Abstract
Motivation
Microsatellites are a widely-used marker system in plant genetics and forensics. The development of reliable microsatellite markers from resequencing data is challenging.
Results
We extended MISA, a computational tool assisting the development of microsatellite markers, and reimplemented it as a web-based application. We improved compound microsatellite detection and added the possibility to display and export MISA results in GFF3 format for downstream analysis.
Availability and Implementation
MISA-web can be accessed under http://misaweb.ipk-gatersleben.de/. The website provides tutorials, usage note as well as download links to the source code.
1 Introduction
Microsatellites arose about 25 years ago (Tautz and Schlotterer, 1994) and still remain a commonly used genetic marker system in plant genetics and breeding (Miah et al., 2013; Matthies et al., 2012)and forensics (Butler, 2005), where they are commonly referred to as simple sequence repeats (SSRs) or short tandem repeats (STR), respectively. The basic building block of a microsatellite is a short sequence motif (usually between one and six base-pairs in length) that is repeated in tandem. These characteristic features can be detected by the in silico analysis of nucleotide sequences obtained by traditional Sanger or high-throughput resequencing data.
The MISA microsatellite finder (Thiel et al., 2003) is a tool for finding microsatellites in nucleotide sequences. In addition to the detection of perfect microsatellites, MISA is also able to find perfect compound microsatellites that are composed multiple occurrences of more than one simple sequence motif. MISA has been widely used over the past ten years, during which two major limitations of MISA have become evident:
The current MISA implementation requires computational expertise and access to a UNIX environment to (i) run the PERL script and (ii) process the results for most downstream applications.
The MISA output contains an overview of identified microsatellites in a proprietary format, which cannot be easily parsed for downstream analysis.
The Generic Feature Format Version 3 (GFF3, https://github.com/The-Sequence-Ontology/Specifications/blob/master/gff3.md) is a commonly used format in genomic data analysis. GFF3 is a tabular format that lists features in nucleotide sequences and provides ontology-based feature classification.
Here, we present the MISA-web, an extension to the command line tool MISA embedded into an easy-to-use web-based graphical user interface available from http://misaweb.ipk-gatersleben.de/.
2 Materials and methods
2.1 Workflow and implementation
A microsatellite analysis with the command line version of MISA requires two input files: (i) a configuration file (‘MISA.ini’) with three input parameters: ‘SSR search parameters’, ‘compound SSR search parameter’ and ‘output file type parameter’; and (ii) a FASTA file containing the nucleotide sequence that is to be mined for microsatellites.
MISA-web runs on a standard Linux server and works in conjunction with several helper scripts and programs in addition to the core MISA PERL script. The outline of the implemented workflow is as follows:
Periodically running scripts in PHP and UNIX shell monitor server load and schedule the execution of MISA analysis requests by users of the web site. Entries from the input fields of the web form are compiled into the two input files. The nucleotide sequences are combined into a single file in FASTA format (<project>.fasta). The other entry fields are written to the MISA.ini file. If no parameters are specified by the user, preset default parameters as shown on the web site will be used.
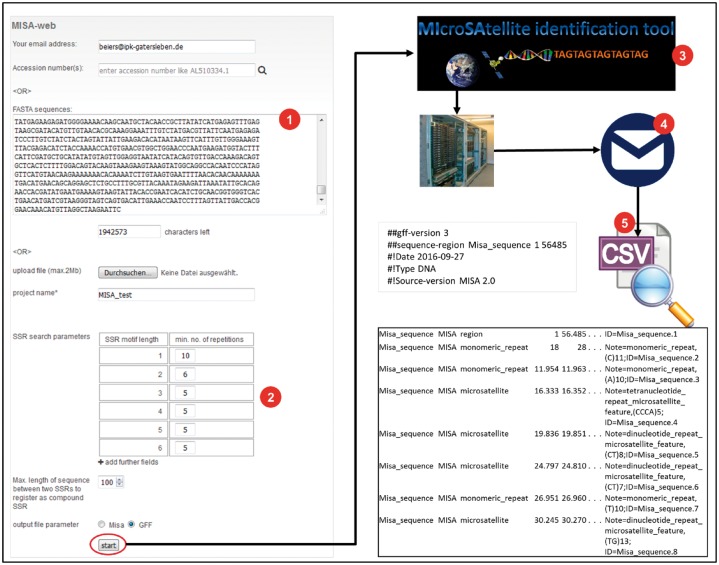
After the conversion of input variables, the core PERL function MISA.pl is called. Upon its successful termination, the result files are compressed with UNIX gzip, and the archive is sent to a user-specified email address. A typical workflow is presented in Figure 1.
