Abstract
Motivation
Text mining has become an important tool for biomedical research. The most fundamental text-mining task is the recognition of biomedical named entities (NER), such as genes, chemicals and diseases. Current NER methods rely on pre-defined features which try to capture the specific surface properties of entity types, properties of the typical local context, background knowledge, and linguistic information. State-of-the-art tools are entity-specific, as dictionaries and empirically optimal feature sets differ between entity types, which makes their development costly. Furthermore, features are often optimized for a specific gold standard corpus, which makes extrapolation of quality measures difficult.
Results
We show that a completely generic method based on deep learning and statistical word embeddings [called long short-term memory network-conditional random field (LSTM-CRF)] outperforms state-of-the-art entity-specific NER tools, and often by a large margin. To this end, we compared the performance of LSTM-CRF on 33 data sets covering five different entity classes with that of best-of-class NER tools and an entity-agnostic CRF implementation. On average, F1-score of LSTM-CRF is 5% above that of the baselines, mostly due to a sharp increase in recall.
Availability and implementation
The source code for LSTM-CRF is available at https://github.com/glample/tagger and the links to the corpora are available at https://corposaurus.github.io/corpora/.
1 Introduction
Text mining is an important tool for many types of large-scale biomedical data analysis, such as network biology (Zhou et al., 2014), gene prioritization (Aerts et al., 2006), drug repositioning (Wang and Zhang, 2013) or creation of curated databases (Li et al., 2015). The most fundamental task in biomedical text mining is the recognition of named entities (called NER), such as proteins, species, diseases, chemicals or mutations. To date, the best performing NER tools rely on specific features to capture the characteristics of the different entity classes. For instance, the suffix ‘-ase’ is more frequent in protein names than in diseases; species names often consist of two tokens and have latin suffixes; chemicals often contain specific syllabi like ‘methyl’ or ‘carboxyl’, and mutations usually are sequences of letters and digits encoding genomic position, type of mutation, and base changes. Feature engineering, i.e. finding the set of features that best helps to discern entities of a specific type from other tokens (or other entity classes) currently is more of an art than a science, incurring extensive trial-and-error experiments. On top of this costly process, high-quality NER tools today need further entity-specific modules, such as whitelist and blacklist dictionaries, which again are difficult to build and maintain. Defining these steps currently takes the majority of time and cost when developing NER tools (Leser and Hakenberg, 2005) and leads to highly specialized solutions that cannot be used for other entity types than the ones they were designed for. On the other hand, the method used to identify entities in a given text based on the defined features nowadays is fairly homogeneous: conditional random fields (CRFs) (Lafferty et al., 2001), a statistical sequential classification method, is the de-facto standard method. Here, we show that an entirely generic NER method based on deep learning and distributional word semantics outperforms such specific high-quality NER methods across different entity types and across different evaluation corpora.
In the general field of information extraction, two recent developments lead to substantial improvements. First, word embeddings have been introduced to represent a single word by a low-dimensional vector capturing—in some way—the frequencies of co-occurring adjacent words. When compared with the bag-of-words approach underlying the conventional methods outlined earlier, word embeddings capture semantic similarities between words (as mathematical similarities between their vectors) that are not visible from their surface; for instance, the words ‘enables’ and ‘allows’ are syntactically very different, yet their meaning is somewhat related, which leads to similar sets of co-occurring words, whereas the co-occurrences of the word ‘swim’ would be completely different. The underlying idea of representing words ‘by the company they keep’ (Mackin, 1978) is an old concept in linguistics, usually called distributional semantics; its recent popularity is based on the novel idea that the embeddings are automatically adjusted such that information extraction tools benefit the most. Second, it has been shown that the application of artificial neural networks (ANNs), which automatically learn non-linear combinations of features, leads to better recognition results than the usage of CRFs, which can only learn (log-)linear combinations of features. Deep neural networks, and especially long short-term memory networks (LSTM), perform this task particularly efficiently and effectively. As with word embeddings, this idea is not new (Hochreiter and Schmidhuber, 1997), but only recent progress in the available data volumes and machine capabilities made it applicable to practically relevant problems (Pascanu et al., 2014).
Following a suggestion from Lample et al. (2016), we combined the power of word embeddings, LSTMs and CRFs into a single method for biomedical NER, called LSTM-CRF. This method is completely agnostic to the type of the entity; all it requires is an entity-annotated gold standard and word embeddings pre-computed on a large, entity-independent corpus (typically all PubMed abstracts). We assessed the performance of LSTM-CRF by performing 33 evaluations on 24 different gold standard corpora (some with annotations for more than one entity type) covering five different entity types, namely chemical names, disease names, species names, genes/protein names, and names of cell lines. These corpora encompass patents and scientific articles and partly consist of abstracts and partly of full texts. We compared the performance of LSTM-CRF with that of best-of-class, entity-specific NER tools and with another generic NER method using a CRF with a typical NER feature set plus the word embeddings as input. LSTM-CRF turned out to have the best F1-score on 28 of the 33 cases; on average, it is 5% better than the entity-specific NER tools and 3% better than the CRF method with word embeddings.
