALB/c-nu | 8 | 7–9 | 16 | 11 | 68.75b |
aThe metastatic ratio has statistical significance between the SiHa-luc-RFP-221-3p group and SiHa-luc-RFP-NC group, (p = 0.011, χ2-test)
bThe metastatic ratio has statistical significance between the HeLa-luc-RFP-221-3p group and HeLa-luc-RFP-NC group, (p = 0.032, χ2-test)
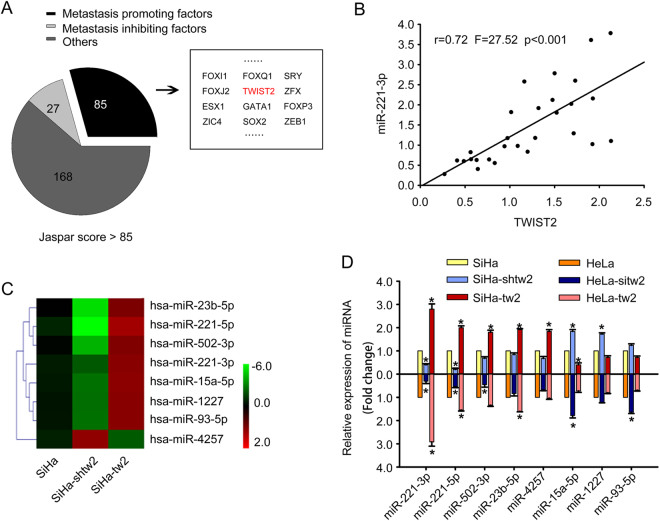
Candidate transcription factors of the miR-221-3p promoter are identified
To investigate the transcription factors (TF) of the miR-221-3p promoter, we scanned the 2000-bp sequence for TF-binding site motifs using the Jaspar database. A total of 290 genes with the Jaspar score > 85 were selected, and further literature searching confirmed that 85 genes were potential metastasis-promoting genes (Fig. 4a). These included TWIST2, a classical EMT promoter. Our previous studies21,22 have verified that the expression of TWIST2 was closely correlated with lymph node metastasis, which could have significant implications for its potential utility as a biomarker of CC prognosis. Then, the expression of miR-221-3p and TWIST2 were detected in a matched collection of human CC tissues with or without lymph node metastasis. qRT-PCR results showed a positive correlation between miR-221-3p and TWIST2 (r = 0.72, F = 27.52, p <0.001, Fig. 4b, Table S2).
Fig. 4. Candidate transcription factors of the miR-221-3p promoter are identified.
a Bioinformatic prediction and screening of potential transcription factors of miR-221-3p. b Correlation analysis of TWIST2 and miR-221-3p expression detected by qRT-PCR in 28 additional SCC tissues. The expression levels of TWIST2 were plotted against the expression levels of miR-221-3p. c The certified result of microarray analysis. Hierarchical clustering of eight significantly dysregulated miRNAs expression profiles of the three groups, including SiHa, SiHa-shtw2, and SiHa-tw2 cells. d qRT-PCR analysis of the eight significantly dysregulated miRNAs expressed in SiHa, SiHa-shtw2, and SiHa-tw2 cells, and in HeLa cells transfected with TWIST2 expression vector or TWIST2 siRNA. MiR-221-3p emerged as a highly upregulated miRNA. Data represent means ± SD of five randomly selected areas (*p < 0.05)
However, it was unclear whether the expression of miR-221-3p was the most significantly changed miRNA, resulting from the different expression levels of TWIST2 in CC cells. Therefore, we used a miRNA microarray to identify the miRNA expression profiles among SiHa, SiHa-tw2, and SiHa-shtw2 (Deep-sequencing data has been deposited in GEO with accession number: GSE102706). By analyzing the microarray data, we obtained a list of differentially expressed miRNAs (p < 0.05).The significantly deregulated miRNAs (changes of more than twofold in expression) included miR-23b-5p, miR-221-3p, miR-502-3p, miR-221-5p, miR-15a-5p, miR-1227, miR-93-5p, and miR-4257 (Fig. 4c). qRT-PCR analysis confirmed the expression levels of the eight significantly deregulated miRNAs among SiHa and HeLa cells with different TWIST2 expression states (Fig. 4d). The expression level of miR-221-3p in SiHa-tw2 (2.80 ± 0.22-fold, p < 0.05) and HeLa-tw2 (2.92 ± 0.19-fold, p < 0.05) cells was much higher than that in SiHa and HeLa cells, respectively, and the expression level of miR-221-3p in SiHa-shtw2 (0.40 ± 0.05-fold, p < 0.05) and HeLa-sitw2 (0.31 ± 0.10-fold, p < 0.05) cells was lower than that in SiHa and HeLa cells, respectively, suggesting that miR-221-3p displayed the most significant difference in both SiHa and HeLa cells with different expression level of TWIST2.