Fig. 1.
MISA-web analysis workflow. MISA-web was updated and set up as a web-application on the IPK server. Users may either paste their nucleotide sequence of interest in the input fields of MISA-web or supply accession numbers to have the corresponding sequences fetched from NCBI (1). Once all input fields have been filled (2), a click on the start button on the bottom of the page starts the analysis. The computation will be conducted on a compute server (3) and the result files will be sent to a user-specified email address (4). Result files can be examined afterwards (5)
MISA-web can retrieve sequences from the NCBI database by specifying the corresponding accession numbers in the input field. MISA-web then communicates with the NCBI servers using PHP (www.php.net) and JQuery (www.jquery.com), downloads the sequences and reports them as FASTA sequence in the textbox. A comma-separated list of accession numbers can be entered to retrieve multiple sequences at once (up to a maximum sequence length of 2 Mb).
2.2 Output formats
MISA-web supports two different output formats: the proprietary MISA output format and generic GFF3.
3 Validation
To compare the performance of MISA-web we analyzed ten sequence assemblies of barley bacterial artificial chromosomes (BACs) published by (Munoz-Amatriain et al., 2015). The assemblies (accession numbers: AC256511.1, AC269605.1, AC265197.1, AC263353.1, AC264961.1, AC266636.1, AC261250.1, AC267178.1, AC259365.1, AC257258.1) were retrieved from the NCBI database. A total of 6,022 microsatellites were identified with the following parameters set: motif length 1 to 6; repetition minimum of 5; 0 base pairs between two microsatellites for compound SSR detection. Almost all of these microsatellites (98%) are simple mononucleotide microsatellites, while 0.16% and 0.03% were di- and trinucleotide microsatellites, respectively. Only two tetranucleotide microsatellites were found.
We evaluated seven other microsatellite detection tools on the same BAC dataset: GMATo (Wang et al., 2013), IMEx (Mudunuri and Nagarajaram, 2007), mreps (Kolpakov et al., 2003), ProGeRF (Lopes et al., 2015), SciRoKo (Kofler et al., 2007), TRF (Benson, 1999) and TROLL (Castelo et al., 2002). The tools IMEx, TRF and ProGeRF are accessible as web application. We disabled compound microsatellite detection used a motif length between 1 and 6 with minimum number of repetition of 5 for all motif lengths. If possible we turned off imperfect microsatellite detection (Table 1).
Table 1.
Comparison of detected microsatellites and execution time (in seconds) of GMATo, TRF, TROLL, mreps, SciRoKo, ProGeRF and MISA-web
| Sequence | GMATo | TRF | TROLL | Mreps | SciRoKo | ProGeRF | MISA-web |
|---|---|---|---|---|---|---|---|
| AC256511.1 (113 kb) | 549 | 580 | 1506 | 56 | 549 | 560 | 549 |
| AC257258.1 (124 kb) | 938 | 943 | 1965 | 85 | 938 | 901 | 938 |
| AC259365.1 (118 kb) | 641 | 666 | 1584 | 76 | 641 | 628 | 641 |
| AC261250.1 (91 kb) | 498 | 457 | 1166 | 60 | 498 | 456 | 498 |
| AC263353.1 (33 kb) | 153 | 173 | 413 | – | 153 | 142 | 153 |
| AC264961.1 (126 kb) | 654 | 620 | 1641 | – | 654 | 605 | 654 |
| AC265197.1 (113 kb) | 505 | 496 | 1407 | 44 | 505 | 503 | 505 |
| AC266636.1 (167 kb) | 839 | 865 | 2174 | 79 | 839 | 811 | 839 |
| AC267178.1 (121 kb) | 517 | 530 | 1524 | 46 | 516 | 496 | 517 |
| AC269605.1 (119 kb) | 728 | 676 | 1711 | 76 | 728 | 700 | 728 |
| Sum | 6022 | 6006 | 15091 | 522 | 6021 | 5802 | 6022 |
| Execute time per batch [sec] | 7.498 | 30.735 | 1.042 | 1.286 | 0.643 | 20.994 | 1.796 |
The tool IMEx generated errors while executing due to operating system incompatibility as reported by (Lopes et al., 2015). The programs mreps and TROLL required the plain nucleotide sequence without a header.