2 Materials and methods
In the following sections, we give a technical explication of the LSTM-CRF approach and describe the competitor NER systems. Furthermore, we describe the corpora we used for evaluation, the different embeddings evaluated, and details regarding text pre-processing and evaluation metrics.
2.1 LSTM-CRF
LSTM-CRF (Lample et al., 2016) is a domain-independent NER method which does not rely on any kind of background knowledge. We first describe LSTM, a specific kind of ANN, and then discuss the architecture of LSTM-CRF in detail.
An LSTM is a special kind of ANN which processes sequences of arbitrary length and is able to model dependencies between sequence elements even if they are far apart (Hochreiter and Schmidhuber, 1997). The input to an LSTM unit is a sequence of vectors of length T, for which it produces an output sequence of vectors of equal length by applying a non-linear transformation learned during the training phase. Each is called the activation of the LSTM at token t. The exact formula to compute one activation of an LSTM unit in the LSTM-CRF model is provided below (Lample et al., 2016):
where all Ws and bs are trainable parameters, σ denotes the element-wise sigmoid function and is the element-wise product.
Such an LSTM-layer processes the input only in one direction and thus can only encode dependencies on elements that came earlier in the sequence. As remedy for this problem, we use another LSTM-layer processing in the reversed direction, which allows detecting dependencies on elements later in the text. The resulting neural network is called a bi-directional LSTM (Graves and Schmidhuber, 2005).
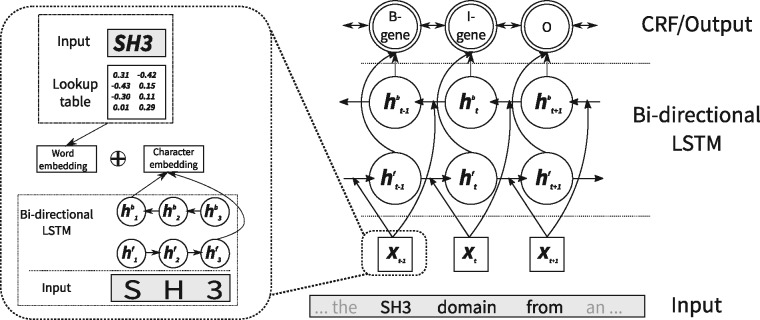
The architecture of LSTM-CRF is shown in Figure 1. It is comprised of three main layers. The first layer is the embedding layer. It receives the raw sentence S made of the sequence of words as its input and produces an embedding (i.e. a dense vector representation) for each word in S. The embedding vector of word wt is a concatenation of a word- and a character-level embedding. The word-level embedding is simply retrieved from a lookup-table (see the example in Fig. 1) of word embeddings trained on a large corpus (see Section 2.3). The character-level embedding is obtained by applying a bi-directional LSTM to the character sequence of each word and then concatenating the last activations of both directions, as exemplified for the word ‘SH3’ in the left side of Figure 1. The resulting sequence of embeddings is fed into another bi-directional LSTM-layer that produces a refined representation of the input, which is the input to a final CRF-layer. The classical Viterbi algorithm is used to obtain the final output from this layer. All components together form a single fully differentiable neural network that can be trained by backpropagation.
Fig. 1.
CRF-LSTM architecture. For instance, for the word ‘SH3’ from the input sentence S, the character-based representation is computed by applying a bi-directional LSTM onto the sequence of its characters ‘S’, ‘H’, ‘3’. The resulting embedding is concatenated with the corresponding word embedding, trained on a huge corpus. This word representation is then processed by another bi-directional LSTM and finally by a CRF layer. The output is the most probable tag sequence, as estimated by the CRF
In our experiments, we used a learning rate of 0.005 for all corpora except for BioSemantics where we set it to 0.0005, because the model did not converge with the default one. Regarding the other hyperparameters, we used the default values from (Lample et al., 2016) except for (i) the tag scheme which we set to IOB instead of IOBES, and (ii) the dropout rate which we set to 0.3 instead of 0.5 because this value was optimal for most corpora evaluated by Lample et al. (2016).
2.2 Competitor systems
We compare the performance of LSTM-CRF against two types of competitors: a CRF using a generic feature set for NER tasks plus word embeddings, and entity-specific NER tools for each class. The former should help to separate the impact of using word embeddings from the impact of using the LSTM-CRF architecture. For the latter we selected the current best-in-class entity-specific NER tools. All trainable baseline systems were retrained using the same corpora and folds.