MiR-221-3p is regulated directly by TWIST2
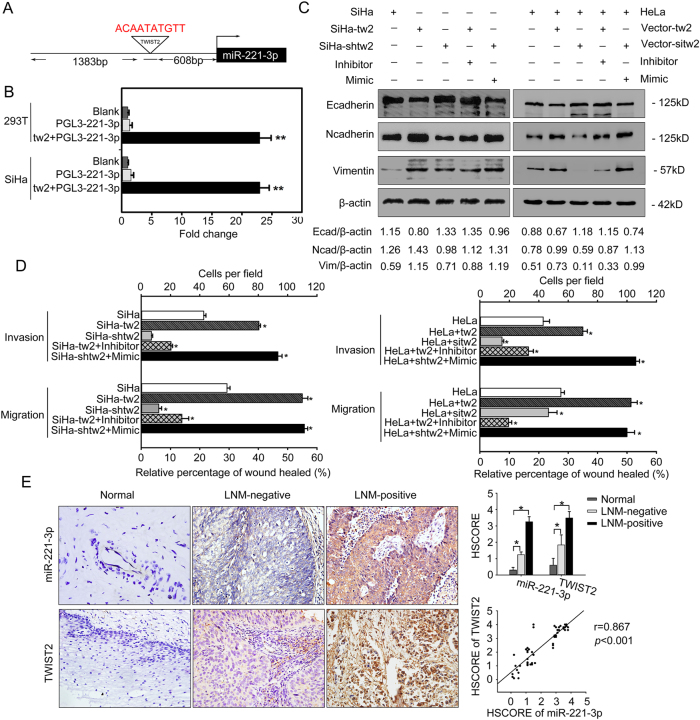
To understand how miR-221-3p expression was regulated by the transcription factor, the miR-221-3p promoter was subcloned into a pGL3-basic vector (Fig. S2), and a dual-luciferase reporter assay was performed to study the functionality of interaction between TWIST2 and miR-221-3p. Transient expression of TWIST2 effectively stimulated transcription of miR-221-3p in SiHa and 293 T-cell lines (p < 0.01; Fig. 5a, b).
Fig. 5. miR-221-3p is regulated directly by TWIST2.
