Abstract
Background
ZWINT is a crucial component of the mitotic checkpoint. However, its possible role in lung cancer is unclear. In this study, we determined its correlation with lung cancer.
Methods
Real-time PCR and immunohistochemistry (IHC) were used to determine 40 collected clinical lung cancer samples. Chi-square test was used to examine possible correlations between ZWINT expression and clinicopathological factors. The prognostic significance of mRNA expression of ZWINT in lung cancer was evaluated using the Kaplan–Meier plotter. Univariate and multivariate Cox proportional hazards regression analysis were performed to determine whether ZWINT is an independent risk factor for overall survival (OS) and disease-free survival (DFS) of lung cancer patients. Additionally, STRING database was used to analyze protein-protein interactions.
Results
In this study, we screened 13 GSE datasets and detected that ZWINT is highly expressed in multiple carcinomas including lung, melanoma, prostate, nasopharyngeal, gastric, pancreatic, colon, esophageal, ovarian, renal, breast and liver cancer. Real-time PCR and IHC results of collected clinical lung cancer samples confirmed that ZWINT is highly expressed in tumor tissues compared with adjacent non-tumor tissues. Additionally, high expression of ZWINT might predict poor OS and DFS in lung cancer patients. Moreover, disease stage and expression level of ZWINT were correlated with recurrence-free survival and OS in lung cancer. Analysis of protein-protein interaction based on STRING database gained 8 top genes which could interact with ZWINT, including PMF1, MIS12, DSN1, ZW10, BUB1, BUB1B, CASC5, NDC80, NSL1 and NUF2.
Conclusion
ZWINT is aberrantly highly expressed in lung tumor tissues and might be involved in the pathogenesis of lung cancer.
Keywords: ZWINT, lung cancer, prognosis, overall survival, recurrence-free survival
Introduction
Lung cancer is the most common cause of cancer-related deaths on a worldwide scale.1,2 More than half of the lung cancer patients are diagnosed at a distant stage, with a 5-year overall survival (OS) rate of 18%.2 The current TNM staging system is widely used as guidance to select initial treatment and evaluate prognosis of patients. However, as high as 40% of lung cancer patients at early TNM stage suffer from relapse after surgical resection,3 suggesting that additional molecular markers in combination with TNM staging system are urgently needed for the prognosis of patients with lung cancer.
ZW10 interacting protein (ZWINT) is a known component of the kinetochore complex required for the mitotic spindle checkpoint.4,5 It has been reported that depletion of ZWINT leads to aberrant premature chromosome segregation.6 Recent studies have suggested that ZWINT might be involved in the pathogenesis of tumor development. The potential of ZWINT in promoting the proliferation of breast, prostate and bladder cancers has been demonstrated.7–11 Moreover, the expression of ZWINT is associated with estrogen regulation in breast cancer and androgen receptor expression in prostate cancer,8,9 suggesting that studies on ZWINT might provide alternative information for cancer treatment. Xu et al have reported that ZWINT was highly expressed in ovarian cancer and higher levels of ZWINT mRNA were associated with worse OS for ovarian cancer patients.12 Using Cox proportional hazards model, investigators selected eight genes (PTK7, CIT, SCNN1A, PGES, ERO1L, ZWINT and two ESTs) that would concomitantly predict the OS of pulmonary adenocarcinoma (ADC) patients.13 A couple of algorithms have been developed to produce multiple predictive models.14–16 However, the complex structure of most algorithms or models has substantially reduced their potential in clinical applications. In this study, we detected the expression level of ZWINT in lung cancer by real-time PCR to identify a marker in association with tumor recurrence. Furthermore, we evaluated the prognostic significance of ZWINT in patients with lung cancer.
Materials and methods
Tissue specimens
The fresh specimens of tumors and matched surrounding non-tumor tissues were obtained from 40 newly diagnosed lung cancer patients who underwent surgical resection. Detailed information of samples is listed in the Supplementary material (Table S1). This study was approved by the Ethics Committees of Renmin Hospital of Wuhan University and Zhongnan Hospital of Wuhan University. Written informed consent to take part in this study was obtained from all participants.
RNA extraction and real-time PCR
Total RNA was extracted from fresh tissues using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s specifications and was quantified by Nano-Drop 2000 (Thermo Fisher Scientific). Total RNA (2 µg) was reverse-transcribed to cDNA with random primers using the RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. To assess the gene expression, cDNAs were amplified with the SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara, Kusatsu, Japan) using the QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). The cycling program used was 50°C for 30 min, 94.5°C for 15 min, and then 40 cycles of 96°C for 30 s and 59.7°C for 1 min. The expression level of each gene was measured in triplicate, and β-actin was used as the reference gene. The average Ct value of each target gene was normalized against the Ct values of the reference gene. Relative gene expression was calculated as 2−ΔΔCt.
The sequences of specific primers were as follows: ZWINT – forward: 5′-CACGTAGAGGCCATCAAAATTGG-3′, reverse: 5′-CGGAGTTGTGTCCGTTTCCT-3′; β-actin – forward: 5′-GAAGAGCTACGAGCTGCCTGA-3′, reverse: 5′-CAGACAGCACTGTGTTGGCG-3′.
Immunohistochemistry
Paraffin-embedded tissue sections were dewaxed and rehydrated, and antigen retrieval was performed by microwaving in 10 mM sodium citrate buffer, pH 6.0, for 20 min. Sections were then incubated with 3% hydrogen peroxide for 30 min at room temperature to block endogenous peroxidase, followed by blocking with 10% normal goat serum (AR1009; Boster, Pleasanton, CA, USA) for 0.5 h. Immunostaining was performed by incubating with anti-ZWINT antibody (1:50, ab197794; Abcam Corp., Cambridge, UK) at 4°C overnight. Slides were then washed in phosphate-buffered saline with Tween 20 and incubated with secondary antibody (anti-rabbit detection system, 1:200, GB23303-1; Wuhan Goodbio Technology, Wuhan, People’s Republic of China) for 30 min at 37°C. Staining was visualized with 3,3-diaminobenzidine, and slides were counterstained with hematoxylin.
The Pannoramic MIDI, an automatic digital slide scanner, was used to record all the information of the tissue sections, and analysis by QuantCenter, the new-generation image analysis application from 3DHISTECH, was optimized for whole-slide quantification. H-SCORE or histochemistry score is a scoring method for the immunohistochemical results. The percentage of immunostaining and the staining intensity (0, negative; 1+, weak; 2+, moderate; 3+, strong) were recorded. An H-SCORE was calculated using the following formula: H-SCORE = (percentage of cells of weak intensity ×1) + (percentage of cells of moderate intensity ×2) + (percentage of cells of strong intensity ×3). The maximum H-SCORE would be 300, corresponding to 100% of cells with strong intensity.