2.2.1 Baseline CRF
We used CRFSuite (http://www.chokkan.org/software/crfsuite/) (Okazaki, 2007) with default settings to train a first-order linear-chain CRF model utilizing identical features for all entity types. These features were defined by the NER feature extractor shipped with CRFsuite and were designed for domain-independent NER. Additionally, we provided as features (in turn) one of the word embeddings described in Section 2.4.
2.2.2 Baseline methods
For each of the five entity classes considered in our evaluation, we chose the presumably best performing and publicly available current NER tool:
Chemicals: we employed tmChem (https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/tmTools/) (Leaman et al., 2015) Model I, considered as the state-of-the-art chemical NER tool also in other recent studies (Habibi et al., 2016; Leaman and Lu, 2016;). tmChem Model I trains a first-order CRF using features from BANNER (Leaman and Gonzalez, 2008) plus further ones obtained through a trial-and-error procedure, including character n-grams, chemical specific identifiers, and the output of ChemSpot (Rocktäschel et al., 2012) (another high-quality chemical NER tool). The output of the model is filtered by several type-specific post-processing steps for abbreviation resolution, enforcing of tagging consistency and balancing of parentheses.
Genes/proteins: we utilized Gimli (http://bioinformatics.ua.pt/software/gimli/) (Campos et al., 2013), an open source retrainable gene/protein NER tool with competitive performance (Campos et al., 2012). Gimli trains both first- and second-order CRF models using a set of specific features, such as orthographic, morphological, linguistic-based, dictionary-based and a conjunction of features from adjacent tokens. Post-processing steps like parentheses correction, abbreviation resolution, and name extension using a domain dictionary are also applied.
Species: we used SPECIES (http://species.jensenlab.org/) (Pafilis et al., 2013), a freely available dictionary-based tool with state-of-the-art tagging quality. Species is the only NER tool in our set which does not build on a CRF, and it is also the only tool which does not train a corpus-specific model.
Diseases: we used DNorm (https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/tmTools/) (Leaman et al., 2013), a freely available tool showing excellent performance in several studies (Leaman and Lu, 2016; Wei et al., 2015). It uses the rich feature set provided by BANNER and a dictionary of disease names created by the authors to train a first-order CRF model. The output of the model is also filtered by manual rules like abbreviation resolution.
Cell Lines: we used a model presented in (Kaewphan et al., 2016), reported to outperform many other available systems in a cross-corpus setting. It is based on the domain-independent CRF-based tool NERsuite (http://nersuite.nlplab.org/). In addition to the pre-defined NERsuite features, it uses a comprehensive dictionary of cell line names from multiple sources.
2.3 Gold standard corpora
We performed our evaluations on five entity types: genes/proteins, chemicals, diseases, cell lines and species. We relied on a total of 24 corpora, each containing manual annotations for one or more of these entity types, such as CHEMDNER patent (Krallinger et al., 2015a,b) for chemicals and genes/proteins, NCBI Disease (Doğan et al., 2014) for disease names, Gellus (Kaewphan et al., 2016) for cell lines and S800 (Pafilis et al., 2013) for species. The corpora encompass two different genres of texts: (i) patent texts from the European Patent Office (EPO) (http://www.epo.org/), World Intellectual Property Organization (WIPO) (http://www.wipo.int/), and United States Patent and Trademark Office (USPTO) (http://www.uspto.gov/) and (ii) scientific articles from PubMed Central (PMC) (http://www.ncbi.nlm.nih.gov/pmc) and PubMed (https://www.ncbi.nlm.nih.gov/pubmed/).Table 1 lists all corpora together with important characteristics like the number of sentences, tokens, and annotated entities per entity class (measured after text pre-processing as described in Section 2.5).
Table 1.