a Schematic structure of the miR-221-3p upstream promoter containing a TWIST2-binding site. b Luciferase activity of PGL3-221-3p construct after transfection of TWIST2 plasmid in SiHa and 293 T cells. c Western blotting analysis of expression of E-cadherin, N-cadherin, and Vimentin proteins in SiHa, SiHa-tw2, SiHa-shwt2, or SiHa-shtw2 cells transfected with miR-221-3p mimic or miR-221-3p inhibitor; and HeLa cells transfected with TWIST2 expression vector with/without miR-221-3p mimic or TWIST2 siRNA with/without miR-221-3p inhibitor. β-actin was used as loading control. d Cell migration was quantified as the percentage of wound-healed area; average number of invaded cells per field from three independent experiments. Data represent means ± SD of five randomly selected areas (*p < 0.05; **p < 0.001). e IHC analysis of TWIST2 and in situ hybridization analysis of miR-221-3p in matched cervical specimens. Human CC tumors with high TWIST2 expression and had strong miR-221-3p expression. Normal cervical tissues with low TWIST2 expression and had weak miR-221-3p expression. Representative images are shown at ×400 magnification. The HSCOREs of miR-221-3p and TWIST2 in different cervical specimens are also shown (*p < 0.05) (upper-right panel). Correlation of miR-221-3p staining and TWIST2 staining was analyzed (lower-right panel)
To further confirm the effects of miR-221-3p on TWIST2-induced EMT in cervical cancer cell lines, SiHa-tw2 cells were transfected with miR-221-3p inhibitor, SiHa-shtw2 cells were transfected with miR-221-3p mimic, and HeLa cells were transfected with TWIST2 expression vector with/without miR-221-3p mimic or TWIST2 siRNA with/without miR-221-3p inhibitor. Western blotting and qRT-PCR results showed that overexpression of TWIST2 sharply decreased the expression of E-cadherin and increased the expression of Vimentin and N-cadherin. However, the combination of the miR-221-3p inhibitor and TWIST2 reversed the epithelial gene suppression and the upregulated expression of mesenchymal genes compared to treatment with TWIST2 alone. As expected, TWIST2 silencing also increased the expression of E-cadherin and decreased the expression of Vimentin and N-cadherin, but results were abrogated when transfected with miR-221-3p mimic (Fig. 4c, S3–4). The results of wound-healing and Boyden chamber assays showed that inhibition of TWIST2 in CC cells reduced the motility of these cells, but this result was reversed by the treatment with miR-221-3p mimic. Overexpression of TWIST2 increased their invasiveness and migration ability, which were reversed by the treatment with the miR-221-3p inhibitor (*p < 0.05, Fig. 5d, Figs. S5–6).
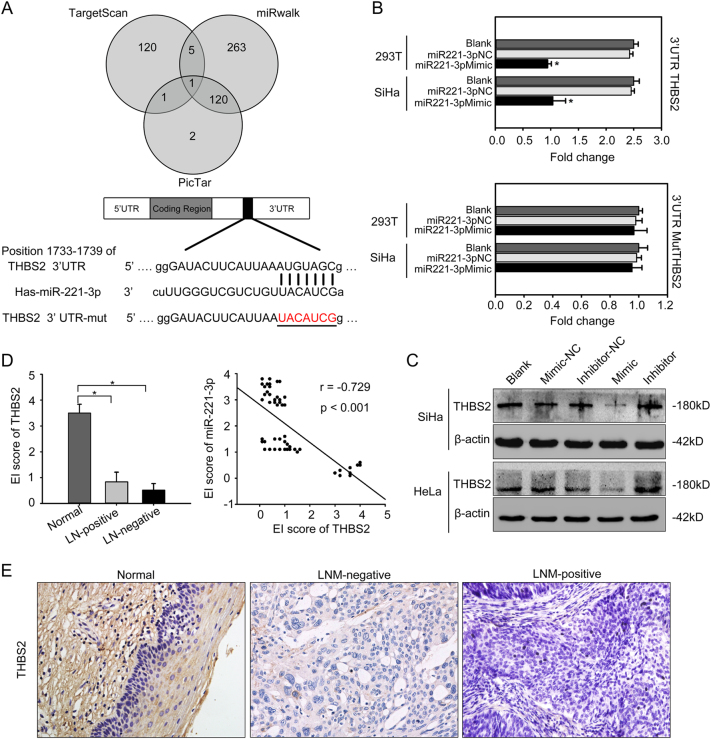
Fig. 6. The 3′UTR region of THBS2 is a direct target of miR-221-3p.
