Abstract
Summary
The CRISPR/Cas System has been shown to be an efficient and accurate genome-editing technique. There exist a number of tools to design the guide RNA sequences and predict potential off-target sites. However, most of the existing computational tools on gRNA design are restricted to small deletions. To address this issue, we present pgRNAFinder, with an easy-to-use web interface, which enables researchers to design single or distance-free paired-gRNA sequences. The web interface of pgRNAFinder contains both gRNA search and scoring system. After users input query sequences, it searches gRNA by 3' protospacer-adjacent motif (PAM), and possible off-targets, and scores the conservation of the deleted sequences rapidly. Filters can be applied to identify high-quality CRISPR sites. PgRNAFinder offers gRNA design functionality for 8 vertebrate genomes. Furthermore, to keep pgRNAFinder open, extensible to any organism, we provide the source package for local use.
Availability and implementation
The pgRNAFinder is freely available at http://songyanglab.sysu.edu.cn/wangwebs/pgRNAFinder/, and the source code and user manual can be obtained from https://github.com/xiexiaowei/pgRNAFinder.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The RNA-guided CRISPR (clustered regularly interspaced short palindrome repeats) with its associated nuclease Cas9 was recently demonstrated to be versatile for genome engineering (Cong et al., 2013; Jinek et al., 2012; Mali et al., 2013a,b). Until now, studies have mainly focused on protein-coding genes, as shown in the genome-wide loss-of-function screening in human (Shalem et al., 2014). Previous studies have also used CRISPR/Cas system to delete large sequence fragments and knock out non-coding genes in mammalian cells (Canver et al., 2014; Fulco et al., 2016; Ho et al., 2015; Liu et al., 2017).
The CRISPR-Cas system delivers the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell. The gRNA guides the Cas9 to cleave a desired DNA site in the cell's genome, allowing existing genes to be removed and/or new ones added. To design a ‘guide’ sequence, the gRNA, which is typically 20 nts in length followed by a PAM sequence such as NGG, it is important to consider gRNA sequence itself and potential off-target sites(Fu et al., 2013; Hsu et al., 2014; Mali et al., 2013a,b; Pattanayak et al., 2013). Many web-delivered and/or standalone software solutions are available for designing highly specific single and paired-gRNA for protein coding genes in vertebrate genomes(Hodgkins et al., 2015; Perez et al., 2017). Still, it is paramount to develop a new tool which can design distance-independent paired-gRNA for non-coding genes.
Here we present pgRNAFinder, a web-based platform for gRNA design, which integrates three different tools for a comprehensive study of gRNA design based on different algorithms for the first time. Our web interface includes 8 vertebrate genomes for gRNA design. Furthermore, we provide a python-based package that allows users to design paired- and single-gRNA for any organism.
2 Materials and methods
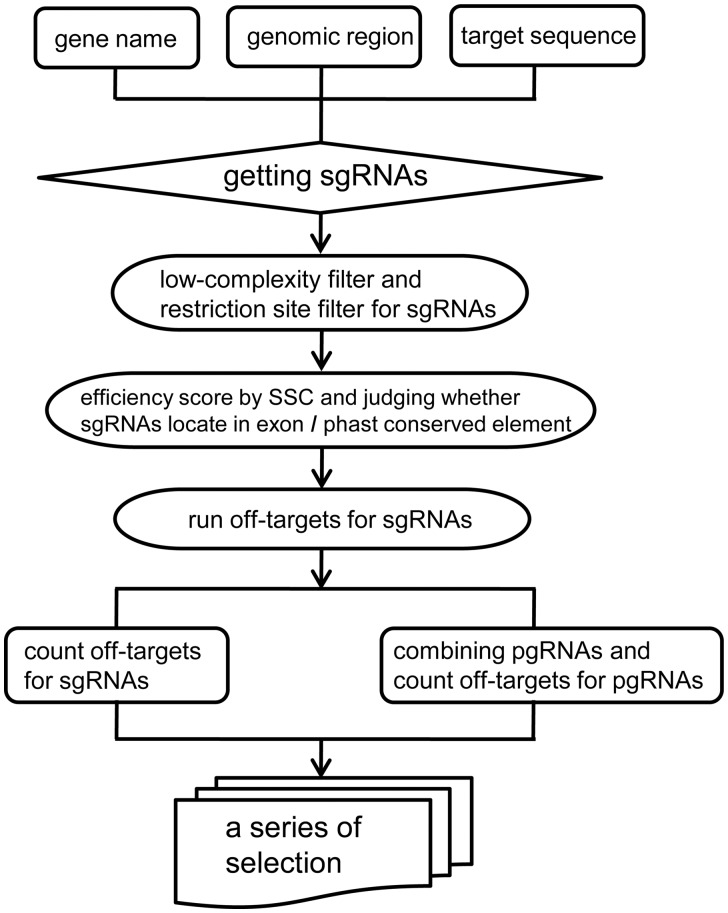
The software package contains eight Python scripts. The main steps of those scripts are listed as follows (Fig. 1): Step1. Searching single gRNA by PAMs and filtering gRNA by sequence features; Step2. Evaluating gRNA efficiency; Step3. Identifying all genomic off-target sites; Step4. For paired-gRNA, combining gRNA pair, counting the total number of off-targets and those in exons, and extracting deleted sequences to calculate conservation scores; Step5. Counting CG content, pair gRNA distance to the target gene and coverage of the deleted sequence.