The details of the gold standard corpora
| Corpora | Text genre | Text type | Entity type | No. sentences | No. tokens | No. unique tokens | No. annotations | No. unique annotations |
|---|---|---|---|---|---|---|---|---|
| CHEMDNER patent (Krallinger et al., 2015a,b) | Patent | Abstract | Chemicals | 35 584 | 1 465 471 | 62 344 | 64 026 | 20 893 |
| Genes/proteins | 35 584 | 1 465 471 | 62 344 | 12 597 | 5835 | |||
| CHEBI (http://chebi.cvs.sourceforge.net/viewvc/chebi/chapati/) | Patent | Full-text | Chemicals | 10 638 | 314 105 | 24 424 | 17 724 | 5109 |
| BioSemantics (Akhondi et al., 2014) | Patent | Full-text | Chemicals | 173 808 | 5 690 518 | 208 326 | 368 091 | 72 756 |
| CHEMDNER (Krallinger et al., 2015a) | Scientific article | Abstract | Chemicals | 89 679 | 2 235 435 | 114 837 | 79 842 | 24 321 |
| CDR (Li et al., 2016) | Scientific article | Abstract | Chemicals | 14 228 | 323 281 | 23 068 | 15 411 | 3629 |
| Diseases | 14 228 | 323 281 | 23 068 | 12 694 | 3459 | |||
| BioCreative II GM (Smith et al., 2008) | Scientific article | Abstract | Genes/proteins | 20 510 | 508 257 | 50 864 | 20 703 | 14 906 |
| JNLPBA (Kim et al., 2004) | Scientific article | Abstract | Genes/proteins | 22 562 | 597 333 | 25 046 | 35 460 | 10 732 |
| Cell Lines | 22 562 | 597 333 | 25 046 | 4332 | 2528 | |||
| CellFinder (Neves et al., 2012) | Scientific article | Full-text | Genes/proteins | 2177 | 65 031 | 7977 | 1340 | 600 |
| Species | 2177 | 65 031 | 7977 | 435 | 51 | |||
| Cell Lines | 2177 | 65 031 | 7977 | 350 | 69 | |||
| OSIRIS (Furlong et al., 2008) | Scientific article | Abstract | Genes/proteins | 1043 | 28 697 | 4669 | 768 | 275 |
| DECA (Wang et al., 2010) | Scientific article | Abstract | Genes/proteins | 5468 | 138 034 | 14 515 | 6048 | 2360 |
| Variome (Verspoor et al., 2013) | Scientific article | Full-text | Genes/proteins | 6471 | 172 409 | 12 659 | 4309 | 596 |
| Diseases | 6471 | 172 409 | 12 659 | 6025 | 629 | |||
| Species | 6471 | 172 409 | 12 659 | 182 | 8 | |||
| PennBioIE (Kulick et al., 2004) | Scientific article | Abstract | Genes/proteins | 14 305 | 357 313 | 21 089 | 17 427 | 4348 |
| FSU-PRGE (Hahn et al., 2010) | Scientific article | Abstract | Genes/proteins | 35 465 | 899 426 | 57 498 | 56 087 | 19 470 |
| IEPA (Ding et al., 2002) | Scientific article | Abstract | Genes/proteins | 243 | 15 174 | 2923 | 1109 | 209 |
| BioInfer (Pyysalo et al., 2007) | Scientific article | Abstract | Genes/proteins | 1141 | 33 832 | 5301 | 4162 | 1150 |
| miRNA (Bagewadi et al., 2014) | Scientific article | Abstract | Genes/proteins | 2704 | 65 998 | 7821 | 1006 | 409 |
| Diseases | 2704 | 65 998 | 7821 | 2123 | 671 | |||
| Species | 2704 | 65 998 | 7821 | 726 | 47 | |||
| NCBI Disease (Doğan et al., 2014) | Scientific article | Abstract | Diseases | 7639 | 174 487 | 12 128 | 6881 | 2192 |
| Arizona Disease (Leaman et al., 2009) | Scientific article | Abstract | Diseases | 2884 | 76 489 | 7358 | 3206 | 1188 |
| SCAI (Gurulingappa et al., 2010) | Scientific article | Abstract | Diseases | 4332 | 104 015 | 12 558 | 2226 | 1048 |
| S800 (Pafilis et al., 2013) | Scientific article | Abstract | Species | 7933 | 195 197 | 20 526 | 3646 | 1564 |
| LocText (Goldberg et al., 2015) | Scientific article | Abstract | Species | 949 | 22 550 | 4371 | 276 | 39 |
| Linneaus (Gerner et al., 2010) | Scientific article | Full-text | Species | 17 788 | 473 148 | 34 396 | 4077 | 419 |
| CLL (Kaewphan et al., 2016) | Scientific article | Abstract, Full-text | Cell Lines | 215 | 6547 | 2468 | 341 | 311 |
| Gellus (Kaewphan et al., 2016) | Scientific article | Abstract, Full-text | Cell Lines | 11 221 | 278 910 | 25 628 | 640 | 237 |
2.4 Word embeddings
We utilized word embedding techniques to capture the semantics of words (and their similarities) based on their surrounding words. We evaluated three different embedding models. The first model, denoted PubMed-PMC, was trained on a combination of PubMed abstracts (nearly 23 million abstracts) and PMC articles (nearly 700, 000 full texts). The second model, denoted Wiki-PubMed-PMC, was trained using these two collections plus approximately four million English Wikipedia articles. The second model thus mixes domain-specific texts with domain-independent ones. Both models were created by Pyysalo et al. (2013) using Google's word2vec (http://bio.nlplab.org/); we use 200D vectors. To also be able to study the impact of text genre, we trained a third model with 50 dimensions, denoted Patent, on a set of roughly 20 000 European patents with biomedical topics using the Gensim toolkit. We optimized the hyper-parameters of Gensim for the F1-score of the CRF model (see Section 2.2) on the development set of the CHEMDNER patent corpus.