GEO DataSets database
The Gene Expression Omnibus (GEO) DataSets is a publicly available database, which provides help with users query and to download experiments and curate gene expression profiles. Several GEO DataSets databases were selected to analyze the expression levels of ZWINT in carcinoma and noncarcinoma in this study. The basic features of the database are summarized in Table 1.
Table 1.
The basic features of the 15 GEO DataSets databases
| Tumor type | Accession number | Sample size (tumor/control) | P-value |
|---|---|---|---|
| Lung cancer | GSE30219 | 293/14 | <0.0001 |
| Lung cancer | GSE31210 | 226/20 | 0.0004 |
| Lung cancer | GSE19188 | 91/65 | <0.0001 |
| Melanoma | GSE3189 | 45/25 | <0.0001 |
| Prostate tumor | GSE6919 | 65/63 | <0.0001 |
| Nasopharyngeal carcinoma | GSE12452 | 31/10 | <0.0001 |
| Gastric cancer | GSE13911 | 38/31 | <0.0001 |
| Pancreatic cancer | GSE15471 | 39/39 | <0.0001 |
| Colon tumor | GSE20916 | 111/34 | 0.0394 |
| Esophageal carcinoma | GSE23400 | 53/53 | <0.0001 |
| Ovarian cancer | GSE26712 | 185/10 | <0.0001 |
| Pediatric T-ALL | GSE26713 | 117/7 | 0.2380 |
| Renal clear cell carcinoma | GSE36895 | 29/23 | <0.0001 |
| Breast cancer | GSE42568 | 104/17 | <0.0001 |
| Liver tumor | GSE14520 | 225/220 | <0.0001 |
Abbreviations: ZWINT, ZW10 interacting protein; T-ALL, T-cell acute lymphoblastic leukemia.
GSE30219 with 293 lung tumor samples and corresponding clinical information were used to analyze the correlation of ZWINT expression with clinicopathological characteristics and whether ZWINT is an independent risk factor for OS and disease-free survival (DFS) of lung cancer patients was evaluated by employing univariate and multivariate Cox proportional hazards regression analysis.17 GSE31210 is a gene expression database for pathological stage I–II primary lung ADCs with 84 cases excluded from prognosis analysis due to incomplete resection or adjuvant therapy and the rest 142 cases enrolled in the univariate and multivariate Cox proportional hazards regression analysis.18 Patients were divided into ZWINT-high-expression (top 50%) and ZWINT-low-expression (remaining 50%) groups.
Oncomine analysis
Oncomine is a cancer microarray database and web-based data mining platform, which aims to stimulate new discoveries from genome-wide expression analyses and compares the transcriptome data in various types of cancers with respective normal tissues.19 The gene expression level of ZWINT was analyzed by Oncomine in this study. We selected 10 datasets containing 1,739 samples with filter set as gene: ZWINT, analysis type: Cancer vs Normal analysis, cancer type: lung cancer. In this study, we selected 2.0-fold change, P-value =0.05 and top 10% gene rank as threshold, and then comparison of ZWINT across 17 analyses was performed based on the threshold. The details of the analyses are listed in Table 2.
Table 2.
Comparison of ZWINT expression across 17 analyses
| Legend | 10 datasets | 17 analyses | P-value | Fold change |
|---|---|---|---|---|
| 1 | Bhattacharjee et al27 | ADC vs normal | 0.001 | 3.074 |
| 2 | SCLC vs normal | 6.18E–6 | 6.775 | |
| 3 | SCC vs normal | 3.42E–8 | 14.518 | |
| 4 | Garber et al28 | LCC vs normal | 2.14E–6 | 4.530 |
| 5 | ADC vs normal | 3.03E–6 | 2.903 | |
| 6 | SCLC vs normal | 0.002 | 5.529 | |
| 7 | SCC vs normal | 1.26E–6 | 3.289 | |
| 8 | Hou et al29 | LCC vs normal | 8.10E–9 | 6.357 |
| 9 | ADC vs normal | 6.03E–18 | 3.961 | |
| 10 | SCC vs normal | 4.07E–30 | 5.798 | |
| 11 | Landi et al30 | ADC vs normal | 1.76E–20 | 2.982 |
| 12 | Okayama et al18 | ADC vs normal | 1.30E–15 | 2.953 |
| 13 | Selamat et al31 | ADC vs normal | 6.18E–16 | 1.762 |
| 14 | Stearman et al32 | ADC vs normal | 2.51E–9 | 3.758 |
| 15 | Su et al33 | ADC vs normal | 5.57E–10 | 2.696 |
| 16 | Talbot et al34 | SCC vs normal | 2.18E–8 | 2.471 |
| 17 | Wachi et al35 | SCC vs normal | 1.64E–4 | 3.455 |
Abbreviations: GEO, Gene Expression Omnibus; ADC, adenocarcinoma; SCLC, small-cell lung cancer; SCC, squamous cell carcinoma; LCC, large-cell carcinoma.
The Kaplan–Meier plotter
The prognostic significance of mRNA expression of ZWINT in lung cancer was evaluated using the Kaplan–Meier plotter (www.kmplot.com),20 an online database including gene expression data and clinical data. The OS of 1,926 patients and the first progression (FP) of 982 patients were collected in lung cancer database with a mean follow-up of 49 months. Briefly, the gene ZWINT was uploaded into the database, and samples were divided into two cohorts according to the median expression of ZWINT (high vs low expression) to obtain the Kaplan–Meier survival plots, in which the number-at-risk was shown below the main plot. Log-rank P-value and hazard ratio (HR) with 95% confidence intervals were calculated and displayed on the web page. In this study, “array quality control” was selected to “exclude biased arrays”.
Protein–protein interaction network construction
The Search Tool for the Retrieval of Interacting Genes (STRING) database aims to construct functional protein association networks by consolidating known and predicted protein–protein association data for a large number of organisms.21 The STRING resource is available at http://string-db.org/. The corresponding protein–protein interaction network of ZWINT was constructed when we selected the interactions pertaining to Homo sapiens and showed minimum interactions with a confidence score >0.9.
Statistical analysis
Statistical analysis was performed using SPSS ver. 18 (SPSS Inc., Chicago, IL, USA). Two-tailed t-test was used to compare the expression of ZWINT in different groups. Chi-square test was used to examine possible correlations between ZWINT expression and clinicopathological factors. Univariate and multivariate analyses of survival were performed using Cox proportional hazards regression model. Factors with prognostic significance in the univariate analysis were included in the subsequent multivariate analysis. A P-value of <0.05 was considered statistically significant.