Fig. 1.
Flowchart of pgRNAFinder
PgRNAFinder incorporates other gRNA-designed tools to search and evaluate gRNA. sgRNAcas9 is a software package for quickly searching for CRISPR gRNA with user-defined parameters and analyzing the potential off-target cleavage sites (Xie et al., 2014).Moreover, sgRNAcas9 searches gRNA without limiting the PAM sequence or the number of mismatching bases. These ideas were applied in gRNA searching in the Setp1 of pgRNAFinder. Step1 is performed by the script 01_get_sequence_for_search.py, 02_search_sgRNA.py and 03_get_sgRNA_candidate.py. SSC is a software package developed for predicting sgRNA efficiency from genomic sequences, and outperforms existing methods. Unlike other off-target evaluated methods that are based on the alignment of spacer sequences to the genome, SSC is based on modeling sequence features (Xu et al., 2015). Step2 includes SSC to predict sgRNA efficiency, which is performed by 04_sgRNA_ efficiency.py. Off-Spotter is a tool that can quickly and exhaustively identify all genomic sites that satisfy the PAM constraint and are identical or nearly identical to the provided gRNA (Pliatsika and Rigoutsos, 2015). Therefore, we included Off-Spotter in Step3, as a genome-wide off-target predictor. Step3 is performed by the script 05_offtarget_for_sgRNA.py.
3 Results and discussion
The web interface of pgRNAFinder is divided into three modules: (i) Single gRNA design module and (ii) short distance paired-gRNA design module. (iii) long distance paired-gRNA design module. Users select or upload a target sequence or gene. There are three options implemented for user input or upload, entering a gene symbol, genome region (by gene structure query), or sequence directly. They will then be prompted to select a species and a gRNA design module. Following sgRNAcas9, users can set the length of gRNA as an optional argument in the website, and the default offset of paired-gRNA ranges from -2 to 32 bp (base pairs) (Ran et al., 2013; Xie et al., 2014). The default offset of distance-free paired-gRNA ranges from 200 to 5 kbp, and the max distance is set as 1 Mb, as 1 Mb was tested to have very low probabilistic of deletion(Canver et al., 2014). Max distance set determines the effective off-target pair number count and deletion frequency is inversely related to the deletion size, thus, those parameters need to be set carefully. The strand of gRNA is reported in the result, sense/antisense or both strand gRNA can be selected form the result. However, there is also an option for the same/different or both strand searching mode. The default strand for short paired-gRNA is different, due to the Cas9nickase system (Ran et al., 2013).
The running time of pgRNAFinder is inversely correlated with gRNA number, and off-target evaluation is the time-consuming step. Off-Spotter is very fast as reported which can complete searching hundreds of thousands of potential off-targets in a few seconds (Pliatsika and Rigoutsos, 2015). For example, we selected human genomic loci chr1:69091-70008 as input and used the long distance paired-gRNA design mode to test the performance of pgRNAFinder. The result was reported in seconds. As shown in Supplementary Figure S1, the results list the paired-gRNA sites of target loci chr1:69091-70008, information of genomic loci, GC content, strand, sequence, SSC score, whether overlap with exon, pair's distance, off-target site number and coverage to the gene. Furthermore, the following information was also included: phastCons score, which can be used to check whether the target loci is evolutionary conserved. Moreover, evaluation of the off-target pair score is reported for paired-gRNA. The score is the sum of all intra-chromosomal off-target pairs that are smaller than the preset maximum distance (10 kb by default). All information would help researchers choose a more functional and unique gRNA pair. The help page contains detailed information about using the web interface and the illustration of output.
In conclusion, our pgRNAFinder tool has a user-friendly interface that speeds up designing single- and paired-gRNA. Meanwhile, PgRNAFinder provides potential off-target paired number and conservation information of target sequence to improve the gRNA selection.
Funding
The research has been supported by National Natural Science Foundation of China (NSFC) (Grant 31171397,31271533, 31301085,81330055, and 91640119), Pearl River Nova Program of Guangzhou (2012T50739 and 201710010044), National Natural Science Funds of Guangdong Province for Distinguished Young Scholar (2014A030306044), CPRIT RP160462, and the Welch Foundation Q-1673.
Conflict of Interest: none declared.
Supplementary Material
References
- Canver M.C. et al. (2014) Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem., 289, 21312–21324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.), 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol., 31, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco C.P. et al. (2016) Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science (New York, N.Y.), 354, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.T. et al. (2015) Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res., 43, e17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkins A. et al. (2015) WGE: a CRISPR database for genome engineering. Bioinformatics, 31, 3078–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D. et al. (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M. et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.), 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J. et al. (2017) CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science (New York, N.Y.), 355, eaah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P. et al. (2013a) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol., 31, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P. et al. (2013b) RNA-guided human genome engineering via Cas9. Science (New York, N.Y.), 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V. et al. (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol., 31, 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A.R. et al. (2017) GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol., 35, 347–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliatsika V., Rigoutsos I. (2015) “Off-Spotter”: very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs. Biology Direct, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A. et al. (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell, 154, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S. et al. (2014) sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One, 9, e100448.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res., 25, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.