Abstract
Motivation
The amount of information available in textual format is rapidly increasing in the biomedical domain. Therefore, natural language processing (NLP) applications are becoming increasingly important to facilitate the retrieval and analysis of these data. Computing the semantic similarity between sentences is an important component in many NLP tasks including text retrieval and summarization. A number of approaches have been proposed for semantic sentence similarity estimation for generic English. However, our experiments showed that such approaches do not effectively cover biomedical knowledge and produce poor results for biomedical text.
Methods
We propose several approaches for sentence-level semantic similarity computation in the biomedical domain, including string similarity measures and measures based on the distributed vector representations of sentences learned in an unsupervised manner from a large biomedical corpus. In addition, ontology-based approaches are presented that utilize general and domain-specific ontologies. Finally, a supervised regression based model is developed that effectively combines the different similarity computation metrics. A benchmark data set consisting of 100 sentence pairs from the biomedical literature is manually annotated by five human experts and used for evaluating the proposed methods.
Results
The experiments showed that the supervised semantic sentence similarity computation approach obtained the best performance (0.836 correlation with gold standard human annotations) and improved over the state-of-the-art domain-independent systems up to 42.6% in terms of the Pearson correlation metric.
Availability and implementation
A web-based system for biomedical semantic sentence similarity computation, the source code, and the annotated benchmark data set are available at: http://tabilab.cmpe.boun.edu.tr/BIOSSES/.
1 Introduction
Semantic text similarity estimation is a research problem that aims to calculate the similarities among texts based on their meanings and semantic content, rather than their shallow or syntactic representation. The measures on semantic text similarity have undertaken a crucial role in many natural language processing (NLP) applications such as machine translation (Finch et al., 2005), automatic summarization (Wang et al., 2008), and question answering (Jeon et al., 2005).
Several approaches for semantic sentence similarity computation have been proposed for generic English. These approaches are in general based on computing word-level similarities and combining these to obtain sentence-level similarity scores. Corpus-based measures such as Latent Semantic Indexing (LSA), knowledge-based measures that utilize general-domain ontologies including WordNet (Miller, 1995), and string-based measures such as edit distance have been effectively used for word-level similarity computation (Li et al., 2006; Liu et al., 2015; Mihalcea et al., 2006). The SemEval Semantic Textual Similarity (STS) task series, which is being conducted annually since 2012 has also boosted research in this area (Agirre et al., 2012, 2013, 2014, 2016; Agirrea et al., 2015). Manually annotated and test datasets provided by STS enabled the development and comparison of different approaches for semantic text similarity estimation. Supervised machine learning methods that integrate different features such as WordNet and corpus-based features, syntactic features, and features based on the distributed dense vector representation of words were shown to be effective for semantic text similarity computation (Han et al., 2013; Šarić et al., 2012; Sultan et al., 2015).
Publicly available tools such as ADW (Align, Disambiguate and Walk) (Pilehvar et al., 2013; Pilehvar and Navigli, 2015) and SEMILAR (Semantic Similarity Toolkit) (Rus et al., 2013) for generic domain sentence semantic similarity computation have also been developed. ADW is a knowledge-based system that uses the Topic-sensitive PageRank algorithm (Haveliwala, 2002) over a graph generated using WordNet to model the similarity between linguistic items of different granularity such as words, sentences, and documents (Pilehvar et al., 2013; Pilehvar and Navigli, 2015). ADW was evaluated on SemEval 2012 data set and was shown to outperform the top three ranked systems (Pilehvar et al., 2013). SEMILAR is a toolkit that implements several measures based on WordNet or LSA (Rus et al., 2013). Different algorithms such as the optimal matching and the quadratic assignment problem algorithm are applied for assessing the similarity of sentence pairs by using the calculated word-level similarities (Rus et al., 2013). The general domain state-of-the-art systems ADW and SEMILAR are considered as baseline models in our study.
Assessing the similarity between two sentences is an important problem in the biomedical domain as well, due to the huge amount of information available in textual format, which renders effective retrieval, extraction and summarization of information vital. The excessive use of domain specific-language along with the rich variety of expressions and inadequate training corpora make measuring sentence similarity in the biomedical domain a difficult task. Therefore, semantic text similarity measures to be used in biomedical NLP studies call for domain-specific approaches including the use of biomedical domain-specific corpora or biomedical knowledge sources. As an example, consider the following two sentences taken from (Wang et al., 2014) and (Fu et al., 2013), respectively.
S1: This form of necrosis, also termed necroptosis, requires the activity of receptor-interacting protein kinase 1 and its related kinase 3.
S2: Moreover, other reports have also shown that necroptosis could be induced via modulating RIP1 and RIP3.
The example sentences S1 and S2 are on the same topic and are similar to each other. The ‘receptor-interacting protein kinase 1’ in S1 is the same concept as ‘RIP1’ in S2; likewise ‘kinase 3’ and ‘RIP3’ refer to the same biomedical term. Domain-independent semantic text similarity measures developed for generic English can neither recognize these concepts nor give high weight to them while estimating the similarity between the sentences.
These examples illustrate that new approaches that can handle both biomedical and domain independent words are needed for sentence similarity computation in the biomedical domain. Garla and Brandt (2012) compared knowledge-based (ontology-based) and distributional (corpus-based) similarity measures and observed that knowledge-based measures are more effective for semantic similarity computation in the biomedical domain. Most previous work on semantic similarity in the biomedical domain focused on computing ontology-based similarity between terms (Aouicha and Taieb, 2016; Harispe et al., 2014; Mabotuwana et al., 2013; Pedersen et al., 2007; Pesquita et al., 2009; SáNchez and Batet, 2011). Several studies showed that the use of biomedical ontologies to measure semantic similarity provided valuable information for a number of tasks performed in this domain such as similarity computation between gene products (Lord et al., 2003), scoring protein–protein interactions (Jain and Bader, 2010) as well as disambiguation of biomedical terms (McInnes and Pedersen, 2013). To the best of our knowledge, there is neither a manually annotated benchmark data set, nor a comprehensive study on sentence-level semantic similarity computation in the biomedical domain. Although sentence-level semantic similarity computation has recently been used as a component in a text-mining system for evidence-based medicine (Hassanzadeh et al., 2015) and for biomedical question answering (Papagiannopoulou et al., 2016), these studies used general domain semantic similarity computation methods and did not perform any domain-specific adaptation.