a Bioinformatic prediction and screening of potential miRNAs targeting miR-221-3p; incomplete complementation of the base of miR-221-3p to the 3′UTR region of THBS2 mRNA. Also shown are nucleotides mutated in THBS2 mutant 3′UTR. b Luciferase activity of wild-3′UTR-THBS2-luc and mutant 3′UTR-THBS2-luc constructs in SiHa or 293 T cells after transfection of miR-221-3p mimic. c Western blotting analysis of the expression of THBS2 in SiHa and HeLa cells transfected with miR-221-3p mimic, miR-221-3p inhibitor, miR-221-3p mimic-nc, and miR-221-3p inhibitor-nc. β-actin was used as loading control. d The HSCOREs of THBS2 in different cervical specimens (left), and correlation of THBS2 staining and miR-221-3p was analyzed (right). e Normal cervical tissues with high THBS2 expression. Human CC tumors with low THBS2 expression. *p < 0.05
The expression of miR-221-3p and TWIST2 were also detected in cervical specimens. A significant increase in miR-221-3p and TWIST2 expression was found at CC tissues with lymph node metastasis (p < 0.05, Fig. 5e). Spearman correlation analysis showed a positive relationship between the miR-221-3p expression level and the TWIST2 protein level (r = 0.867; p < 0.001). These data verify that upregulation of miR-221-3p and TWIST2 may facilitate lymph node metastasis, and the increased expression of TWIST2 can be one of the causes of high expression of miR-221-3p in CC.
The 3'UTR region of THBS2 is a direct target of miR-221-3p
Bioinformatics analysis using the TargetScan, PicTar, and miRwalk database predicted that THBS2 was the possible downstream target gene of miR-221-3p (Fig. 6a). Based on TargetScan, there existed one suitable binding site with perfect matches for miR-221-3p (mirSVR score: −0.3494) in the 3′-UTR sequence of THBS2. miRanda was also used to check and re-deduce the putative binding sites from position 1733 to 1739 of the THBS2 3′UTR region. To confirm this speculation, a 1187-bp fragment of the 3′-UTR region of THBS2 mRNA that include the predicted miR-221-3p recognition site was subcloned and then inserted into a luciferase reporter plasmid (Fig. S7). MiR-221-3p-binding sites in the 3′UTR region of THBS2 were mutated to obtain the 3′-UTR-MutTHBS2-luc plasmid. Transient transfection of wild-type THBS2-luc reporter with miR-221-3p mimic into 293T and SiHa cells led to a significant decrease in luciferase activity compared with NC or blank control (*p < 0.05; Fig. 6b). However, miR-221-3p could not decrease the luciferase activity of the mutant construct-3′UTR-MutTHBS2-luc in the miR-221-3p-binding site compared with NC or blank (p > 0.05; Fig. 6b). The results make evident that miR-221-3p affects THBS2 expression by directly binding to the 3′-UTR region of THBS2 and validate that THBS2 is a direct downstream target of miR-221-3p.
Transfection of SiHa and HeLa cells with miR-221-3p mimic caused decreased THBS2 protein expression. Conversely, THBS2 expression was elevated after transfection of miR-221-3p inhibitor in SiHa or HeLa cells (Fig. 6c). The expression of THBS2 was detected in the same cervical specimens used in previous study (Table S2). The results showed that the THBS2 expression decreased in CC tissues but not in normal cervix (p < 0.05, Fig. 6d, e). The expression level of THBS2 was significantly lower in CC with lymph node metastasis compared with those without lymph node metastasis (*p < 0.05). Spearman correlation analysis showed a negative relationship between the THBS2 and miR-221-3p expression (r = −0.729; p < 0.001). These data verify that the deletion of THBS2 protein was related to poor outcome of CC patients, and upregulation of miR-221-3p is responsible for the aberrant expression of THBS in CC.
Discussion
Metastasis is the key hallmark of malignance24. In CC, lymph node metastasis is recognized as the major route for tumour metastasis, which is one of the most common poor prognostic factors in patients27. Our study investigated the involvement of a miRNA-mediated mechanism in tumour metastasis in CC. In the current study, miR-221-3p represents a significantly upregulated miRNA in human cervical squamous carcinoma tissues with lymph node metastasis compared with those without lymph node metastasis. Recently, miR-221-3p upregulation has been found in several different tumour types such as breast cancer, renal cell carcinoma, hepatocellular carcinoma28–32 and may represent a more aggressive phenotype. Moreover, silencing of miR-221-3p blocks hepatocellular carcinoma and promotes survival31. However, the association of miR-221-3p with tumour invasion and metastasis has not yet been clarified. Loss-of-function and gain-of-function assays were performed and confirmed that miR-221-3p induces CC cells to invade and metastasize in vitro and in vivo. In agreement with a previous report that miR-221 could be EMT-related miRNAs in aggressive cancers33, our results also showed that miR-221-3p upregulates mesenchymal markers (such as Vimentin and N-cadherin), downregulates epithelial markers (such as E-cadherin), and strengthens cell invasion and migration in CC cells. Furthermore, miR-221-3p level correlates with lymphatic metastasis in CC patients, which provides new insight into this area of research by identifying miR-221-3p as a clinically relevant promoter of cancer metastasis.