Results
ZWINT was highly expressed in multiple carcinomas in GEO DataSets databases
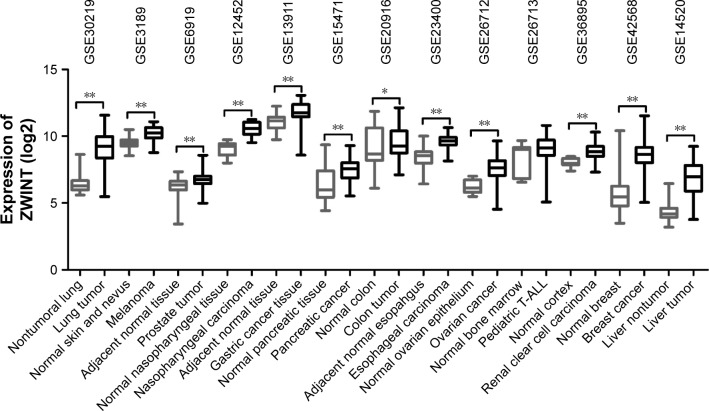
To assess the difference in the expression level of ZWINT between multiple cancerous and their noncancerous tissues, we analyzed 13 GEO DataSets databases and observed that ZWINT was highly expressed in various carcinomas including lung, melanoma, prostate, nasopharyngeal, gastric, pancreatic, colon, esophageal, ovarian, renal, breast and liver carcinomas, but not in pediatric T-cell acute lymphatic leukemia (Figure 1 and Table 1). The expression levels of ZWINT gene were upregulated in multiple tumors, indicating that it might be involved in the biology of tumorigenesis.
Figure 1.
The expression levels of ZWINT in multiple tumors. ZWINT expression levels were significantly elevated in cancerous, relative to noncancerous tissues, identified in 13 publicly available GEO DataSets databases. ZWINT expression was measured as log2 (probe intensities).
Notes: *P<0.05; **P<0.001.
Abbreviations: ZWINT, ZW10 interacting protein; GEO, Gene Expression Omnibus; T-ALL, T-cell acute lymphoblastic leukemia.
ZWINT was abundantly expressed in lung cancer and closely correlated with clinicopathological features
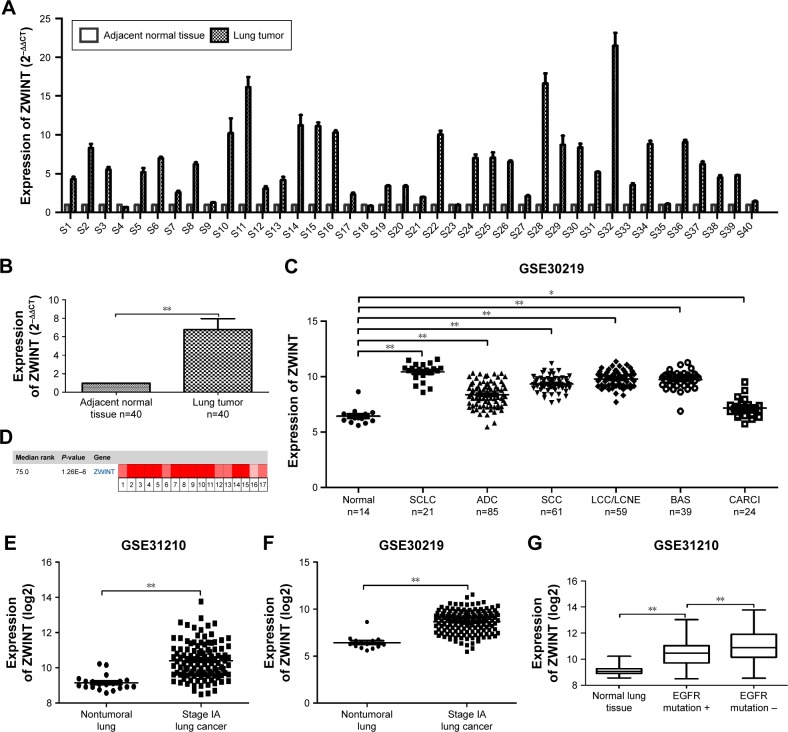
We determined the expression level of ZWINT in 40 lung cancer tissues collected from our hospital. As expected, we found using real-time PCR that ZWINT mRNA level in lung cancer tissues was significantly higher than relative adjacent nontumor tissues (P<0.001) (Figure 2A and B).
Figure 2.
ZWINT was highly expressed in lung cancer patients. (A and B) Real-time PCR analysis of ZWINT expression in 40 lung cancer specimens and adjacent nontumor tissues. (C) Analysis of ZWINT expression in normal lung and different histological subtypes of lung cancer in GSE30219. (D) Oncomine data showing ZWINT expression in lung cancer. (E and F) Analysis of ZWINT expression in normal lung and stage IA lung cancer tissue in GSE31210 and GSE30219. (G) Analysis of ZWINT expression in EGFR-mutated and EGFR-non-mutated lung cancer tissue. β-Actin was used as the internal control for real-time PCR. Experiments were done in triplicate. *P<0.05; **P<0.01.
Abbreviations: ZWINT, ZW10 interacting protein; SCLC, small-cell lung cancer; ADC, adenocarcinoma; SCC, squamous cell carcinoma; LCC, large-cell carcinoma; LCNE, large-cell neuroendocrine tumor; BAS, basaloid; CARCI, carcinoid tumor.
Next, we explored the expression of ZWINT in different histological subtypes of lung cancer using published data from GEO DataSets and Oncomine. As shown in Figure 2C, ZWINT was highly expressed in several histological subtypes including small-cell lung cancer (SCLC), squamous cell carcinoma (SCC), ADC and large-cell carcinoma, large-cell neuroendocrine tumor and carcinoid tumor compared with nontumoral lung tissues (all P<0.05) in GSE30219. Consistently, comparison of ZWINT gene expression across 17 analyses by Oncomine demonstrated the same result that ZWINT was significantly highly expressed in non-small-cell lung cancer (NSCLC) and SCLC (Figure 2D and Table 2). Furthermore, the expression of ZWINT was significantly upregulated in stage IA lung cancer tissue (Figure 2E and F). In comparison with EGFR-mutated lung cancer tissue, the expression of ZWINT was elevated in EGFR-non-mutated lung cancer tissue (Figure 2G).