In this study, we show that general domain state-of-the-art sentence similarity computation systems fail to effectively model sentence similarity in the biomedical domain. We propose new approaches specifically adapted for the biomedical domain that can be categorized into four areas: string similarity measures, ontology based measures, a distributional vector model and a supervised method combining these different measures. Besides a general domain ontology, namely WordNet (Miller, 1995), we also exploit a biomedical ontology, UMLS (Unified Medical Language System) (Bodenreider, 2004). The distributional vector representations of sentences are learned using a large biomedical corpus of full text articles. In addition, we present a manually annotated benchmark data set for biomedical sentence similarity estimation, which can be used for training and evaluation in future studies in this area.
2 System and methods
2.1 BIOSSES dataset
Since there are no suitable datasets that comprise sentence pairs from the biomedical domain, we created a benchmark dataset for biomedical sentence similarity estimation. The dataset comprises 100 sentence pairs, in which each sentence was selected from the TAC (Text Analysis Conference) Biomedical Summarization Track Training Dataset containing articles from the biomedical domain. TAC dataset consists of 20 articles (reference articles) and citing articles that vary from 12 to 20 for each of the reference articles. We selected the BIOSSES sentence pairs from citing sentences, i.e. sentences that have a citation to a reference article, instead of choosing random sentence pairs, majority of which would be unrelated. Our motivation to use the TAC data set was that both semantically related and irrelevant sentence pairs occur in the annotation files. Some of the citing sentences cite the same reference articles because of similar reasons such as referring to a recent study on protein–protein interactions. Sentences citing the same reference article for a similar reason, in general have some degree of semantic similarity. On the other hand, there are also some citing sentences that cite reference article that are written about different topics or research fields (e.g. one refers to a study on microbiology, the other mentions research on embryology). Such citing sentences are expected to have lower or no semantic similarity. Therefore, it was possible to obtain sentence pairs with different similarity degrees by using this approach over the TAC dataset.
The sentence pairs were evaluated by five different human experts that judged their similarity and gave scores ranging from 0 (no relation) to 4 (equivalent). The score range was described based on the guidelines of SemEval 2012 Task 6 on STS (Agirre et al., 2012). Besides the annotation instructions, example sentences from the biomedical literature were provided to the annotators for each of the similarity degrees. These example sentence pairs that are scored between 0 and 4 are shown in Table 1.
Table 1.
Example annotations
| Sentence 1 | Sentence 2 | Comment | Score |
|---|---|---|---|
| Here we show that both C/EBPα and NFI-A bind the region responsible for miR-223 upregulation upon RA treatment. | Isoleucine could not interact with ligand fragment 44, which contains amino group. | The two sentences are on different topics. | 0 |
| Membrane proteins are proteins that interact with biological membranes. | Previous studies have demonstrated that membrane proteins are implicated in many diseases because they are positioned at the apex of signaling pathways that regulate cellular processes. | The two sentences are not equivalent, but are on the same topic. | 1 |
| This article discusses the current data on using anti-HER2 therapies to treat CNS metastasis as well as the newer anti-HER2 agents. | Breast cancers with HER2 amplification have a higher risk of CNS metastasis and poorer prognosis. | The two sentences are not equivalent, but share some details. | 2 |
| We were able to confirm that the cancer tissues had reduced expression of miR-126 and miR-424, and increased expression of miR-15b, miR-16, miR-146a, miR-155 and miR-223. | A recent study showed that the expression of miR-126 and miR-424 had reduced by the cancer tissues. | The two sentences are roughly equivalent, but some important information differs/missing. | 3 |
| Hydrolysis of β-lactam antibiotics by β-lactamases is the most common mechanism of resistance for this class of antibacterial agents in clinically important Gram-negative bacteria. | In Gram-negative organisms, the most common β-lactam resistance mechanism involves β-lactamase-mediated hydrolysis resulting in subsequent inactivation of the antibiotic. | The two sentences are completely or mostly equivalent, as they mean the same thing. | 4 |
Table 2 shows the Pearson correlation of the scores of each annotator with respect to the average scores of the remaining four annotators. It is observed that there is strong association among the scores of the annotators. The lowest correlations are 0.902, which can be considered as an upper bound for an algorithmic measure evaluated on this dataset.
Table 2.
Correlation scores among annotators
| Correlation r | |
|---|---|
| Annotator A | 0.952 |
| Annotator B | 0.958 |
| Annotator C | 0.917 |
| Annotator D | 0.902 |
| Annotator E | 0.941 |
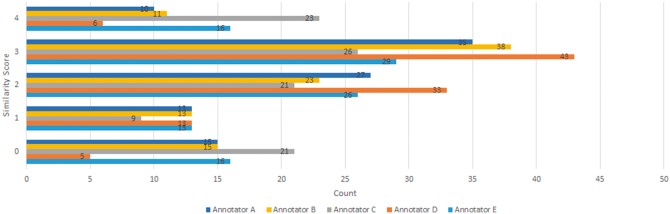
The distribution of the scores by each of the annotators is illustrated in Figure 1. The distribution suggests that there are enough instances for each of the similarity degrees in our dataset.
Fig. 1.
Distribution of the similarity scores in the dataset
The BIOSSES dataset of sentence pairs and the annotators’ scores are publicly available at http://tabilab.cmpe.boun.edu.tr/BIOSSES/DataSet.html.
2.2 String similarity measures
We evaluated the character- and term-based string similarity approaches briefly described in the following subsections using the annotated dataset. Simple pre-processing steps consisting of removal of the punctuation marks (Dot, Comma, Colon, Exclamation Mark, Semicolon, Slash Mark, Dash, Question Mark) and stop-words (http://www.ranks.nl/stopwords) were applied to the sentence pairs before applying the similarity algorithms. The implementations of the string similarity methods in the SimMetrics Library (https://github.com/Simmetrics/simmetrics) were used.
2.2.1. Qgram similarity
Qgram similarity (Ukkonen, 1992) is typically used in approximate string matching by ‘sliding’ a window of length q over the characters of a string to create ‘q’ length grams for matching. A match is then rated as the number of q-gram matches within the second string over the possible q-grams obtained from the first string.