Apart from TROLL and mreps all tools found about 6000 microsatellites in the ten BAC sequences. TROLL detected more than 15,000 microsatellites because it also reports degenerated (imperfect) microsatellites by default. Mreps detected the lowest amount of SSRs due to a hardcoded minimum output sequence length that prevented the identification of small microsatellites. Mreps did not report results for BACs AC263353.1 and AC264961.1 because of an excessive number of ‘N’ characters in their sequences. TRF reported spurious microsatellites as a result of substituting ‘N’ bases with random nucleotides which in turn increased the amount of reported microsatellites. In order to get comparable results for TRF the user needs to manually remove every microsatellites that includes at least a single ‘N’ character. Among the evaluated tools here, only ProGeRF is able to detect microsatellites in protein sequences.
The execution time of MISA-web is comparable to that of the other tools. SciRoKo and TRF were the fastest and slowest programs, respectively.
4 Conclusion
We developed the web-application MISA-web as an extension of the microsatellite finder MISA with a user-friendly GUI and improved output formatting options. The GFF3 output format facilitates the integration of MISA-web search results in downstream analysis pipelines.
Acknowledgements
We thank Jens Bauernfeind and Heiko Miehe for administration of UNIX servers. We greatly acknowledge input and motivation of Andreas Graner for implementing MISA-web.
Funding
This work was supported by the German Federal Ministry of Education and Research (BMBF) grant FKZ 0315954A in the frame of project TRITEX to US.
Conflict of Interest: None declared.
References
- Benson G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res., 27, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.M. (2005) Forensic DNA typing: biology, technology, and genetics of STR markers. Academic Press, Burlington, MA, USA. [Google Scholar]
- Castelo A.T. et al. (2002) TROLL–tandem repeat occurrence locator. Bioinformatics, 18, 634–636. [DOI] [PubMed] [Google Scholar]
- Kofler R. et al. (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics, 23, 1683–1685. doi:10.1093/bioinformatics/btm157 [DOI] [PubMed] [Google Scholar]
- Kolpakov R. et al. (2003) mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res., 31, 3672–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes R.D. et al. (2015) ProGeRF: proteome and genome repeat finder utilizing a fast parallel hash function. Biomed. Res. Int., 2015, doi:10.1155/2015/394157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies I.E. et al. (2012) Population structure revealed by different marker types (SSR or DArT) has an impact on the results of genome-wide association mapping in European barley cultivars. Mol. Breed., 30, 951–966. [Google Scholar]
- Miah G. et al. (2013) A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int. J. Mol. Sci., 14, 22499–22528. doi:10.3390/ijms141122499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudunuri S.B., Nagarajaram H.A. (2007) IMEx: imperfect microsatellite extractor. Bioinformatics, 23, 1181–1187. doi:10.1093/bioinformatics/btm097 [DOI] [PubMed] [Google Scholar]
- Munoz-Amatriain M. et al. (2015) Sequencing of 15 622 gene-bearing BACs clarifies the gene-dense regions of the barley genome. Plant J. Cell Mol. Biol., 84, 216–227. doi:10.1111/tpj.12959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Schlotterer, (1994) Simple sequences. Curr. Opin. Genet. Dev., 4, 832–837. [DOI] [PubMed] [Google Scholar]
- Thiel T. et al. (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). TAG Theoretical and Applied Genetics Theoretische Und Angewandte Genetik, 106, 411–422. doi:10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- Wang X. et al. (2013) GMATo: a novel tool for the identification and analysis of microsatellites in large genomes. Bioinformation, 9, 541–544. doi:10.6026/97320630009541. [DOI] [PMC free article] [PubMed] [Google Scholar]