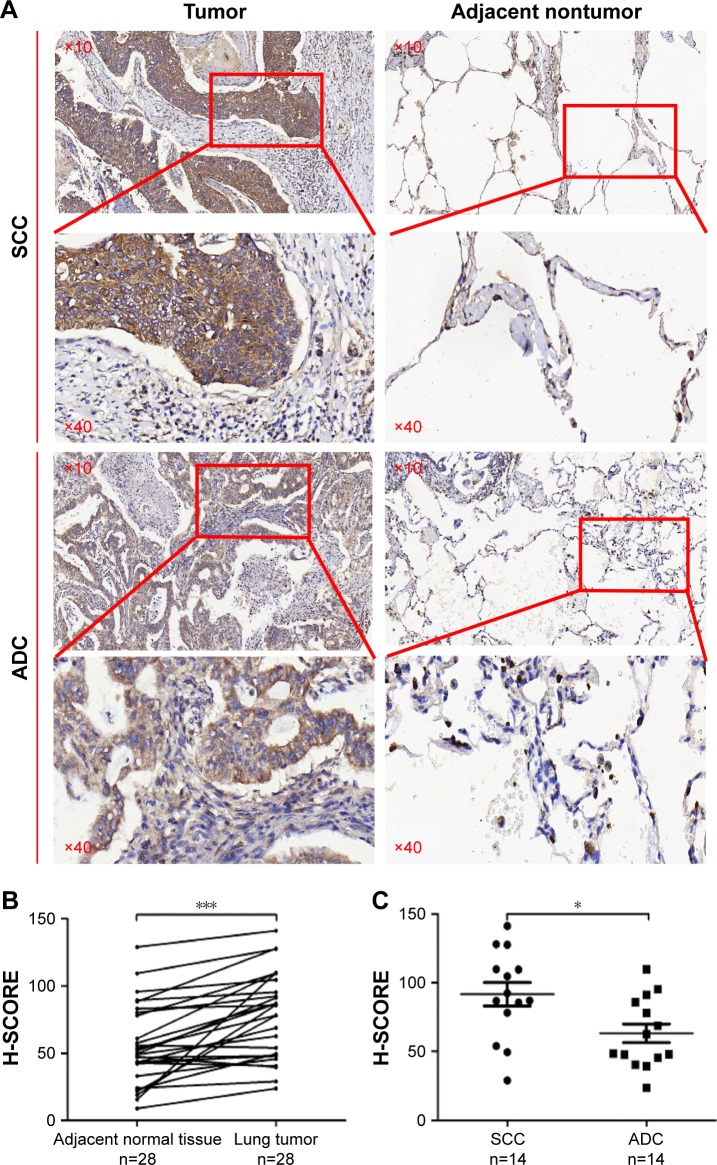
We detected the expression levels of ZWINT in 28 pairs of NSCLC specimens (tumor and adjacent nontumor tissues) which included 14 SCCs and 14 ADCs, by immunohistochemistry (Figure 3). The results showed that the H-SCORE of tumor was significantly higher than adjacent nontumor tissues (P<0.0001) (Figure 3A). Notably, we found that the expression level of ZWINT in SCC was significantly higher than that in ADC (Figure 3B).
Figure 3.
Immunohistochemistry for lung cancer tissues. (A) Representative images of lung cancer patients by immunohistochemistry (magnification: ×10, ×40). (B) H-SCORE of lung cancer and adjacent nontumor tissues. (C) H-SCORE of ADC and SCC.
Note: *P<0.05; ***P<0.001.
Abbreviations: SCC, squamous cell carcinoma; ADC, adenocarcinoma.
GSE30219 and GSE31210 were chosen to confirm the correlation between ZWINT expression and the clinicopathological parameters of lung cancer. As shown in Tables 3 and 4, ZWINT expression was closely associated with T stage (P<0.0001), N stage (P<0.0001), pathological stage (P=0.002), early recurrence (all P<0.05) and later recurrence (P=0.026). These results strongly indicated that high ZWINT expression showed significant correlation with higher stage and earlier recurrence of lung cancer.
Table 3.
Correlation between ZWINT expression and the clinicopathological features in GSE30219 with 293 lung cancer patients
| Characteristics | No. of patients | ZWINT expression
|
Chi-square value | P-value | |
|---|---|---|---|---|---|
| High | Low | ||||
| Age (years) | 0.314 | 0.575 | |||
| ≤55 | 80 | 42 | 38 | ||
| >55 | 213 | 104 | 109 | ||
| Gender | 2.136 | 0.144 | |||
| Male | 250 | 129 | 121 | ||
| Female | 43 | 17 | 26 | ||
| T stage | 25.000 | <0.0001 | |||
| 1 | 166 | 62 | 104 | ||
| 2 | 69 | 42 | 27 | ||
| 3 | 31 | 24 | 7 | ||
| 4 | 27 | 18 | 9 | ||
| N stage | 34.803 | <0.0001 | |||
| 0 | 198 | 76 | 122 | ||
| 1 | 53 | 35 | 18 | ||
| 2–3 | 42 | 35 | 7 | ||
| M stage | 2.379 | 0.122 | |||
| 0 | 282 | 138 | 144 | ||
| 1 | 11 | 8 | 3 | ||
| Early recurrencea | 15.155 | <0.0001 | |||
| No | 164 | 64 | 100 | ||
| Yes | 81 | 53 | 28 | ||
| Later recurrence | 1.770 | 0.183 | |||
| No | 164 | 64 | 100 | ||
| Yes | 33 | 17 | 16 | ||
Note:
Using 2 years as the cutoff, tumor recurrence was classified as either early recurrence or late recurrence.
Abbreviation: ZWINT, ZW10 interacting protein.
Table 4.
Correlation between ZWINT expression and the clinicopathological features in GSE31210 with 142 stage I–II primary lung adenocarcinomas patients
| Characteristics | No. of patients | ZWINT expression
|
Chi-square value | P-value | |
|---|---|---|---|---|---|
| High | Low | ||||
| Age (years) | 2.961 | 0.085 | |||
| ≤55 | 37 | 23 | 14 | ||
| >55 | 105 | 48 | 57 | ||
| Gender | 0.466 | 0.495 | |||
| Male | 58 | 31 | 27 | ||
| Female | 84 | 40 | 44 | ||
| Pathological stage | 12.073 | 0.002 | |||
| IA | 82 | 35 | 47 | ||
| IB | 34 | 15 | 19 | ||
| II | 26 | 21 | 5 | ||
| Early recurrencea | 6.727 | 0.009 | |||
| No | 110 | 47 | 63 | ||
| Yes | 17 | 13 | 4 | ||
| Later recurrence | 4.972 | 0.026 | |||
| No | 110 | 47 | 63 | ||
| Yes | 15 | 11 | 4 | ||
Note:
Using 2 years as the cutoff, tumor recurrence was classified as either early recurrence or late recurrence.