2.2.2. Block distance
Block distance (Krause, 1987), also known as Manhattan Distance, computes the distance between two points by summing the differences of their corresponding components. The Equation for block distance between a point A = (A1,A2,…,An) and a point B = (B1,B2,…,Bn) in n-dimensional space is:
| (1) |
In our case, Ai refers to the count of term i in sentence A and Bi refers to the count of term i in sentence B.
2.2.3. Jaccard similarity
Jaccard similarity (Jaccard, 1908) measures the similarity between two sets and is computed as the number of common terms over the number of unique terms in both sets (Equation 2). In our case, set A consists of the unique words in the first sentence and set B consists of the unique words of the second sentence.
| (2) |
2.2.4. Overlap coefficient
Overlap coefficient (Lawlor, 1980) is a similarity measure that differs from Jaccard similarity with being divided by the size of the smaller sized of the two sets (Equation 3).
| (3) |
2.2.5. Levenshtein distance
Levenshtein distance (Levenshtein, 1966) is a simple edit distance, which consists of the operations for transforming one of the given strings to the other, where an operation is defined as an insertion, deletion, substitution or copying of a character. The distance is defined as the minimum number of the required operations to change one string into another. The Levenshtein distance and block distance values are converted into similarity values by subtracting from 1.
2.3 Distributional vector model
2.3.1. Paragraph vector model
The word2vec model (Mikolov et al., 2013), which constructs distributed representations of words, has been widely adopted to many recent NLP tasks including the biomedical domain (Aydin et al., 2017; Chiu et al., 2016; Moen and Ananiadou, 2013; Muneeb et al., 2015). In this model, a large amount of unlabeled text data is used in training to represent words in a new low-dimensional space as real-valued vectors. The model’s ability of considering the word context allows us to easily relate word vectors in a semantic way (e.g. similar words have similar vectors). Word2vec is an unsupervised neural network based learning model based on two approaches, namely Skip-Gram and Continuous-Bag-of-Words (CBOWs). In the CBOW approach, the words are predicted based on their surrounding words ignoring the word order, whereas in Skip-Gram, a word is used to predict its surrounding words while considering how distant they are in the text.
Paragraph vector is presented following the word2vec model as a way to describe sentences (Le and Mikolov, 2014). The paragraph vector method was utilized to capture semantic information from the texts. The difference of this model from the word2vec model is that the paragraphs are also mapped to distributed vector representations and used to predict the next word in the given context together with the distributed vector representations of the words in the paragraph. We trained a paragraph vector model by using a subset of the Open Access Subset of PubMed Central (http://www.ncbi.nlm.nih.gov/pmc/) dataset, which comprises ∼4G text data of ∼37K articles. The size of the output sentence vectors was set to 100 and the Skip-Gram approach was employed.
2.4 Ontology-based similarity
Ontologies are widely used for measuring semantic similarity between concepts/terms, since their representation links terms semantically. Due to the fact that a sentence consists of a set of words, we can utilize ontology-based word-level similarity measures to compute semantic similarity scores between sentences. To make our proposed algorithms clearer, we first briefly introduce the WordNet (Section 2.4.1) and the UMLS ontologies (Section 2.4.2), then describe the ontology-based word-level similarity algorithms (Section 2.4.3). Finally, we present our proposed approaches (Section 2.4.4), which exploit the word-level algorithms described in Section 2.4.3 to obtain sentence-level similarity scores.
2.4.1. WordNet
WordNet (Miller, 1995) is a large English lexical thesaurus that has been widely used for computing semantic similarity by using the measures described in Section 2.4.3. According to the structure of WordNet, each word consists of a form ‘f’ which is a string and a sense ‘s’ represented by a set of synonyms that have that meaning. Words in WordNet are categorized according to their syntactic categories such as verb, noun, adjective, and adverb. Since the same words can be interpreted as having different part-of-speech (POS) tags according to the contexts they occur in, this syntactic categorization allows to save the same word with each possible POS tags separately in a taxonomy. In addition, words and word senses are connected to each other with various types of relationships. The types of relationships most commonly used for measuring semantic similarity are listed below:
Synonymy is the basic relation type in WordNet, since sets of synonyms (synsets) are used to represent word senses.
Hyponymy and hypernymy represent the hierarchical relations between a word and its sub-name and super-name, respectively.
Antonymy represents the relation between a name and its opposite-name.
2.4.2 UMLS
UMLS (Bodenreider, 2004) is a comprehensive thesaurus consisting of >1.7 million biomedical concepts. It comprises of the vocabulary sources on specialized topics such as MeSH consisting of medical subject headings, OMIM containing genetic knowledge bases, and SnomedCT which consists of the concepts belonging to clinical repositories. Since UMLS consists of various terminology sources, some concepts can overlap. In other words, the same concept can belong to different sources. To be able to use multiple sources as a single resource in the UMLS Metathesaurus, concept unique identifiers are assigned to the concepts.
2.4.3. Word-level similarity methods
The rich semantic information carried by ontologies enables the computation of semantic similarity scores among concepts. In this subsection, we briefly describe the ontology based path-based and information content (IC)-based similarity metrics that are employed in our proposed sentence-level similarity computation method. Path-based approaches utilize the structure of the taxonomy, whereas IC-based approaches use extra information that is learned from corpus statistics.
The Path algorithm (Rada et al., 1989) measures the semantic similarity of two concepts by calculating the shortest path between them in taxonomy. The intuition behind the algorithm is that the shorter the path between concepts in a hierarchy the more similar they are.
| (4) |
In Equation 4, the len function computes the shortest path between concepts c1–c2, and depthmax refers to the maximum depth of the taxonomy. For example, given the sample taxonomy provided in Figure 2, the semantic distance between the terms ‘protein’ and ‘beta-lactams’ is computed as:
| (5) |
The shortest path between c1 and c2 counts all nodes between them—including themselves. Since the maximum depth of the taxonomy is constant, this measure does not take into consideration the specificity of the concepts. According to the definition, len(c1,c2) is equal to 4 and depthmax is 5.
Fig. 2.