2.5 Text pre-processing
All corpora first were converted into a simple uniform format to ease further processing. In this format, all texts and all annotations are stored in one single file. Each document is represented by a document identifier, a tab separator and the entire document text in one line. Annotations are given in the following lines, one annotation per line, with start character position end character position, entity mention text, and entity type. An empty line indicates the end of a document, and the next document starts after this empty line. We converted this file into the specific input format defined by each baseline NER tool. Moreover, we used Apache OpenNLP (https://opennlp.apache.org/) to split documents into sentences, sentences into tokens, and to assign part-of-speech tags. Finally, we created a file in CoNLL2003 (Tjong Kim Sang and De Meulder, 2003) format as input for the LSTM-CRF model and the CRFSuite tool.
2.6 Evaluation metrics
We randomly divided each corpus into three disjoint subsets. 60% of the samples were used for training, 10%, as the development set for the training of methods, and 30% for the final evaluation. We compared all methods in terms of precision, recall and F1-score on the test sets. We performed exact matching to compute these performance values. We also performed an error analysis by comparing the sets of false positives (FPs) and false negatives (FNs) of the different NER methods. To this end, we measured the number of FP and FN counts for each mention by each method and then calculated the overlap between sets of FP or FN using fuzzy set operations that take into account the frequency of mistakes per entity mention (Thole et al., 1979).
3 Results
We assessed the performance of a novel method for entity-type independent NER, namely LSTM-CRF, on 33 different evaluation sets covering five different types of biomedical entities. LSTM-CRF uses as features only low-dimensional representations of the words in the vicinity of the to-be-classified tokens, created by mixing word-level embeddings created in an unsupervised fashion with character-level embeddings trained on the respective corpus. Results were compared with a traditional CRF using typical NER features and the same word embeddings, and to type-specific baselines representing the state-of-the-art in biomedical NER.
3.1 Impact of different word embeddings
We first studied the impact of using different word embeddings. We compared the results of three models, differing only in the unsupervised part, i.e. the text collections used for computing word-level embeddings. For evaluation, we considered the LSTM-CRF and the pure CRF approach. The macro averaged performance values over all corpora in terms of precision, recall and F1-score are provided in Table 2; detailed performance values are given in Appendix A. In five out of six cases, Wiki-PubMed-PMC achieves the best performance, and this model is also very close to the best one in the sixth case. Based on this data, we used the Wiki-PubMed-PMC embeddings in all further evaluations.
Table 2.
Macro averaged performance values in terms of precision, recall and F1-score for CRF and LSTM-CRF methods with word embedding features: (i) Patent, (ii) PubMed-PMC and (iii) Wiki-PubMed-PMC
| Precision (%) |
Recall (%) |
F1-score (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| (i) | (ii) | (iii) | (i) | (ii) | (iii) | (i) | (ii) | (iii) | |
| CRF | 82.71 | 84.55 | 84.49 | 71.98 | 73.07 | 73.26 | 76.36 | 77.98 | 78.04 |
| LSTM-CRF | 80.10 | 81.39 | 81.77 | 81.04 | 80.72 | 81.08 | 80.26 | 80.79 | 81.11 |
The highest values for each method are represented in bold.
3.2 Performance of LSTM-CRF, CRF, and baselines
We compared the performance of five baseline NER tools, the CRF, and the LSTM-CRF using Wiki-PubMed-PMC embeddings on 33 evaluation sets. Results in terms of F1-score for each entity type and each corpus are shown Figure 2; exact precision, recall, and F1 values are given in Appendix A. LSTM-CRF achieves the best performance for 28 out of 33 evaluations and is very close to the best method in the remaining five cases. On average (macro average), F1-scores are 81.11% for the generic LSTM-CRF method, 78.04% for the generic CRF method and 76.61% for the baselines. The improvements are mainly due to a strong increase in recall (mean 81.08%, 73.26%, 75.13%) at the cost of decrease in precision (mean 81.77%, 84.49%, 80.38%).
Fig. 2.
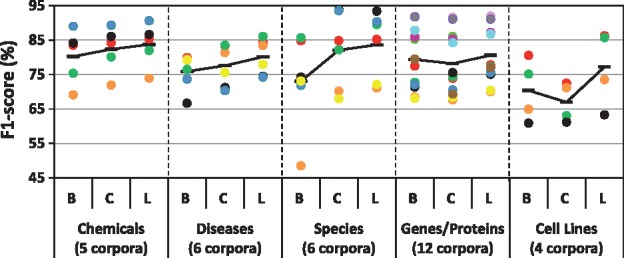
F1-scores of baseline (B), generic CRF (C) and generic LSTM-CRF (L) for five entity types, each measured within 4–12 corpora. The score for each corpus per entity type is depicted by a specific colored circle
We also computed performance metrics aggregated per entity type to see if methods are more suitable for some types than for the others. Both macro and micro averaged performance values in Table 3 reveal that this does not seem to be the case; LSTM-CRF achieves the best average F1-score and recall for all entity types.