Abbreviation: ZWINT, ZW10 interacting protein.
High expression of ZWINT predicted poor OS and FP in lung cancer
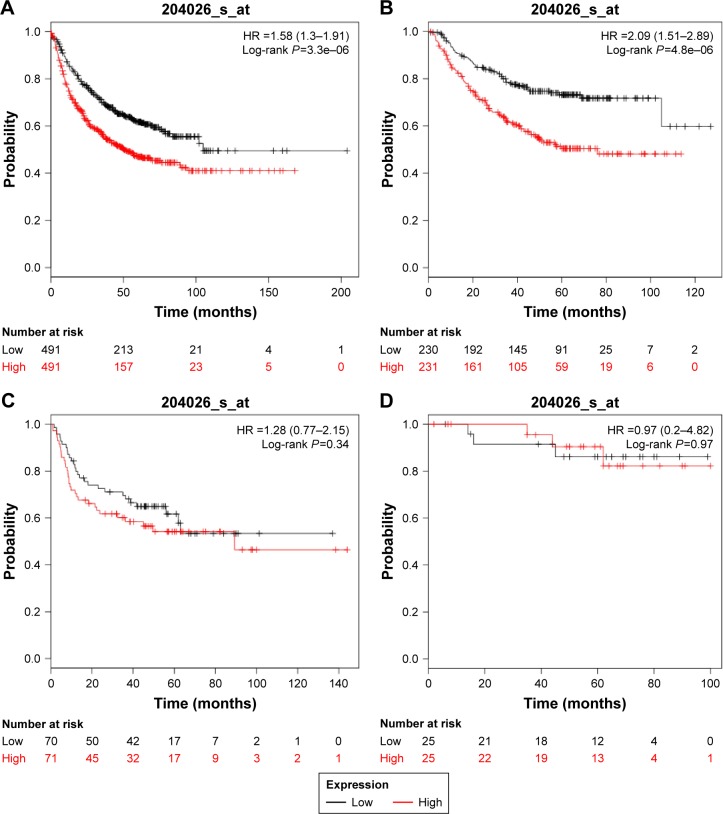
We explored the prognostic value of the expression of ZWINT using Kaplan–Meier plotter (www.kmplot.com). The desired Affymetrix ID was valid: 204026_at (ZWINT), and patients were split into low- and high-expression groups by the median of ZWINT expression values. A high expression of ZWINT mRNA was related to significantly shorter FP for all lung cancer patients (n=982, HR 1.58 [1.3–1.91], P=3.3e–06) (Figure 4A). In addition, high ZWINT expression predicted shorter FP in ADC patients (n=461, HR 2.09 [1.51–2.89], P=4.8e–06) (Figure 4B), but not in SCC patients (n=141, HR 1.28 [0.77–2.15], P=0.34) (Figure 4C). In subgroup analysis, among patients with the American Joint Committee on Cancer (AJCC) T1N0M0 stage (n=50), no significant difference in FP was observed between ZWINT-mRNA-high- and ZWINT-mRNA-low-expression groups (HR 0.97 [0.2–4.82], P=0.97) (Figure 4D).
Figure 4.
Kaplan–Meier survival curves showing the first progression survival based on the expression of ZWINT.
Notes: Survival curves were plotted for (A) all lung cancer patients (n=982), (B) ADC patients (n=461), (C) SCC patients (n=141) and (D) patients with AJCC T1N0M0 stage (n=50). The desired Affymetrix ID was valid: 204026_at (ZWINT). Data were analyzed using Kaplan–Meier plotter (www.kmplot.com).
Abbreviations: ZWINT, ZW10 interacting protein; ADC, adenocarcinoma; SCC, squamous cell carcinoma; AJCC, American Joint Committee on Cancer; HR, hazard ratio.
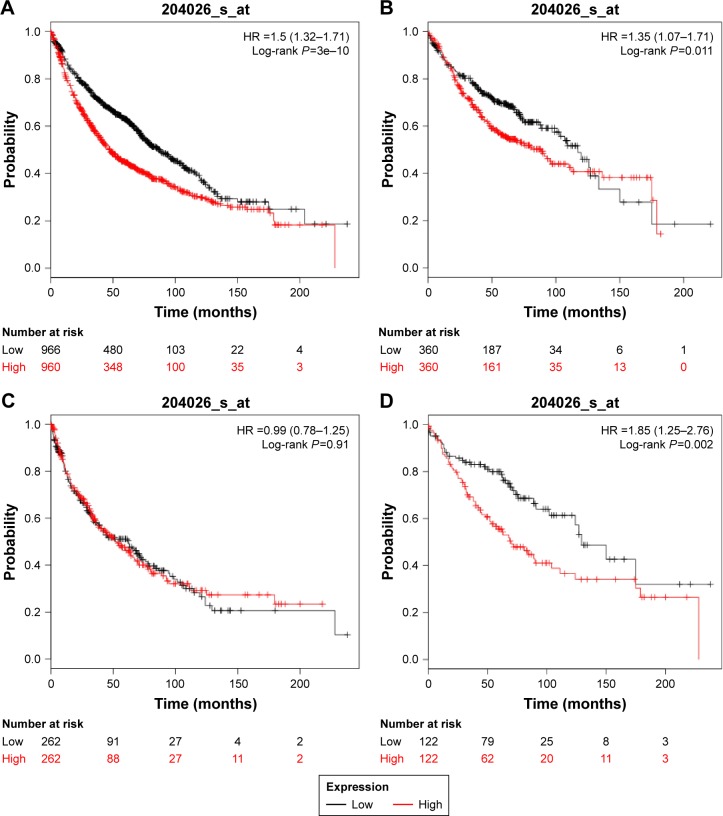
As shown in Figure 4, high ZWINT expression tended to show an unfavorable effect on OS for all lung cancer patients (n=1,926, HR 1.5 [1.32–1.71], P=3e–10) (Figure 5A). In addition, high ZWINT expression predicted worse OS in ADC patients (n=720, HR 1.35 [1.07–1.71], P=0.011) (Figure 5B), but not in SCC patients (n=524, HR 0.99 [0.78–1.28], P=0.91) (Figure 5C). In further analysis, among patients with AJCC T1N0M0 stage (n=244), significant difference in OS was observed between ZWINT-mRNA-high- and ZWINT-mRNA-low-expression groups (HR 1.85 [1.25–2.76], P=0.0002) (Figure 5D).
Figure 5.
Kaplan–Meier survival curves indicating overall survival based on the expression of ZWINT.