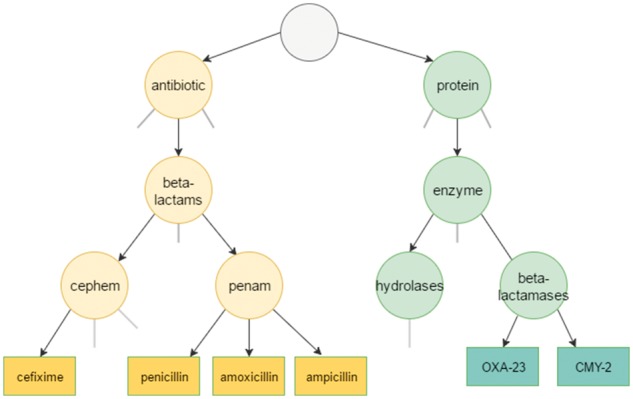
Hierarchical relationships among a small subset of proteins and antibiotics
Similarly, the Leacock and Chodorow (LCH) measure (Leacock and Chodorow, 1998) takes the maximum depth of the taxonomy into account and the similarity is determined as:
| (6) |
Unlike the Path and LCH measures, Wu and Palmer (WP) (Wu and Palmer, 1994) measure accounts for the specificity of the concepts, due to the concept depth feature. WP similarity between concepts c1 and c2 is measured as twice the depth of the lowest common subsumer of the given concepts over the sum of the depths of c1 and c2.
| (7) |
The following example based on the sample taxonomy in Figure 2 illustrates the effect of concept depth using the WP and the Path metrics.
| (8) |
| (9) |
| (10) |
| (11) |
Although the Path algorithm gives the same semantic similarity score for the two pairs, which have different specificity, WP estimates that cephem and ampicillin are more similar than antibiotic and enzyme. The result of the WP metric is reasonable for this example, since the path between deeper concepts causes less semantic distance.
Both the concept depth feature and the frequency of the concept in a corpus give an idea about the specificity of the concept. With the motivation of these facts, IC is used for measuring the semantic similarity between concepts. IC of a concept is defined as the negative log likelihood of encountering concept c in a given corpus.
| (12) |
The probability of encountering concept c is given as,
| (13) |
In Equation (13), N denotes the total number of words in the corpus used, while freq(c) is the number of occurrences of concept c in the corpus.
The Resnik (Resnik, 1995) similarity measure is determined as the IC of the lowest common subsumer of concepts c1 and c2.
| (14) |
The Lin (Lin, 1998) similarity between concepts c1 and c2 is calculated as twice the IC of the lowest common subsumer of the concepts over the sum of ICs of c1 and c2.
| (15) |
Jiang and Conrath (JCN) (Jiang and Conrath, 1997) measures the semantic similarity between concepts c1 and c2 as in Equation (16), which uses the ICs of the concepts and their lowest common subsumer.
| (16) |
2.4.4. Sentence-level ontology-based methods
In this section, we introduce our sentence-level ontology-based methods namely WordNet-based Similarity Method (WBSM), UMLS-based Similarity Method (UBSM) and combined ontology method (COM). The general design of these approaches is shown in Figure 3. There are two main tasks in the general flow; calculation of word-level similarities (Section 2.4.3), adapting word-level similarities to obtain sentence-level score (sentence-level similarity method). Although the proposed three methods use the same algorithms for these tasks, they differ from each other by using different ontologies for word-level similarity calculation.
Fig. 3.
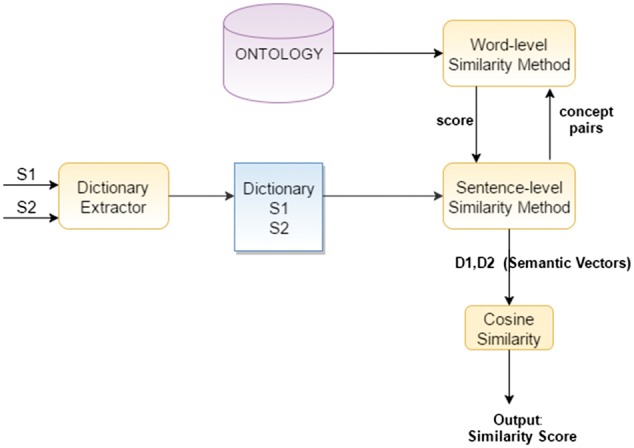
Sentence-level similarity module
Inspired by the study of Li et al. (2006), we developed a sentence-level similarity method, which is an algorithm to adapt word-level similarities to sentence-level. The algorithm is explained below using a walk-through example.
A walk-through example
S1: Necroptosis requires the activity of RIP1 and RIP3.
S2: Necroptosis could be induced via modulating RIP1 and RIP3.
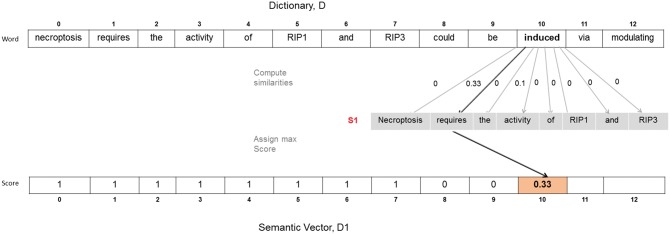
Given two sentences S1 and S2, dictionary D is constructed, which consists of the union of the unique words from the two sentence. D for the example sentences S1 and S2 is:
D: {Necroptosis, requires, the, activity, of, RIP1, and, RIP3, could, be, induced, via, modulating}
D is used to build the semantic vectors D1 and D2 for S1 and S2, respectively, which have the same dimension as the dictionary. For instance, in order to build a semantic vector for S1, each word in the dictionary is compared with every word in S1 and the highest similarity score is assigned for the corresponding dimension index in the semantic vector. As shown in Figure 4, D is obtained by using all distinct words in S1 and S2. For determining the score of the 10th dimension of the semantic vector D1, the ontology-based word-level similarity scores between each word in S1 and the 10th dimension of D are computed. Since the highest score is 0.33 among all similarity scores, the score of the 10th index of D1 is set as 0.33. This process is repeated for the remaining indexes of the semantic vector D1. Then, the same algorithm is applied to create the semantic vector D2. Finally, the cosine similarity between D1 and D2 gives the semantic similarity score between the two sentences S1 and S2.
