Abstract
Summary
Arrayed high-throughput screens (HTS) cover a broad range of applications using RNAi or small molecules as perturbations and specialized software packages for statistical analysis have become available. However, exploratory data analysis and integration of screening results has remained challenging due to the size of the data sets and the lack of user-friendly tools for interpretation and visualization of screening results. Here we present HTSvis, a web application to interactively visualize raw data, perform quality control and assess screening results from single to multi-channel measurements such as image-based screens. Per well aggregated raw and analyzed data of various assay types and scales can be loaded in a generic tabular format.
Availability and implementation
HTSvis is distributed as an open-source R package, downloadable from https://github.com/boutroslab/HTSvis and can also be accessed at http://htsvis.dkfz.de.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Arrayed high-throughput screens (HTS) in high-density multiwell plates are a powerful method for small molecule screening and target discovery (Macarron et al., 2011; Sundberg, 2000). Automated technologies allow to screen tens of thousands of genetic or chemical perturbations, resulting in very large datasets. HTS experiments can range in complexity from univariate cell viability measurements (Whitehurst et al., 2007), to multichannel fluorescence-activated cell sorting (FACS) (Björklund et al., 2006) or multiparametric image based screens (Bray et al., 2016; Fischer et al., 2015). A range of statistical analysis methods have been developed for processing, normalization and quality control of HTS data to robustly identify and annotate significant perturbations (Birmingham et al., 2009). Open-source software for integrated statistical analysis using statistical languages, such as cellHTS has been developed previously (Boutros et al., 2006; Dutta et al., 2016; List et al., 2016). Although commercial desktop software, e.g. TIBCO Spotfire, exist for visualization and exploratory data analysis, few open-source options, in particular for multiparametric screens, are available (Antal et al., 2015; Dao et al., 2016). Thus, there is a need for lightweight software packages that are easy to install and use to aid the interpretation and evaluation of HTS data without requiring extensive programming skills.
2 The HTSvis application
We developed HTSvis, an application for the visualization of data from arrayed HTSs. After installation as an R package, data input and all user interactions are controlled via a user interface requiring no programming skills. Input data can be in commonly used formats to store raw- and analyzed data, such as delimited files (.txt, .csv, .xlsx) or RData stores. In addition, we provide a web service to access HTSvis (http://htsvis.dkfz.de). HTSvis accepts data in a generic tabular format, providing flexibility towards the assay type (e.g. multiparametric data) and scale (Fig. 1A). In particular, data that have been statistically analyzed with the R/Bioconductor package cellHTS (Boutros et al., 2006) can be imported directly into HTSvis for exploratory data analysis.
Fig. 1.
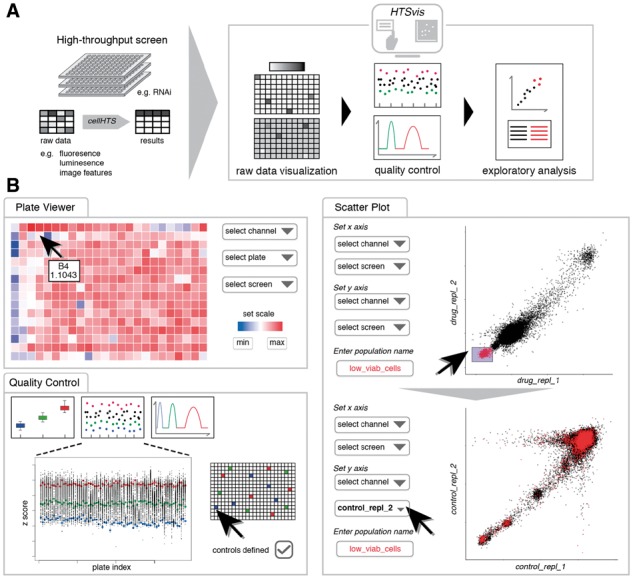
Workflow diagram and functionalities for visualization and exploratory data analysis of arrayed HTSs. (A) Raw data or statistically analyzed data of various assay formats, ranging from single-channel readouts to image features in 6- to 384-well plates, in tabular formats can be loaded into the application to facilitate interactive data visualizations. (B) The user can switch between four pages for performing, e.g. the identification of experimental artifacts (see plate viewer), perform quality control checks and the identification of hits by brushing and comparing measurements
2.1 Local installation and data structure
HTSvis can be installed on local computers from GitHub (https://github.com/boutroslab/HTSvis). After loading the package in R, a single command launches the app in any default web browser. Further instructions, also how to deploy HTSvis in a local shiny server, are documented on the GitHub repository. Input data can be in common tabular formats (.txt, .csv, .xlsx) and requires a certain structure and annotation, such as well and plate annotations and measured variables in distinct, named columns. Specifics about input formats are detailed in Supplementary Material. When data were analyzed with cellHTS, the summary table (‘topTable.txt’) provides all required information and can be uploaded directly. The number of parameters per well is not limited. This allows to load multiparametric datasets from various assay types. More detailed help can be found within the application.