Notes: Survival curves were plotted for (A) all lung cancer patients (n=1,926), (B) ADC patients (n=720), (C) SCC patients (n=524) and (D) patients with AJCC T1N0M0 stage (n=244). The desired Affymetrix ID was valid: 204026_at (ZWINT). Data were analyzed using Kaplan–Meier plotter (www.kmplot.com).
Abbreviations: ZWINT, ZW10 interacting protein; ADC, adenocarcinoma; SCC, squamous cell carcinoma; AJCC, American Joint Committee on Cancer; HR, hazard ratio.
High expression of ZWINT is an independent predictor of both shorter recurrence-free survival (RFS) and OS for early-stage ADC patients
Identification of subpopulations with poor prognosis for early-stage lung cancer is critical to the optimization of personalized treatment. ZWINT expression may be helpful in prediction of survival rate in patients with early-stage lung cancer after curative pulmonectomy. GSE31210 with 142 stage I–II primary lung ADCs was applied to explore whether higher expression level of ZWINT could be an independent predictor of shorter RFS and OS for ADC patients by univariate and multivariate Cox regression analyses.
In univariate Cox regression analyses, the higher stage and high ZWINT mRNA expression exhibited unfavorable effects on both RFS (P<0.001 and P=0.003, respectively) and OS (P=0.001 and P=0.008, respectively) (Table 5). In multivariate analysis, the higher stage and high ZWINT mRNA expression were independent predictors of both shorter RFS (P=0.001 and P=0.029, respectively) and OS (P=0.007 and P=0.032, respectively). Patients with high ZWINT expression were more likely to suffer from recurrence and death than cohorts with low ZWINT expression (HR 2.524 [1.099–5.793] and HR 5.233 [1.154–23.719], respectively) (Table 6).
Table 5.
Univariate analyses of clinicopathological parameters and ZWINT expression for recurrence-free survival and overall survival in GSE31210
| Variable | Recurrence-free survival
|
Overall survival
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (≤55 vs >55 years) | 0.907 (0.420–1.962) | 0.805 | 0.579 (0.217–1.645) | 0.319 |
| Gender (female vs male) | 1.048 (0.517–2.123) | 0.896 | 1.969 (0.733–5.290) | 0.179 |
| Smoking (never vs ever) | 1.314 (0.657–2.630) | 0.440 | 2.238 (0.813–6.162) | 0.119 |
| Pathological stage (IA vs IB vs II) | 2.419 (1.595–3.669) | <0.001 | 2.862 (1.563–5.243) | 0.001 |
| ZWINT expression (low vs high) | 3.430 (1.540–7.637) | 0.003 | 7.343 (1.668–32.330) | 0.008 |
Abbreviations: ZWINT, ZW10 interacting protein; HR, hazard ratio; CI, confidence interval.
Table 6.
Multivariate analyses of clinicopathological parameters and ZWINT expression for recurrence-free survival and overall survival in GSE31210
| Variable | Recurrence-free survival
|
Overall survival
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Pathological stage (IA vs IB vs II) | 2.069 (1.354–3.162) | 0.001 | 2.308 (1.263–4.218) | 0.007 |
| ZWINT expression (low vs high) | 2.524 (1.099–5.793) | 0.029 | 5.233 (1.154–23.719) | 0.032 |
Abbreviations: ZWINT, ZW10 interacting protein; HR, hazard ratio; CI, confidence interval.
Interaction networks of ZWINT
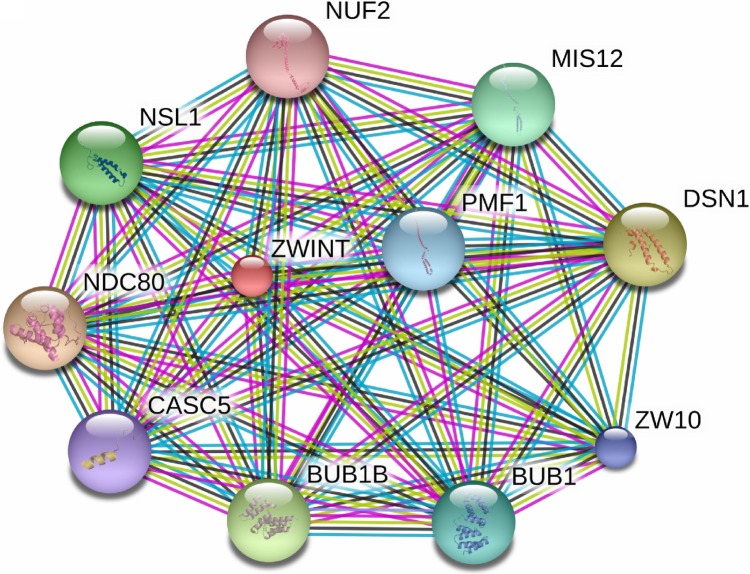
The STRING database was used to consolidate known and predicted protein–protein association with ZWINT. As shown in Figure 6, the top 10 predicted functional partners were as follows: NDC80 (score =0.997), DSN1 (score =0.997), BUB1B (score =0.997), NSL1 (score =0.996), MIS12 (score =0.995), BUB1 (score =0.994), PMF1 (score =0.993), ZW10 (score =0.993), CASC5 (score =0.993) and NUF2 (score =0.992). Function enrichment analysis against gene ontology in this network showed that for biological processes, this network is most enriched in cell division, small GTPase-mediated signal transduction and chromosome segregation, while for cellular components, it is significantly enriched in condensed chromosome kinetochore.
Figure 6.
The interaction network of ZWINT protein with other proteins.
Note: The interaction network was gained from STRING database.
Abbreviations: ZWINT, ZW10 interacting protein; STRING, Search Tool for the Retrieval of Interacting Genes.
Discussion
The majority of lung cancer patients are diagnosed in advanced or metastatic stages, which are largely inoperable.22 Lung cancer may be diagnosed at an earlier stage through the use of screening with gene signature.23 Lung cancer screening has demonstrated a reduction in lung cancer mortality by 20%.24 In this study, we found that ZWINT expression was significantly increased in stage IA lung cancer tissue compared with normal lung tissue. Thus, ZWINT can have high sensitivity for screening lung cancer at an earlier stage.
Although TNM staging system has been widely used to evaluate the prognosis of lung cancer, clinical results have shown that the relapse rate of early-stage lung cancer patients receiving potentially curative treatment is still high.3 Therefore, we still need to recognize those early-stage lung cancer patients receiving surgical management who are at high risk to recurrence. We found that among AJCC T1N0M0 stage patients, those with high expression of ZWINT showed significant propensities for poor OS compared with ZWINT-low-expression cohorts. Therefore, ZWINT might be a useful marker to predict OS for early-stage lung cancer patients.