Fig. 4.
Illustration of the proposed sentence-level ontology-based similarity algorithm which constructs semantic vectors of sentences
WBSM. WBSM takes two sentences to be compared as inputs and returns the semantic similarity score by exploiting WordNet. We used the WS4J library (https://github.com/Sciss/ws4j) for calculating the similarities between words by utilizing the WordNet ontology. The algorithms described in Section 2.4.3 were evaluated for WBSM. These measures were calculated using the Is-A relations in the WordNet ontology. Then, the sentence-level similarity method was used to combine word-level similarity scores to sentence-level.
UBSM. Differently from WBSM, UBSM uses METAMAP (Aronson, 2001), which is a tool for extracting medical concepts from text rather than assuming each word as a concept. This approach is more reliable, since concepts can consist of more than one word. The METAMAP tool is run on both sentences S1 and S2 and a dictionary is constructed from the unique mapped concepts/phrases in the two sentences. Therefore, the word-level similarity method utilizing UMLS takes concepts mapped by METAMAP as inputs. The rest of the methodology for constructing the sentence-level vectors is the same as WBSM.
Umls:Similarity (McInnes et al., 2009) web interface was used to calculate the similarity of the concepts, which were mapped by METAMAP. The scope of Umls:Similarity is limited to the OMIM (Online Mendelian Inheritance in Man) and MeSH (Medical Subject Headings) ontologies, which are subsets of the UMLS ontology. Parent/Child (PAR/CHD) relationship was used as the relationship parameter in the UMLS:Similarity web interface. The algorithms described in Section 2.4.3 were evaluated for UBSM.
COM. The major motivation behind the COM was to benefit from both biomedical domain and general domain ontologies, since sentences in biomedical text consist of both general terms and biomedical-specific terms. To utilize the knowledge from both UMLS and WordNet ontologies, we propose a new approach in this section. Our method performs combination of different approaches on sentence-level. As shown in Figure 5, the sentence-level COM takes the similarity scores of WBSM and UBSM for a sentence pair, then combines these scores by using Equation 17, where λ represents the weight parameter. When λ is set to 0.5, equal weight is given to the similarity scores obtained from the WordNet and UMLS ontologies. When λ is set to a value >0.5, higher weight is given to the similarity score obtained from WordNet, and when it is set to a value smaller than 0.5, higher weight is given to the similarity score obtained from UMLS.
| (17) |
Fig. 5.
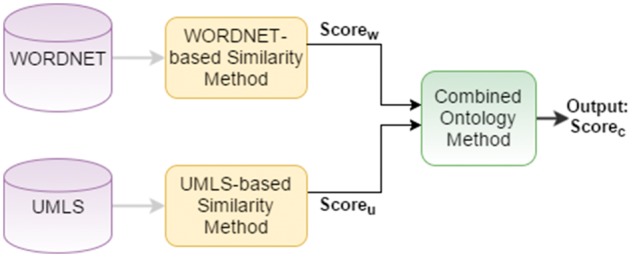
Sentence-level COM
If a word does not occur in either of the ontologies (UMLS and/or WordNet), the similarity score between the word and any other word with respect to the corresponding ontology is considered to be 0.
2.5 Supervised combination of similarity measures
We combined our unsupervised semantic similarity measures within a supervised method. We used the similarity scores computed by the unsupervised COM, Paragraph Vector and Qgram similarity as features in a supervised regression model. Linear Regression implemented in the Weka library (Hall et al., 2009) was used as the supervised model. A linear regression model can be expressed as in Equation (18) (Alpaydin, 2014; Buckley and James, 1979; Raftery et al., 1997),
| (18) |
where y is the dependent variable, each xj is an input variable, and k equals to the number of predictors (input variables). βjs correspond to the parameters of the linear regression model, which are estimated from the training data. Therefore, in our supervised similarity model, the predicted sentence similarity score (y) is calculated through the similarity scores (xj) that were obtained by the unsupervised methods.
The supervised system exploiting the results of the unsupervised similarity computation methods is illustrated in Figure 6. The pre-processed sentences are given to each unsupervised system as inputs. Then, the output score of each system, which is the semantic similarity score for the given pair, is used as a feature in our supervised system.
Fig. 6.
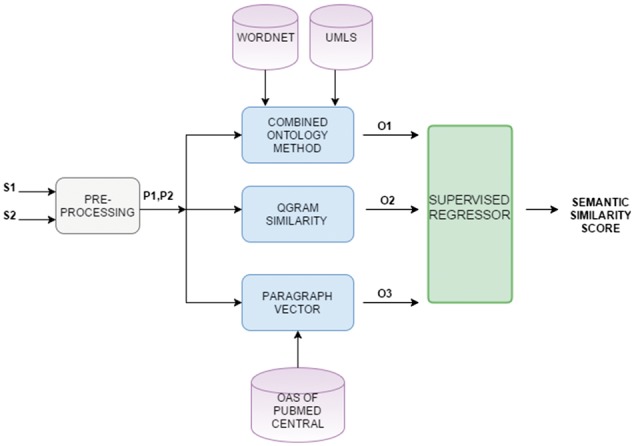
Supervised combination of similarity measures
3 Experimental results
The proposed sentence-level semantic similarity estimation algorithms are evaluated using the manually annotated dataset described in Section 2.1. For each sentence pair in the dataset, the mean of the scores assigned by the five human annotators was taken as the gold standard. The Pearson correlation (Pearson, 1895) between the gold standard scores and the scores estimated by the algorithms was used as the evaluation metric. The strength of correlation can be assessed by the general guideline proposed by Evans (1996) as follows:
very strong: 0.80–1.00
strong: 0.60–0.79
moderate: 0.40–0.59
weak: 0.20–0.39
very weak: 0.00–0.19
Since there is no previous study on sentence semantic similarity computation developed specifically for the biomedical domain, we considered the domain-independent state-of-the-art approaches ADW (Pilehvar et al., 2013; Pilehvar and Navigli, 2015) and SEMILAR (Rus et al., 2013) introduced in Section 1 as our baseline models. According to the results shown in Table 3, both ADW and SEMILAR obtain moderate correlation based on Evans’ definition (Evans, 1996). The poor results of these generic-domain similarity estimation systems demonstrate the need for new approaches for this domain-specific research field.
