Abstract
Motivation
Along with the increasing accessibility to comprehensive sequence information, such as whole genomes and transcriptomes, the demand for assessing their quality has been multiplied. To this end, metrics based on sequence lengths, such as N50, have become a standard, but they only evaluate one aspect of assembly quality. Conversely, analyzing the coverage of pre-selected reference protein-coding genes provides essential content-based quality assessment, but the currently available pipelines for this purpose, CEGMA and BUSCO, do not have a user-friendly interface to serve as a uniform environment for assembly completeness assessment.
Results
Here, we introduce a brand-new web server, gVolante, which provides an online tool for (i) on-demand completeness assessment of sequence sets by means of the previously developed pipelines CEGMA and BUSCO and (ii) browsing pre-computed completeness scores for publicly available data in its database section. Completeness assessments performed on gVolante report scores based on not just the coverage of reference genes but also on sequence lengths (e.g. N50 scaffold length), allowing quality control in multiple aspects. Using gVolante, one can compare the quality of original assemblies between their multiple versions (obtained through program choice and parameter tweaking, for example) and evaluate them in comparison to the scores of public resources found in the database section.
Availability and implementation
gVoalte is freely available at https://gvolante.riken.jp/.
1 Introduction
As the accessibility to comprehensive sequence information increases, the demand for assessing their continuity and completeness has been multiplied. Comprehensive sequence information currently emerging is mostly prepared by de novo sequence assembly. Products of genome and transcriptome assemblies are often not thoroughly assessed because of the time-consuming nature of assembly program executions, and it is also not straightforward to assess them on a uniform criterion. Metrics based on sequence lengths, such as N50, have become a standard for assessing de novo assemblies, but they can overestimate the completeness upon overassembly and obviously cannot evaluate compositional aspects.
The program pipeline CEGMA (Parra et al., 2009) was recommended for completeness assessment of genome assemblies based on the coverage of pre-selected reference protein-coding genes by the project Assemblathon2 (Bradnam et al., 2013). More recently, the program pipeline BUSCO was introduced for the same purpose (Simao et al., 2015). These pipelines are becoming more frequently used, but they are executable only from Unix command-line, and do not have a user-friendly interface.
2 Implementation
CEGMA ver. 2.5 and BUSCO ver. 1.22 and ver. 2.0.1 (equivalent to v3.0.1 as the latest modification was only a refactoring), as well as all the programs required by these pipelines, were built on a Linux server, which provides a web interface using Secure Socket Layer (SSL) encryption. The user information and the file uploaded by the user are used only for completeness assessment and are erased after a minimal time to ensure anonymity. The web server also hosts pages for user instruction including a step-by-step manual. Base compositions and length-based metrics are reported, based on the script assemblathon_stats.pl available at https://github.com/ucdavis-bioinformatics/assemblathon2-analysis/.
3 Functions
3.1 Executing an assessment
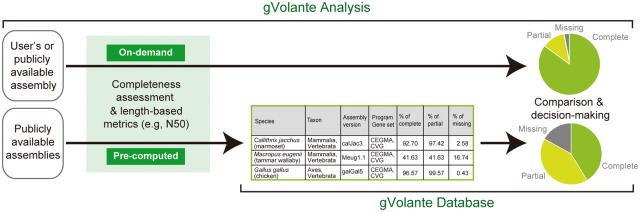
gVolante is a web server designed to make the best use of the reference gene set CVG previously introduced for more accurate completeness assessment for vertebrates (Hara et al., 2015). It achieves a method of handy analysis execution without command line operation and allows the standardized scoring of completeness on a uniform computational environment. In addition, an analysis on gVolante gives a concise report of length-based metrics and base compositions (Fig. 1).
Fig. 1.
Functions of gVolante. The web server provides two functions, ‘Analysis’ in the upper row and ‘Database’ in the lower row. Using gVolante, one can compare the quality of original assemblies and evaluate them in comparison to the scores of public resources found in the database section, for content-based decision-making for more comprehensive downstream analyses
In the ‘Analysis’ page, the user is guided to first upload a sequence file and enter an arbitrary project name and one’s email address. The file to be uploaded is expected to, but does not have to, be a de novo assembly product of no more than 10 GB. Compressed files are also accepted. Additional inputs include a cut-off length for computing N50 lengths. Because typical de novo assembly products contain a number of short sequences, e.g. shorter than 500 bp, the length cut-off can have a large impact on N50 values and should thus be deliberately set.
3.2 Gene search pipeline: CEGMA or BUSCO
Only BUSCO is designed to assess transcriptome assemblies and peptide sequences, which is usually completed within an hour. For assessing genome assemblies though, one needs to carefully choose which search pipeline is used. An assessment of a genome assembly with CEGMA and BUSCO can take longer than a day.
When CEGMA is selected, the user needs to choose a taxonomic property of the species of interest (mammal, vertebrate or other). This specifies the maximum lengths of intronic and flanking sequences of the candidate genic regions in the CEGMA pipeline (Parra et al., 2007).
3.3 Choice of a reference gene set
Completeness assessment should be performed with a careful consideration for the compatibility of a reference gene set with the taxonomic position of the species of interest. gVolante provides a choice between the reference gene set ‘CEG’ associated with CEGMA, our original gene set for vertebrates ‘CVG’ (Hara et al., 2015), and some of the gene sets provided with BUSCO—the latter applies only when BUSCO is chosen as a gene search pipeline. CVG is designed to prevent (i) overestimation of assembly completeness, which can be caused by an expanded gene repertoire owing to whole genome duplication in the vertebrate lineage and (ii) underestimation caused by confusion of lineage-specific gene loss with missing from an assembly (Hara et al., 2015).
3.4 Browsing pre-computed completeness scores
We uniformly employed CEGMA and the reference gene set CVG which showed more accurate assessment than other configurations (Hara et al., 2015) and executed completeness assessments on selected public sequence resources. This combination was necessitated by the vast range of targets covering the whole taxon Vertebrata. They consisted of 73 genomes and 17 transcriptome nucleotide sequences of diverse vertebrates including cyclostomes and cartilaginous fishes.
The obtained completeness scores are tabulated in the ‘Database’ page on the web server. One can click on the row of an individual analysis result to view it in detail (Fig. 1), and further explore which CVG components were identified or missing in the assessment. When one is urged to carefully verify the presence or absence of a particular CVG component, he or she is guided to perform molecular phylogeny inference using the aLeaves-MAFFT online suite (Kuraku et al., 2013).
Acknowledgements
The authors thank other members of Phyloinformatics Unit, RIKEN CLST for daily discussion about demands in genomics and Federico G. Hoffmann for his suggestion about the name of the web server.
Funding
This work has been supported by the RIKEN Center for Life Science Technologies. Funding for open access charge: RIKEN Center for Life Science Technologies.
Conflict of Interest: none declared.
References
- Bradnam K.R. et al. (2013) Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience, 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y. et al. (2015) Optimizing and benchmarking de novo transcriptome sequencing: from library preparation to assembly evaluation. BMC Genomics, 16, 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S. et al. (2013) aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res., 41, W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G. et al. (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics, 23, 1061–1067. [DOI] [PubMed] [Google Scholar]
- Parra G. et al. (2009) Assessing the gene space in draft genomes. Nucleic Acids Res., 37, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao F.A. et al. (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31, 3210–3212. [DOI] [PubMed] [Google Scholar]