Although ZWINT in combination with other genes might predict OS in ADC,13 the prognostic significance of ZWINT alone in OS and DFS of lung cancer is still unclear. In this study, we demonstrated that high ZWINT expression was an independent predictor of shorter OS and RFS for early-stage (pathological stage I–II) ADC patients.
Ho et al claimed that ZWINT was upregulated during bladder cancer pathogenesis in the FGFR3-non-mutated tumor pathway.11 Incidence of thanatophoric dysplasia mutations in FGFR3 gene was significantly higher in low-grade or superficial tumors than high-grade or muscle-invasive tumors.25 In this study, we found by GEO31210 analysis that the expression level of gene ZWINT was higher in the EGFR-non-mutated group than in the EGFR-mutated group. Lung ADC with EGFR-activating mutations responded well to gefitinib.26 For the EGFR-non-mutated lung cancer patients, high expression of ZWINT may be a potential therapeutic target. Endo et al found that the growth rate of breast cancer MCF7 cells was significantly increased by stable expression of ZWINT.7 Moreover, aromatase inhibitor treatment of breast cancer is associated with changes in the expression of ZWINT.8 The study demonstrated that the expression of ZWINT was increased in castration-resistant prostate containing AR gene amplification with high AR expression,9 and COX-2 inhibitors arrested prostate cancer cell cycle progression by down-regulation of kinetochore/centromere ZWINT proteins.10
Thus, ZWINT might be used as a prognostic marker for lung cancer. Clinical investigation of ZWINT on large number of lung cancer samples is needed in future. Furthermore, functional detection of ZWINT in the pathogenesis of lung cancer is still needed.
Supplementary material
Table S1.
Clinical information of 40 lung cancer samples
| Sample no | Gender | Age (years) |
Subtype | TNM | ||
|---|---|---|---|---|---|---|
| s1 | F | 44 | Adenocarcinoma | T2 | N2 | M0 |
| s2 | M | 66 | Adenocarcinoma | T2 | N0 | M0 |
| s3 | M | 74 | Adenocarcinoma | T2 | N0 | M0 |
| s4 | F | 64 | Adenocarcinoma | T2 | N2 | M0 |
| s5 | F | 63 | Adenocarcinoma | T1 | N0 | M0 |
| s6 | M | 68 | Adenocarcinoma | T3 | N2 | M0 |
| s7 | M | 74 | Adenocarcinoma | T3 | N2 | M0 |
| s8 | F | 47 | Adenocarcinoma | T3 | N2 | M0 |
| s9 | M | 64 | Squamous carcinoma | T2 | N0 | M0 |
| s10 | M | 63 | Squamous carcinoma | T2 | N1 | M0 |
| s11 | M | 74 | Squamous carcinoma | T3 | N2 | M0 |
| s12 | M | 65 | Squamous carcinoma | T2 | N2 | M0 |
| s13 | M | 55 | Squamous carcinoma | T3 | N2 | M0 |
| s14 | M | 75 | Squamous carcinoma | T2 | N2 | M0 |
| s15 | M | 56 | Squamous carcinoma | T3 | N2 | M0 |
| s16 | F | 57 | Adenocarcinoma | T3 | N2 | M0 |
| s17 | F | 48 | Adenocarcinoma | T2 | N2 | M0 |
| s18 | F | 53 | Adenocarcinoma | T1 | N0 | M0 |
| s19 | M | 58 | Squamous carcinoma | T2 | N2 | M0 |
| s20 | M | 59 | Adenocarcinoma | T2 | N2 | M0 |
| s21 | F | 62 | Squamous carcinoma | T1 | N0 | M0 |
| s22 | M | 51 | Squamous carcinoma | T3 | N1 | M0 |
| s23 | F | 53 | Adenocarcinoma | T3 | N2 | M0 |
| s24 | M | 65 | Squamous carcinoma | T2 | N2 | M0 |
| s25 | F | 45 | Adenocarcinoma | T2 | N2 | M0 |
| s26 | F | 52 | Adenocarcinoma | T1 | N0 | M0 |
| s27 | M | 75 | Squamous carcinoma | T1 | N0 | M0 |
| s28 | F | 54 | Adenocarcinoma | T1 | N0 | M0 |
| s29 | M | 69 | Squamous carcinoma | T2 | N0 | M0 |
| s30 | M | 67 | Squamous carcinoma | T3 | N2 | M0 |
| s31 | M | 68 | Squamous carcinoma | T4 | N2 | M0 |
| s32 | M | 70 | Squamous carcinoma | T3 | N2 | M0 |
| s33 | F | 65 | Squamous carcinoma | T2 | N0 | M0 |
| s34 | F | 75 | Adenocarcinoma | T2 | N0 | M0 |
| s35 | M | 56 | Adenocarcinoma | T2 | N0 | M0 |
| s36 | M | 53 | Squamous carcinoma | T4 | N2 | M0 |
| s37 | M | 63 | Squamous carcinoma | T4 | N2 | M0 |
| s38 | F | 41 | Adenocarcinoma | T1 | N0 | M0 |
| s39 | F | 68 | Adenocarcinoma | T2 | N2 | M0 |
| s40 | F | 74 | Adenocarcinoma | T1 | N0 | M0 |
Acknowledgments
This project was supported by National Natural Foundation of China (No: 81500151, 81400121, 81270607, 81541027, 81501352).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 4.Kasuboski JM, Bader JR, Vaughan PS, et al. Zwint-1 is a novel Aurora B substrate required for the assembly of a dynein-binding platform on kinetochores. Mol Biol Cell. 2011;22(18):3318–3330. doi: 10.1091/mbc.E11-03-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Hu X, Ding X, et al. Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J Biol Chem. 2004;279(52):54590–54598. doi: 10.1074/jbc.M407588200. [DOI] [PubMed] [Google Scholar]
- 6.Lin YT, Chen Y, Wu G, Lee WH. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene. 2006;25(52):6901–6914. doi: 10.1038/sj.onc.1209687. [DOI] [PubMed] [Google Scholar]
- 7.Endo H, Ikeda K, Urano T, Horie-Inoue K, Inoue S. Terf/TRIM17 stimulates degradation of kinetochore protein ZWINT and regulates cell proliferation. J Biochem. 2012;151(2):139–144. doi: 10.1093/jb/mvr128. [DOI] [PubMed] [Google Scholar]
- 8.Miller WR. Clinical, pathological, proliferative and molecular responses associated with neoadjuvant aromatase inhibitor treatment in breast cancer. J Steroid Biochem Mol Biol. 2010;118(4–5):273–276. doi: 10.1016/j.jsbmb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Urbanucci A, Sahu B, Seppälä J, et al. Overexpression of androgen receptor enhances the binding of the receptor to the chromatin in prostate cancer. Oncogene. 2012;31(17):2153–2163. doi: 10.1038/onc.2011.401. [DOI] [PubMed] [Google Scholar]