Table 3.
Experimental results of the presented approaches
| Methods | Pearson correlation |
|---|---|
| Domain-independent systems | |
| ADW | 0.586 |
| SEMILAR | 0.419 |
| String similarity measures | |
| Qgram | 0.754 |
| Jaccard | 0.710 |
| Block | 0.752 |
| Levenshtein | 0.592 |
| Overlap coefficient | 0.695 |
| Word Embeddings based Similarity | |
| Paragraph Vector | 0.787 |
| Ontology-based similarity | |
| WBSM-Path | 0.644 |
| WBSM-Resnik | 0.234 |
| WBSM-Lin | 0.495 |
| WBSM-WP | 0.354 |
| WBSM-JCN | 0.623 |
| WBSM-LCH | 0.287 |
| UBSM-Path | 0.651 |
| UBSM-Resnik | 0.473 |
| UBSM-Lin | 0.645 |
| UBSM-WP | 0.576 |
| UBSM-JCN | 0.624 |
| UBSM-LCH | 0.333 |
| COM ([λ = 0.5]) | 0.710 |
| Supervised semantic similarity system | |
| Linear regression | 0.836 |
We evaluated several string similarity measures on our dataset. We experimented with performing preprocessing as described in Section 2.2 and without performing preprocessing for all string-based methods as well as for the other evaluated methods. Pre-processing improved the performances of all methods. Therefore, in Table 3 we report the results when preprocessing was performed. Our experiments showed that the application of preprocessing methods contributed more to the performance of the string similarity measures compared with the other methods. The range of increase in Pearson correlation varies between 10 and 31% for the string similarity measures. This result is expected, as string-based approaches are highly sensitive to small changes, since they do not take into consideration the semantic information of text.
Paragraph vector is an unsupervised approach, which we used with a large unlabeled corpus of biomedical text to learn semantic information. The strong correlation result obtained by the Paragraph Vector method shows that it is a promising method for representing sentences as vectors while capturing semantics.
For both WBSM and UBSM, using the path algorithm as the word-level similarity approach yielded the best performance with Pearson correlation scores of 0.644 and 0.651, respectively. Therefore, for the combined ontology approach, we used the path algorithm both for computing the WordNet- and the UMLS-based scores. Then, the weighted sum of the similarity scores obtained from the WordNet- and UMLS-based methods was assigned as the final similarity score. The best combination was achieved when the weight parameter lambda was set to 0.5 (λ = 0.5) in Equation (17). The comparison between the COM and the methods that use a single ontology show that the efficient unification of the available biomedical information coming from a biomedical ontology with general domain information increased the overall performance. The results of the combined ontology approach justify our hypothesis, which was based on exploiting both general-domain and domain-specific ontologies for domain-specific text. The significant increase in the correlation performance of the combined model, compared with the individual correlation scores, indicate that the combination is useful.
The evaluation of the supervised model was performed using stratified 10-fold cross-validation over all the sentence pairs, due to the small size of the dataset. The final result for the supervised semantic similarity system was obtained by averaging the individual correlation results of each fold. As the learning model, Linear Regression implemented in the Weka library (Hall et al., 2009) was employed.
The experimental results indicate that the supervised combination of the similarity scores computed by the different methods outperforms the individual performance of each unsupervised method. This shows that these unsupervised system scores complement each other. Although each unsupervised method obtained strong association with the gold standard, combination of these approaches by a supervised algorithm led to very strong correlation. The Supervised Semantic Similarity System exploiting the scores of the unsupervised systems as features produced the best correlation of 83.6% among the others.
4 Discussion
In this study, we presented and compared several approaches to measure semantic sentence similarity in the biomedical domain. We demonstrated the need for adapted or new approaches for domain-specific semantic sentence-level similarity, since our results showed that state-of-the-art domain-independent semantic similarity measures are inadequate when applied to biomedical text. Another important contribution of this research is that we provide a strong baseline as well as a hand-crafted benchmark dataset for further studies due to attempting the first methods in this unexplored research area of biomedical sentence-level semantic similarity computation.
Thanks to the ontologies that enable the computation of semantic distances between concepts, ontology-based measures have been used in our semantic similarity computation study. Since the sentences in our dataset are selected from biomedical articles, we utilized WordNet as the general domain ontology and UMLS as the biomedical domain-specific ontology. The evaluations indicated that the COM, which utilizes both the WordNet and UMLS ontologies, accomplished better results on estimating the similarity among biomedical sentences compared with the methods where a single ontology was utilized. This outcome is reasonable, since sentences in the biomedical domain comprise both biomedical and general concepts. Thus, the knowledge extracted from both WordNet and UMLS complements each other and contributes to the overall performance of the system.
Besides UMLS, there are various biomedical ontologies specialized on different subtopics in the biomedical domain such as the ChEBI ontology focusing on chemical entities (Degtyarenko et al., 2008), the Interaction Network Ontology specializing in the domain of molecular interactions (Özgür et al., 2016), and the Human Phenotype Ontology providing controlled vocabulary for phenotypic features related to human diseases (Köhler et al., 2017). Integrating the semantic similarity scores computed by using different biomedical ontologies might contribute to the performance of the COM. As future work, we aim to make use of the knowledge obtained from different biomedical ontologies, in order to enhance our system to respond to a wider range of concepts and relationships.
Our results revealed that the unsupervised Paragraph Vector approach based on a biomedical corpus to learn the distributional vector representations of sentences is a promising method for biomedical semantic similarity computation.
Finally, we presented a supervised semantic similarity estimation system based on a linear regression model, which exploits high-level features. The high-level features consist of the similarity scores of the best performing unsupervised systems, namely Qgram, Paragraph Vector and the COM. Combining the unsupervised methods with the help of a supervised learning model increased the overall performance of the system. Experiments showed that using different approaches to estimate the similarity contributes to the overall performance of the system.
The manually annotated dataset and the developed semantic similarity estimation systems are publicly available. We believe that our biomedical-domain specific semantic sentence-level similarity measures can be used in various applications of biomedical NLP such as automatic summarization, question answering, text categorization and text retrieval.