MiRNAs have been shown to be regulated by the upstream transcription factors34,35. When we analyzed the promoter region of miR-221-3p, TWIST2 was focused on as a potential transcription regulator that might contribute to metastasis in cervical cancer, which has been found to be an indicator of metastasis potential in CC patients, and functions as a tumour promoter to accelerate tumour metastasis by promoting EMT21,22. A study of Shi J et al.36 also reported that TWIST2 is a key activator of EMT and is closely correlated with metastasis. In addition, miR-221-3p appears to be the most strikingly upregulated miRNA among different expression level of TWIST2, which clearly validates that TWIST2 might have a role in miR-221-3p expression of CC. We proposed that TWIST2 might be a positive regulator for miR-221-3p. Luciferase activity verified that TWIST2 stimulated the transcription activity and expression of miR-221-3p by directly binding to the promoter, and confirmed miR-221-3p as a major downstream effector of TWIST2 in its target network.
Up to present, researches showed that TWIST2 inhibits apoptosis and facilitates EMT correlated with poor outcomes in cancer patients36–38. However, inhibition of miR-221-3p could depress the TWIST2-induced EMT process, as indicated by decreased migration and invasion abilities in SiHa-tw2 and HeLa-tw2 cells treated with miR-221-3p inhibitors. On the contrary, restoration of miR-221-3p can promote such biological functions of CC cells, which recapitulated the TWIST2 knockdown effects. Thus, miR-221-3p is a target regulated by the transcription factor TWIST2, and TWIST2 may promote EMT by regulating miR-221-3p in CC.
Most miRNAs are believed to function by inhibiting translation of their mRNA targets39. To identify downstream effectors of miR-221-3p, we used miRNA-predicting algorithms (TargetScan, miRanda, and PicTar) based on the presence of binding sites in the 3′-UTR. By bioinformatics prediction, THBS2 was found to be an important target gene of miR-221-3p in our study. THBS2, a matricellular glycoprotein, participants in multiple roles which including bone growth, cell adhesion, extracellular matrix modeling, inflammatory responses, development, and pathological angiogenesis40. THBS2 expression in tumour has been associated with decreased vascularity, progression, and metastasis41. We proposed that miR-221-3p might be a novel negative regulator of THBS2 in CC tissues. Results confirmed that miR-221-3p directly targets the 3′UTR of THBS2, and suppressed its 3′-UTR sequence. As we expected, the THBS2 mutant 3′-UTR abolished the miR-221-3p-mediated suppression of the THBS2. Over the past decade, researches have shown that decreased THBS2 expression is associated with the poor outcome of gastric cancer42. In agreement with this study, we have also found that THBS2 deletion contributes to cervical cancer progression. Taken together, our data further indicate that THBS2, which is a functional target of miR-221-3p, is a vital suppressor factor in CC. Although Bornstein P et al.43 have reported that the inhibitory role of THBS2 in tumors is related to its multiple interactions with cell surface receptors (LRP, CD36, CD47, and numerous integrins), ECM components, growth factors (TGF-β, FGF2), enzymes (MMPs, elastase), and calcium binding, the effect and biological mechanism of THBS2 during lymph node metastasis in CC have not been reported. In-depth study is needed to better understand the complex regulatory networks on TWIST2/miR-221-3p/THBS2.
In conclusion, we showed a novel regulatory mechanism by which, under the regulation of TWIST2 transcription, miR-221-3p accelerated the invasion and metastasis of CC via targeting THBS2, although future studies are required to further expand the analysis of THBS2-mediated inhibition of cell migration and invasion. Identification of metastasis-specific miRNAs and their targets is important to understand their roles and define new therapeutic strategies for the relapse and metastasis of CC.