2.2 Interactive data exploration
2.2.1 Spatial plate analysis: plate viewer tab
Plate plots show the data in the format it was measured (e.g. 384-well plates, Fig. 1B). By interactively comparing different plates and measurements, spatial distribution of values can be assessed. This allows to interactively browse the dataset and facilitates the identification of experimental artifacts, such as edge effects (Fig. 1B). The color scale for each plate plot can be adapted for comparisons between plates, e.g. biological replicates. A tooltip on each well provides quick information of the numeric value and annotation (e.g. perturbation reagent) per well.
2.2.2 Assessing screening quality: quality control tab
Screen quality and integrity is commonly assessed based on control perturbations, for which a known phenotypic effect is expected (Birmingham et al., 2009). Up to three control populations (positive, negative and non-targeting) can be defined by selecting wells on a plate map (Fig. 1B). A scatter plot of values vs. plates, a box and a density plot (Kernel-density estimation) of controls are shown. The box and density plot summarize how well controls are separated and allow to estimate effect size and performance of the assay (Z’-factor). The scatter plot adds information about measured values of individual plates over the entire experiment.
2.2.3 Data interpretation: scatter plot tab
The scatter plot tab is a visual tool for quality control and exploratory data analysis. To evaluate the correlation between replicates and to judge the experiment reproducibility, two experiments are plotted against each other. Experiment and measured variable are chosen ad hoc. Users can also brush data points by box selection. Brushed data points can be assigned to a subpopulation with a user-defined name and color (Fig. 1B). Multiple populations can be created and compared. Hypotheses, e.g. how measurements of interest behave in different experimental conditions can be tested accordingly. Brushing of data points is linked to the well and plate position, hence is persistent when measured variable or experiment is changed. This way differential effects between conditions (e.g. between control and drug treatment) can be identified.
2.3 Conclusions
HTSvis is a locally deployable web application to explore and visualize data of arrayed screens with various readouts and scales. Interactive plots and tables provide an advantage compared to the handling of individual files and programming scripts, e.g. one for each plate or plot. Ease-of-use from installation to data input and visualization via the user interface is the main characteristic of HTSvis. Reactive data representations that can be readily accessed provide a versatile tool for exploratory data analysis filling a yet unmet need in the HTS community.
Supplementary Material
Acknowledgements
We thank Oliver Pelz for IT support and Luisa Henkel, Benedikt Rauscher and Jan Winter for helpful suggestions and comments on the article and the Boutros lab for discussions.
Funding
This work was supported in part by an ERC Advanced Grant.
Conflict of Interest: none declared.
References
- Antal B. et al. (2015) Mineotaur: a tool for high-content microscopy screen sharing and visual analytics. Genome Biol., 16, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A. et al. (2009) Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods, 6, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund M. et al. (2006) Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature, 439, 1009–1013. [DOI] [PubMed] [Google Scholar]
- Boutros M. et al. (2006) Analysis of cell-based RNAi screens. Genome Biol., 7, R66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M.-A. et al. (2016) Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc., 11, 1757–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao D. et al. (2016) CellProfiler Analyst: interactive data exploration, analysis and classification of large biological image sets. Bioinformatics, 32, 3210–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta B. et al. (2016) An interactive web-based application for Comprehensive Analysis of RNAi-screen Data. Nat. Commun., 7, 10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B. et al. (2015) A map of directional genetic interactions in a metazoan cell. Elife, 4, e05464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List M. et al. (2016) Comprehensive analysis of high-throughput screens with HiTSeekR. Nucleic Acids Res., 44, 6639–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macarron R. et al. (2011) Impact of high-throughput screening. Nature, 10, 188–195. [DOI] [PubMed] [Google Scholar]
- Sundberg S.A. (2000) High-throughput and ultra-high-throughput screening: Solution- and cell-based approaches. Curr. Opin. Biotechnol., 11, 47–53. [DOI] [PubMed] [Google Scholar]
- Whitehurst A.W. et al. (2007) Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature, 446, 815–819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