The upper bound in this study can be considered as the performance of a typical human, which is 90.2% according to the correlations between the human annotators. Although our best performing system achieved high correlation with human annotations (83.6%), there is still room for improvement for biomedical domain-specific semantic sentence similarity estimation.
Acknowledgements
We would like to thank Ecem Soğancıoğlu, Jesus Lago Garcia, Kübra Eren and Onur Yanar for annotations. We also thank Bridget T. McInnes for her kind help about the UMLS interface. We respectfully acknowledge the TUBITAK (BIDEB 2210-A and 2211-E) scholarship programmes (to G.S. and H.O.) and the BAGEP Award of the Science Academy (to A.O.).
Conflict of Interest: none declared.
References
- Agirre E. et al. (2014). Semeval-2014 task 10: Multilingual semantic textual similarity. In Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014), Dublin, Ireland, pp. 81–91. [Google Scholar]
- Agirre E. et al. (2016). Semeval-2016 task 1: semantic textual similarity, monolingual and cross-lingual evaluation. Proceedings of SemEval, San Diego, CA, pp. 497–511. [Google Scholar]
- Agirre E. et al. (2013). sem 2013 shared task: Semantic textual similarity, including a pilot on typed-similarity. In *SEM 2013: The Second Joint Conference on Lexical and Computational Semantics. Association for Computational Linguistics, Atlanta, Georgia.
- Agirre E. et al. (2012). Semeval-2012 task 6: A pilot on semantic textual similarity. In Proceedings of the First Joint Conference on Lexical and Computational Semantics-Volume 1: Proceedings of the Main Conference and the Shared Task, and Volume 2: Proceedings of the Sixth International Workshop on Semantic Evaluation, pp. 385–393. Association for Computational Linguistics, Montreal, Canada.
- Agirrea E. et al. (2015). Semeval-2015 task 2: Semantic textual similarity, english, spanish and pilot on interpretability. In Proceedings of the 9th international Workshop on Semantic Evaluation (SemEval 2015), Denver, Colorado, pp. 252–263. [Google Scholar]
- Alpaydin E. (2014). Introduction to Machine Learning. MIT press, Cambridge. [Google Scholar]
- Aouicha M.B., Taieb M.A.H. (2016) Computing semantic similarity between biomedical concepts using new information content approach. J. Biomed. Informatics, 59, 258–275. [DOI] [PubMed] [Google Scholar]
- Aronson A.R. (2001). Effective mapping of biomedical text to the umls metathesaurus: the metamap program. In Proceedings of the AMIA Symposium, p. 17. American Medical Informatics Association, Washington, DC. [PMC free article] [PubMed]
- Aydin F. et al. (2017) Automatic query generation using word embeddings for retrieving passages describing experimental methods. Database, doi: 10.1093/database/baw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O. (2004) The unified medical language system (umls): integrating biomedical terminology. Nucleic Acids Res., 32(Suppl. 1), D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley J., James I. (1979) Linear regression with censored data. Biometrika, 429–436. [Google Scholar]
- Chiu B. et al. (2016). How to train good word embeddings for biomedical nlp. ACL 2016, Vol. 66, pp. 166. [Google Scholar]
- Degtyarenko K. et al. (2008) Chebi: a database and ontology for chemical entities of biological interest. Nucleic Acids Res., 36(Suppl. 1), D344–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.D. (1996). Straightforward Statistics for the Behavioral Sciences. Brooks/Cole, Pacific Grove, CA, USA. [Google Scholar]
- Finch A. et al. (2005). Using machine translation evaluation techniques to determine sentence-level semantic equivalence. In Proceedings of the Third International Workshop on Paraphrasing (IWP2005), pp 17–24.
- Fu Z. et al. (2013) The anti-tumor effect of shikonin on osteosarcoma by inducing rip1 and rip3 dependent necroptosis. BMC Cancer, 13, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garla V.N., Brandt C. (2012) Semantic similarity in the biomedical domain: an evaluation across knowledge sources. BMC Bioinformatics, 13, 261.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. et al. (2009) The weka data mining software: an update. ACM SIGKDD Expl. Newslett., 11, 10–18. [Google Scholar]
- Han L. et al. (2013). Umbc ebiquity-core: Semantic textual similarity systems. In Proceedings of the Second Joint Conference on Lexical and Computational Semantics, Vol. 1, pp. 44–52. Association for Computational Linguistics, Atlanta, Georgia.
- Harispe S. et al. (2014) A framework for unifying ontology-based semantic similarity measures: A study in the biomedical domain. J. Biomed. Inform., 48, 38–53. [DOI] [PubMed] [Google Scholar]
- Hassanzadeh H. et al. (2015) A supervised approach to quantifying sentence similarity: with application to evidence based medicine. PloS One, 10, e0129392.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haveliwala T.H. (2002). Topic-sensitive pagerank. In Proceedings of the 11th International Conference on World Wide Web, pp. 517–526. ACM, Honolulu, Hawaii, USA.
- Jaccard P. (1908) Nouvelles recherches sur la distribution florale. Bull. Soc. Vaudoise Sci. Nat., 44, 223–270. [Google Scholar]
- Jain S., Bader G.D. (2010) An improved method for scoring protein-protein interactions using semantic similarity within the gene ontology. BMC Bioinformatics, 11, 562.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J. et al. (2005). Finding similar questions in large question and answer archives. In Proceedings of the 14th ACM International Conference on Information and Knowledge Management, Bremen, Germany, pp. 84–90. ACM.
- Jiang J.J., Conrath D.W. (1997). Semantic similarity based on corpus statistics and lexical taxonomy, In Proceedings of the 10th International Conference on Research on Computational Linguistics, page 19–33.
- Köhler S. et al. (2017) The human phenotype ontology in 2017. Nucleic Acids Res., 45, D865.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause E.F. (1987). Taxicab geometry. Dover Publications, New York.
- Lawlor L.R. (1980) Overlap, similarity, and competition coefficients. Ecology, 61, 245–251. [Google Scholar]
- Le Q.V., Mikolov T. (2014). In Proceedings of the 31st International Conference on Machine Learning, PMLR, 32, 1188–1196.