Electronic supplementary material
Acknowledgments
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers: 81672589, 81372781, 81072132), the Shenzhen Science and Technology Program (grant number: JCYJ20160429161218745), and the National Key Research and Development Program of China (2016YFC1302900).
Competing interests
The authors declare that they have no competing financial interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-022-05469-2
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wen-Fei Wei, Chen-Fei Zhou, and Xiang-Guang Wu contributed equally to this work
Edited by Y. Shi
Change history
12/2/2022
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41419-022-05469-2
Contributor Information
Li Liang, Email: lli@fimmu.com.
Wei Wang, Email: smugowwang@126.com.
Supplementary information
The online version of this article (10.1038/s41419-017-0077-5) contains supplementary material.
References
- 1.Jain RK, Lahdenranta J, Fukumura D. Targeting PDGF signaling in carcinoma-associated fibroblasts controls cervical cancer in mouse model. PLoS. Med. 2008;5:e24. doi: 10.1371/journal.pmed.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lea JS, Lin KY. Cervical cancer. Obstet. Gynecol. Clin. North. Am. 2012;39:233–253. doi: 10.1016/j.ogc.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Haggar FA, Preen DB, Pereira G, Holman CD, Einarsdottir K. Cancer incidence and mortality trends in Australian adolescents and young adults, 1982-2007. BMC. Cancer. 2012;12:151. doi: 10.1186/1471-2407-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Nagura M, et al. Invasion of uterine cervical squamous cell carcinoma cells is facilitated by locoregional interaction with cancer-associated fibroblasts via activating transforming growth factor-beta. Gynecol. Oncol. 2015;136:104–111. doi: 10.1016/j.ygyno.2014.11.075. [DOI] [PubMed] [Google Scholar]
- 6.Gallup, D. G. The spread and staging of cervical cancer. Glob. Libr. Women’s Med. 117–131 (2008) doi:10.3843/GLOWM.10231.
- 7.Wang W, et al. Long-age IA2 to IIA2 cervical cancer: a matched cohort study. Int. J. Gynecol. Cancer. 2016;26:1264–1273. doi: 10.1097/IGC.0000000000000749. [DOI] [PubMed] [Google Scholar]
- 8.Crea F, Clermont PL, Parolia A, Wang Y, Helgason CD. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev. 2014;33:1–16. doi: 10.1007/s10555-013-9455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo BH, et al. Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer. Mol. Cancer. 2011;10:1. doi: 10.1186/1476-4598-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spano D, Heck C, De Antonellis P, Christofori G, Zollo M. Molecular networks that regulate cancer metastasis. Semin. Cancer Biol. 2012;22:234–249. doi: 10.1016/j.semcancer.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Sleeman JP, Thiery JP. SnapShot: the epithelial-mesenchymal transition. Cell. 2011;145:162.e1. doi: 10.1016/j.cell.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thum T, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 16.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 17.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulaiman SA, Ab Mutalib NS, Jamal R. miR-200c regulation of metastases in ovarian cancer: potential role in epithelial and mesenchymal transition. Front. Pharmacol. 2016;7:271. doi: 10.3389/fphar.2016.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ru P, et al. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol. Cancer Ther. 2012;11:1166–1173. doi: 10.1158/1535-7163.MCT-12-0100. [DOI] [PubMed] [Google Scholar]
- 20.Logan M, Hawkins SM. Role of microRNAs in cancers of the female reproductive tract: insights from recent clinical and experimental discovery studies. Clin. Sci. 2015;128:153–180. doi: 10.1042/CS20140087. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, et al. Correlation of TWIST2 up-regulation and epithelial–mesenchymal transition during tumorigenesis and progression of cervical carcinoma. Gynecol. Oncol. 2012;124:112–118. doi: 10.1016/j.ygyno.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Wang T, et al. Twist2, the key Twist isoform related to prognosis, promotes invasion of cervical cancer by inducing epithelial-mesenchymal transition and blocking senescence. Hum. Pathol. 2014;45:1839–1846. doi: 10.1016/j.humpath.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Mathelier A, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhar DK, et al. Downregulation of KiSS‐1 expression is responsible for tumor invasion and worse prognosis in gastric carcinoma. Int. J. Cancer. 2004;111:868–872. doi: 10.1002/ijc.20357. [DOI] [PubMed] [Google Scholar]