Table 3.
Macro and micro averaged performance values in terms of precision, recall and F1-score for the baselines (B), the generic CRF method (C) and the generic LSTM-CRF method (L) over the corpora per each entity type
| a) Macro averaged performance | |||||||||
| Precision (%) | Recall (%) | F1-score (%) | |||||||
| B | C | L | B | C | L | B | C | L | |
| Chemicals | 79.8 | 85.81 | 82.82 | 80.73 | 79.26 | 84.77 | 80.22 | 82.32 | 83.71 |
| Diseases | 78.54 | 82.48 | 81.66 | 73.67 | 73.47 | 78.70 | 75.89 | 77.56 | 80.11 |
| Species | 72.75 | 89.59 | 80.84 | 79.63 | 76.15 | 87.64 | 73.03 | 82.09 | 83.60 |
| Gene/protein | 83.77 | 82.54 | 81.57 | 75.87 | 74.67 | 80.06 | 79.35 | 78.15 | 80.58 |
| Cell lines | 85.14 | 84.04 | 82.65 | 61.32 | 56.91 | 73.22 | 70.35 | 66.96 | 77.2 |
| Average | 80.38 | 84.49 | 81.77 | 75.13 | 73.26 | 81.08 | 76.61 | 78.04 | 81.11 |
| b) Micro averaged performance | |||||||||
| Precision (%) | Recall (%) | F1-score (%) | |||||||
| B | C | L | B | C | L | B | C | L | |
| Chemicals | 75.16 | 83.45 | 81.81 | 79.7 | 79.12 | 83.86 | 77.36 | 81.23 | 82.82 |
| Diseases | 79.29 | 83.62 | 82.56 | 75.98 | 77.17 | 81.68 | 77.60 | 80.26 | 82.12 |
| Species | 77.33 | 88.92 | 83.84 | 67.74 | 76.68 | 83.33 | 72.22 | 82.35 | 83.59 |
| Gene/protein | 81.34 | 81.79 | 81.50 | 77.45 | 78.14 | 82.71 | 79.35 | 79.92 | 82.10 |
| Cell lines | 71.82 | 70.20 | 68.59 | 57.20 | 57.60 | 66.84 | 63.68 | 63.28 | 67.70 |
| Average | 76.4 | 83.11 | 81.72 | 78.87 | 78.71 | 83.45 | 77.62 | 80.85 | 82.58 |
The highest values for each entity class are highlighted in bold.
An interesting observation is that the pure CRF method often performs better than the entity-specific baseline algorithms. There are two explanations for this apparent contradiction to our introductory words, claiming that the best methods to date are CRF with type-specific feature sets. First, NER methods are often developed for a very specific sub-problem, and often only have excellent performance on a particular evaluation corpus. Our evaluation on a much larger set of corpora reveals that such corpus-specific advantages cannot be simply extrapolated to other settings. Second, by using word embeddings our CRF model represents words in a context-encoding space, which enables it to recognize semantic similarity between words. This feature is missing in the word-based baseline tools.
3.3 Error analysis
We compared the errors made by the three methods by computing intersections of the sets of FPs and FNs for each method and each entity type. Results per entity class are shown in Figure 3.
Fig. 3.
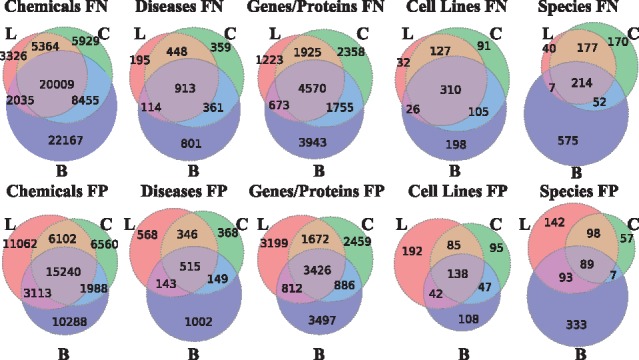
Venn diagrams demonstrating the area of overlap among the FP sets or the FN sets of the three methods: the baseline (B), the generic CRF method (C) and the generic LSTM-CRF method (L) per entity type
The Venn diagrams of FP sets or FN sets for the different entity types follow a similar pattern. Generally, error sets of CRF and LSTM-CRF are more similar to each other than to errors of the baseline methods, probably due to the strong reliance of all baselines on entity type-specific dictionaries, creating their own specific errors. Relying on dictionaries carries two types of dangers: first, they are notoriously incomplete, leading to FNs; second, dictionary matching disregards context which leads to FP matches in case of ambiguous terms. This is particularly visible for species, where the baseline only uses dictionary matching, leading to the worst precision among all entity types.