- Leacock C., Chodorow M. (1998) Combining local context and wordnet similarity for word sense identification. In WordNet, MIT Press, Cambridge, pp. 265–283. [Google Scholar]
- Levenshtein V.I. (1966). Binary codes capable of correcting deletions, insertions, and reversals. In Soviet Physics Doklady, Vol. 10, pp. 707–710. [Google Scholar]
- Li Y. et al. (2006) Sentence similarity based on semantic nets and corpus statistics. IEEE Trans.Knowl. Data Eng. 18, 1138–1150. [Google Scholar]
- Lin D. (1998). An information-theoretic definition of similarity. In ICML, Vol. 98, pp. 296–304. Morgan Kaufmann Publishers Inc, Madison, Wisconsin, USA. [Google Scholar]
- Liu Y. et al. (2015). Computing semantic text similarity using rich features. 29th Pacific Asia Conference on Language, Information and Computation, Shanghai, China, Vol. 1, pp. 44–52. [Google Scholar]
- Lord P.W. et al. (2003) Investigating semantic similarity measures across the gene ontology: the relationship between sequence and annotation. Bioinformatics, 19, 1275–1283. [DOI] [PubMed] [Google Scholar]
- Mabotuwana T. et al. (2013) An ontology-based similarity measure for biomedical data–application to radiology reports. J. Biomed. Inform., 46, 857–868. [DOI] [PubMed] [Google Scholar]
- McInnes B.T., Pedersen T. (2013) Evaluating measures of semantic similarity and relatedness to disambiguate terms in biomedical text. J. Biomed. Inform., 46, 1116–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes B.T. et al. (2009). Umls-interface and umls-similarity: open source software for measuring paths and semantic similarity. In AMIA Annual Symposium Proceedings, Vol. 2009, p. 431. American Medical Informatics Association, San Francisco, CA. [PMC free article] [PubMed]
- Mihalcea R. et al. (2006). Corpus-based and knowledge-based measures of text semantic similarity. In AAAI, Vol. 6, pp. 775–780. [Google Scholar]
- Mikolov T. et al. (2013). Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems, Vol. 26, pp. 3111–3119. [Google Scholar]
- Miller G.A. (1995) Wordnet: a lexical database for english. Commun. ACM, 38, 39–41. [Google Scholar]
- Moen S., Ananiadou T.S.S. (2013). Distributional semantics resources for biomedical text processing. In Proceedings of the 5th International Symposium on Languages in Biology and Medicine, Tokyo, Japan, pp. 39–43.
- Muneeb T. et al. (2015). Evaluating distributed word representations for capturing semantics of biomedical concepts. In Proceedings of the 2015 Workshop on Biomedical Natural Language Processing (BioNLP 2015), Association for Computational Linguistics, Beijing, China, p. 158.
- Özgür A. et al. (2016) The interaction network ontology-supported modeling and mining of complex interactions represented with multiple keywords in biomedical literature. BioData Mining, 9, 41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannopoulou E. et al. (2016). Large-scale semantic indexing and question answering in biomedicine. In Proceedings of the 4th BioASQ Workshop, ACL 2016, Berlin, Germany, pp. 50–54.
- Pearson K. (1895) Note on regression and inheritance in the case of two parents. Proc. R Soc. Lond., 58, 240–242. [Google Scholar]
- Pedersen T. et al. (2007) Measures of semantic similarity and relatedness in the biomedical domain. J. Biomed. Inform., 40, 288–299. [DOI] [PubMed] [Google Scholar]
- Pesquita C. et al. (2009) Semantic similarity in biomedical ontologies. PLoS Comput. Biol., 5, e1000443.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilehvar M.T. et al. (2013). Align, disambiguate and walk: A unified approach for measuring semantic similarity. In ACL (1), Sofia, Bulgaria, pp. 1341–1351. [Google Scholar]
- Pilehvar M.T., Navigli R. (2015). An open-source framework for multi-level semantic similarity measurement. In Proceedings of NAACL-HLT, Denver, Colorado, Association for Computational Linguistics, pp. 76–80. [Google Scholar]
- Rada R. et al. (1989) Development and application of a metric on semantic nets. IEEE Trans.Syst. Man Cybernet., 19, 17–30. [Google Scholar]
- Raftery A.E. et al. (1997) Bayesian model averaging for linear regression models. J. Am. Stat. Assoc., 92, 179–191. [Google Scholar]
- Resnik P. (1995). Using information content to evaluate semantic similarity in a taxonomy. In Proceedings of the 14th International Joint Conference on Artificial Intelligence, Vol. 1, IJCAI'95, pp. 448–453.
- Rus V. et al. (2013). Semilar: The semantic similarity toolkit. In ACL (Conference System Demonstrations), pp. 163–168. Citeseer, Sofia, Bulgaria.
- SáNchez D., Batet M. (2011) Semantic similarity estimation in the biomedical domain: An ontology-based information-theoretic perspective. J. Biomed. Inform., 44, 749–759. [DOI] [PubMed] [Google Scholar]
- Šarić F. et al. (2012). Takelab: Systems for measuring semantic text similarity. In Proceedings of the First Joint Conference on Lexical and Computational Semantics-Volume 1: Proceedings of the Main Conference and the Shared Task, and Volume 2: Proceedings of the Sixth International Workshop on Semantic Evaluation, pp. 441–448. Association for Computational Linguistics, Stroudsburg, PA, USA.
- Sultan M.A. et al. (2015). Dls@ cu: sentence similarity from word alignment and semantic vector composition. In SemEval-2015, p. 148.
- Ukkonen E. (1992) Approximate string-matching with q-grams and maximal matches. Theor. Comput. Sci., 92, 191–211. [Google Scholar]
- Wang D. et al. (2008). Multi-document summarization via sentence-level semantic analysis and symmetric matrix factorization. In Proceedings of the 31st annual International ACM SIGIR Conference on Research and Development in Information Retrieval, pp. 307–314. ACM, New York, NY, USA.
- Wang H. et al. (2014) Mixed lineage kinase domain-like protein mlkl causes necrotic membrane disruption upon phosphorylation by rip3. Mol. Cell, 54, 133–146. [DOI] [PubMed] [Google Scholar]
- Wu Z., Palmer M. (1994). Verbs semantics and lexical selection. In Proceedings of the 32nd annual meeting on Association for Computational Linguistics, pp. 133–138. Association for Computational Linguistics, Stroudsburg, PA, USA.