- 25.Gombos Z, Xu X, Chu CS, Zhang PJ, Acs G. Peritumoral lymphatic vessel density and vascular endothelial growth factor C expression in early-stage squamous cell carcinoma of the uterine cervix. Clin. Cancer Res. 2005;11:8364–8371. doi: 10.1158/1078-0432.CCR-05-1238. [DOI] [PubMed] [Google Scholar]
- 26.Liu D, et al. SIX1 promotes tumor lymphangiogenesis by coordinating TGFβ signals that increase expression of VEGF-C. Cancer Res. 2014;74:5597–5607. doi: 10.1158/0008-5472.CAN-13-3598. [DOI] [PubMed] [Google Scholar]
- 27.Gouy S, et al. Prospective multicenter study evaluating the survival of patients with locally advanced cervical cancer undergoing laparoscopic para-aortic lymphadenectomy before chemoradiotherapy in the era of positron emission tomography imaging. J. Clin. Oncol. 2013;31:3026–3033. doi: 10.1200/JCO.2012.47.3520. [DOI] [PubMed] [Google Scholar]
- 28.Roscigno G, et al. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget. 2015;7:580–592. doi: 10.18632/oncotarget.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu GJ, et al. miRNA-221 promotes proliferation, migration and invasion by targeting TIMP2 in renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:5224–5229. [PMC free article] [PubMed] [Google Scholar]
- 30.Park JK, et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011;71:7608–7616. doi: 10.1158/0008-5472.CAN-11-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ihle MA, et al. miRNA‐221 and miRNA‐222 induce apoptosis via the KIT/AKT signalling pathway in gastrointestinal stromal tumors. Mol. Oncol. 2015;9:1421–1433. doi: 10.1016/j.molonc.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howe EN, Cochrane DR, Richer JK. The miR-200 and miR-221/222 microRNA families: opposing effects on epithelial identity. J. Mammary Gland. Biol. Neoplasia. 2012;17:65–77. doi: 10.1007/s10911-012-9244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah MY, Calin GA. MicroRNAs miR-221 and miR-222: a new level of regulation in aggressive breast cancer. Genome Med. 2011;3:56. doi: 10.1186/gm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma L, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryu S, et al. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell. 2013;23:63–76. doi: 10.1016/j.ccr.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 36.Shi J, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnes RM, Firulli AB. A twist of insight-the role of Twist-family bHLH factors in development. Int. J. Dev. Biol. 2009;53:909. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu AY, et al. Twist2 promotes self-renewal of liver cancer stem-like cells by regulating CD24. Carcinogenesis. 2014;35:537–545. doi: 10.1093/carcin/bgt364. [DOI] [PubMed] [Google Scholar]
- 39.Liang L, et al. MicroRNA-137, an HMGA1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology. 2013;144:624–635. doi: 10.1053/j.gastro.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 40.Meng H, Zhang X, Hankenson KD, Wang MM. Thrombospondin 2 potentiates notch3/jagged1 signaling. J. Biol. Chem. 2009;284:7866–7874. doi: 10.1074/jbc.M803650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calabro NE, Kristofik NJ, Kyriakides TR. Thrombospondin-2 and extracellular matrix assembly. Biochim. Biophys. Acta. 2014;1840:2396–2402. doi: 10.1016/j.bbagen.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R, et al. Down regulation of Thrombospondin2 predicts poor prognosis in patients with gastric cancer. Mol. Cancer. 2014;13:225. doi: 10.1186/1476-4598-13-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bornstein P, Armstrong LC, Hankenson KD, Kyriakides TR, Yang Z. Thrombospondin 2, a matricellular protein with diverse functions. Matrix. Biol. 2000;19:557–568. doi: 10.1016/S0945-053X(00)00104-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.