Fig. 3.
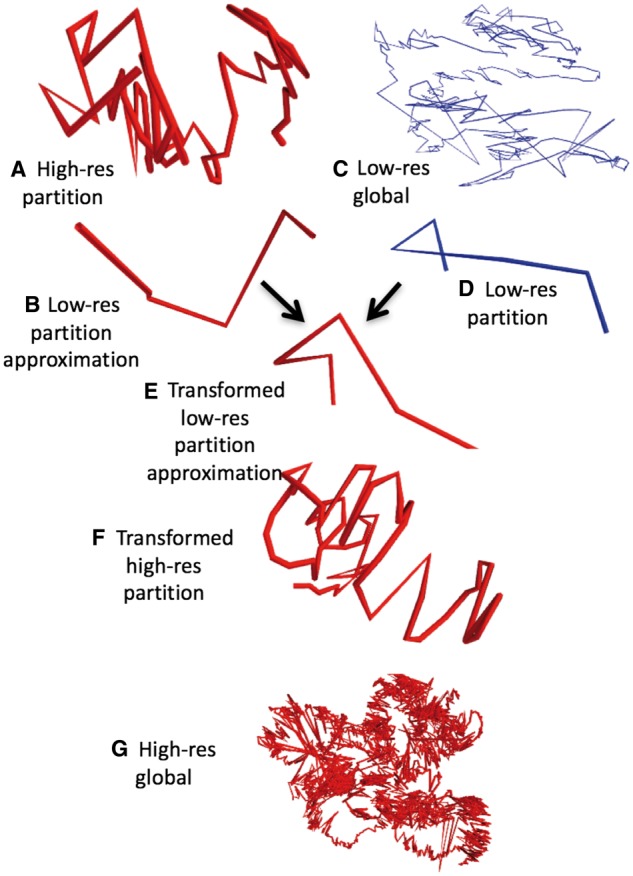
Overview of the miniMDS algorithm, demonstrated on GM12878 chr22 at 10-kbp-resolution. MDS is applied to each partition individually, creating high-resolution local structures, such as the structure shown in (A). The structure is approximated at low-resolution (B). MDS is performed on a low-resolution dataset, creating a global low-resolution structure (C). The low-resolution partition has an analog in the global structure (D). The optimal transformation to align the low-resolution approximation with its analog is calculated. For illustration, (E) shows the transformation applied to the low-resolution approximation. The transformation is applied to the high-resolution partition (F). When this process is repeated for all partitions, a global high-resolution structure is created (G)