We also speculated that the length of an entity might have an impact on the performance of approaches. Very short entities are often abbreviations which are notoriously ambiguous; entities consisting of multiple tokens often produce errors at the border tokens, which are severely punished in any exact matching evaluation (one FP and one FN). To this end, we measured the average length of a FP or FN mention in terms of non-space characters for the three methods per entity class. Furthermore, we computed the F1-scores of single- and multi-token mentions separately. Results are shown in Table 4. First, LSTM-CRF is the best method regardless whether an entity name consists of only one or of multiple tokens. Only for species, the dictionary-based baseline method has a slightly better F1-score for multi-token entities, which is, however, outweighed by a much worse F1-score for single entity names in the overall evaluation. This result shows that the LSTM-CRF method with word embeddings manages to tag precisely also multi-token entities, without relying on any post-processing rules. However, we also observe that the performance of CRF and LSTM-CRF on single-token mentions is considerably better than on multi-token entities, showing that there is still room for improvement regarding such entity names. Second, there is an interesting tendency that FPs tagged by LSTM-CRF are slightly shorter than those found by the CRF, while FNs are slightly longer, indicating that LSTM-CRF seems to be biased towards shorter entity names.
Table 4.
The average length of errors from the lists of FPs and FNs, and the F1-scores of single-token and multi-token entities measured for baselines (B), generic CRF methods (C) and generic LSTM-CRF methods (L) per entity type
| F1-score (%) |
Mention length |
||||
|---|---|---|---|---|---|
| Single-Token | Multi-Token | FP | FN | ||
| Chemicals | L | 84.02 | 79.90 | 19.07 | 20.69 |
| C | 82.53 | 78.07 | 25.1 | 20.11 | |
| B | 76.77 | 76.84 | 19.13 | 16.49 | |
| Diseases | L | 86.13 | 76.54 | 14.51 | 15.58 |
| C | 84.20 | 74.92 | 15.09 | 14.66 | |
| B | 80.42 | 73.80 | 13.90 | 14.36 | |
| Species | L | 86.11 | 79.10 | 10.68 | 12.34 |
| C | 85.57 | 76.75 | 11.55 | 11.52 | |
| B | 67.72 | 79.80 | 8.00 | 9.15 | |
| Genes/Proteins | L | 86.49 | 72.64 | 12.85 | 14.13 |
| C | 84.77 | 69.32 | 13.50 | 13.52 | |
| B | 83.21 | 70.99 | 11.73 | 11.98 | |
| Cell Lines | L | 72.94 | 64.93 | 20.02 | 15.65 |
| C | 64.77 | 62.52 | 19.01 | 15.24 | |
| B | 62.55 | 64.24 | 18.41 | 13.91 | |
The highest F1-scores are emphasized in bold.
3.4 Precision-recall trade-off
The results in Table 3 show that, on average, LSTM-CRF significantly outperforms CRF and baselines in terms of recall at the expense of a less strong decrease in precision. Since, for some applications, obtaining high precision is more important than high recall, we also implemented and evaluated a method which assigns confidence scores to the predictions made by LSTM-CRF, followed by an entity-type independent filter for low confidence predictions with the aim to reduce the number of FP entities. The filter removes a percentage of entities with the lowest confidence values from the output of the generic LSTM-CRF method. First, the labels for all the tokens of a sentence are predicted by Viterbi decoding using the trained model (as described in Section 2.1). Then, all tokens labeled as entity are assigned a confidence score which estimates the probability (given the LSTM-CRF model) of the predicted entity label being part of the true labels of the sentence. The score for a given entity is obtained by summing the scores of all possible label sequences of the sentence which produce this entity label normalized by the sum of scores of all the possible label sequences. These scores can be obtained by the constrained forward-backward algorithm (Culotta and McCallum, 2004) (The implementation is available online at “https://github.com/leonweber/tagger”.).