- 10.Bieniek J, Childress C, Swatski MD, Yang W. COX-2 inhibitors arrest prostate cancer cell cycle progression by down-regulation of kinetochore/centromere proteins. Prostate. 2014;74(10):999–1011. doi: 10.1002/pros.22815. [DOI] [PubMed] [Google Scholar]
- 11.Ho JR, Chapeaublanc E, Kirkwood L, et al. Deregulation of Rab and Rab effector genes in bladder cancer. PLoS One. 2012;7(6):e39469. doi: 10.1371/journal.pone.0039469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Zhou Y, Cao Y, Dinh TL, Wan J, Zhao M. Identification of candidate biomarkers and analysis of prognostic values in ovarian cancer by integrated bioinformatics analysis. Med Oncol. 2016;33(11):130. doi: 10.1007/s12032-016-0840-y. [DOI] [PubMed] [Google Scholar]
- 13.Endoh H, Tomida S, Yatabe Y, et al. Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J Clin Oncol. 2004;22(5):811–819. doi: 10.1200/JCO.2004.04.109. [DOI] [PubMed] [Google Scholar]
- 14.Girard L, Rodriguez-Canales J, Behrens C, et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin Cancer Res. 2016;22(19):4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang P, Cheng CL, Chang YH, et al. Molecular gene signature and prognosis of non-small cell lung cancer. Oncotarget. 2016;7(32):51898–51907. doi: 10.18632/oncotarget.10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahid M, Choi TG, Nguyen MN, et al. An 8-gene signature for prediction of prognosis and chemoresponse in non-small cell lung cancer. Oncotarget. 2016;7(52):86561–86572. doi: 10.18632/oncotarget.13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousseaux S, Debernardi A, Jacquiau B, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5(186):186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstraw P, Crowley J, Chansky K, et al. International Association for the Study of Lung Cancer International Staging Committee; Participating Institutions. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2(8):706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 23.Der SD, Sykes J, Pintilie M, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol. 2014;9(1):59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 24.Dhanasopon AP, Kim AW. Lung cancer screening and its impact on surgical volume. Surg Clin North Am. 2017;97(4):751–762. doi: 10.1016/j.suc.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Kimura T, Suzuki H, Ohashi T, Asano K, Kiyota H, Eto Y. The incidence of thanatophoric dysplasia mutations in FGFR3 gene is higher in low-grade or superficial bladder carcinomas. Cancer. 2001;92(10):2555–2561. doi: 10.1002/1097-0142(20011115)92:10<2555::aid-cncr1607>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 26.Chou TY, Chiu CH, Li LH, et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11(10):3750–3757. doi: 10.1158/1078-0432.CCR-04-1981. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98(24):13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5(4):e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3(2):e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22(7):1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stearman RS, Dwyer-Nield L, Zerbe L, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167(6):1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su LJ, Chang CW, Wu YC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talbot SG, Estilo C, Maghami E, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65(8):3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 35.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21(23):4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Clinical information of 40 lung cancer samples
| Sample no | Gender | Age (years) |
Subtype | TNM | ||
|---|---|---|---|---|---|---|
| s1 | F | 44 | Adenocarcinoma | T2 | N2 | M0 |
| s2 | M | 66 | Adenocarcinoma | T2 | N0 | M0 |
| s3 | M | 74 | Adenocarcinoma | T2 | N0 | M0 |
| s4 | F | 64 | Adenocarcinoma | T2 | N2 | M0 |
| s5 | F | 63 | Adenocarcinoma | T1 | N0 | M0 |
| s6 | M | 68 | Adenocarcinoma | T3 | N2 | M0 |
| s7 | M | 74 | Adenocarcinoma | T3 | N2 | M0 |
| s8 | F | 47 | Adenocarcinoma | T3 | N2 | M0 |
| s9 | M | 64 | Squamous carcinoma | T2 | N0 | M0 |
| s10 | M | 63 | Squamous carcinoma | T2 | N1 | M0 |
| s11 | M | 74 | Squamous carcinoma | T3 | N2 | M0 |
| s12 | M | 65 | Squamous carcinoma | T2 | N2 | M0 |
| s13 | M | 55 | Squamous carcinoma | T3 | N2 | M0 |
| s14 | M | 75 | Squamous carcinoma | T2 | N2 | M0 |
| s15 | M | 56 | Squamous carcinoma | T3 | N2 | M0 |
| s16 | F | 57 | Adenocarcinoma | T3 | N2 | M0 |
| s17 | F | 48 | Adenocarcinoma | T2 | N2 | M0 |
| s18 | F | 53 | Adenocarcinoma | T1 | N0 | M0 |
| s19 | M | 58 | Squamous carcinoma | T2 | N2 | M0 |
| s20 | M | 59 | Adenocarcinoma | T2 | N2 | M0 |
| s21 | F | 62 | Squamous carcinoma | T1 | N0 | M0 |
| s22 | M | 51 | Squamous carcinoma | T3 | N1 | M0 |
| s23 | F | 53 | Adenocarcinoma | T3 | N2 | M0 |
| s24 | M | 65 | Squamous carcinoma | T2 | N2 | M0 |
| s25 | F | 45 | Adenocarcinoma | T2 | N2 | M0 |
| s26 | F | 52 | Adenocarcinoma | T1 | N0 | M0 |
| s27 | M | 75 | Squamous carcinoma | T1 | N0 | M0 |
| s28 | F | 54 | Adenocarcinoma | T1 | N0 | M0 |
| s29 | M | 69 | Squamous carcinoma | T2 | N0 | M0 |
| s30 | M | 67 | Squamous carcinoma | T3 | N2 | M0 |
| s31 | M | 68 | Squamous carcinoma | T4 | N2 | M0 |
| s32 | M | 70 | Squamous carcinoma | T3 | N2 | M0 |
| s33 | F | 65 | Squamous carcinoma | T2 | N0 | M0 |
| s34 | F | 75 | Adenocarcinoma | T2 | N0 | M0 |
| s35 | M | 56 | Adenocarcinoma | T2 | N0 | M0 |
| s36 | M | 53 | Squamous carcinoma | T4 | N2 | M0 |
| s37 | M | 63 | Squamous carcinoma | T4 | N2 | M0 |
| s38 | F | 41 | Adenocarcinoma | T1 | N0 | M0 |
| s39 | F | 68 | Adenocarcinoma | T2 | N2 | M0 |
| s40 | F | 74 | Adenocarcinoma | T1 | N0 | M0 |