We removed 5, 10 and 15% of the entities tagged by the generic LSTM-CRF model with lower confidence values and, on average, obtained precision (recall) scores of 83.62% (78.97%), 85.16% (76.22%) and 86.45% (73.05%), respectively; detailed performance values are given in Appendix A. Across all evaluations, the generic LSTM-CRF model without the 10% lowest confidence predictions obtains, on average, higher precision and higher recall values than the generic CRF model and the baselines. In Figure 4, we compare precision and recall values of the LSTM-CRF method before and after applying filters together with CRF and baselines, macro-averaged by entity type. For four out of five entity types, precision and recall values of this configuration is very close to or higher than those of the generic CRF method and of the baselines. The only exception is species, where even the most stringent filtering that we applied cannot make the LSTM-CRF method reach a precision higher than that of the CRF method. We inspected these evaluations in more detail. The lower precision values on average are mostly due to the two corpora CellFinder and Variome. However, both corpora miss some of the species annotations. For instance, the 41 FP predictions of LSTM-CRF for Variome are 21 times the term ‘mouse’, 4 times ‘mice’ and 16 times ‘human’—all of which are valid species names, and all of which achieve high prediction confidence. The situation is similar for CellFinder, with tokens like ‘goat’, ‘human’ or ‘EColi’ being not annotated as species.
Fig. 4.
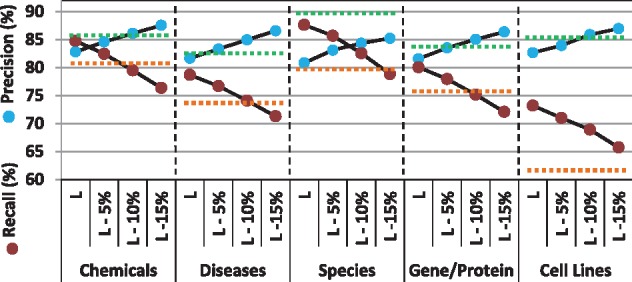
Aggregated precision and recall of the generic LSTM-CRF method before (L) and after applying filters, removing 5 (L—5%), 10 (L—10%) and 15 (L—15%) of entities, per entity type. The highest averaged precision and recall per entity type obtained by baselines or the generic CRF model are represented by the green and the orange dash line, respectively
4 Discussion
4.1 Genre and word embeddings
The word embeddings we used for our study were either derived from patents, from scientific articles or from a mixture of scientific articles and Wikipedia entries. Since also some of our corpora are based on patents and others on scientific articles, we wondered if the use of patent-derived embeddings is advantageous for patent corpora yet less adequate for scientific articles and vice versa. Indeed, our patent-derived embeddings slightly outperform the others on patent-derived corpora in terms of F1-score (recall) (Patent: 78.52% (82.75%), PubMed-PMC: 78.48% (81.85%), Wiki-PubMed-PMC: 77.84% (81.00%)) and the model derived from Wiki-PubMed-PMC achieves a better F1-score (recall) on corpora consisting of scientific articles (Patent: 80.50% (80.81%), PubMed-PMC: 81.11% (80.57%) and Wiki-PubMed-PMC: 81.56% (81.09%)). This observation again shows that patents are different from scientific articles (Habibi et al., 2016) and that their analysis calls for specific resources.
4.2 Corpus size and word embeddings
The performance values obtained in Section 3.1 show that both CRF and LSTM-CRF achieve the best performance using Wiki-PubMed-PMC embeddings in most of the cases. Notably, this model is derived from a collection of domain-specific texts (PubMed, PMC) mixed with domain-unspecific texts (Wikipedia). A possible explanation for its superiority is that it uses the largest text base among the three models; previous works have shown that word embeddings tend to be the more effective the larger the text base is (Stenetorp et al., 2012). Furthermore, the use of general domain corpora in addition to the domain-specific ones may add more out-of-domain information to the embeddings. However, more investigations are required to define optimal corpora for derivation of word embeddings for concrete tasks.
4.3 Related work
Traditional biomedical NER methods have relied on rule- or dictionary-based approaches. The rule-based techniques recognize biomedical entities using several rules manually defined based on the textual patterns of entities (Narayanaswamy et al., 2003; Eltyeb and Salim, 2014). These patterns vary depending on the specific textual properties of an entity class. The definition of such entity-specific patterns is time consuming and requires domain-expert knowledge. Dictionary-based methods extract named entities by searching them in dictionaries constructed for each entity type. For instance, Hettne et al. (2009) employ a dictionary-based approach for the extraction of drugs, and Gerner et al. (2010) use a dictionary-based approach for species names extraction. Again, building such dictionaries is time consuming and challenging (Liu et al., 2015a). Moreover, the recall obtained using these methods is generally low due to the inherent difficulty of the methods in capturing new entities; a strong advantage of dictionary-based methods is that they directly solve the named entity normalization (NEN) problem, i.e. they can output a database identifier for each recognized entity.
Over the last years, pattern- and dictionary- based methods have been superseded by approaches relying on supervised machine learning, especially sequential classification algorithms, such as Hidden Markov Models (Rabiner, 1989) and CRFs (Lafferty et al., 2001). CRFs have become the de-facto standard model, being the method of choice for essentially all tools winning recent NER competitions, such as BioCreative IV (Krallinger et al., 2013) or i2b2 (Uzuner et al., 2011). Popular biomedical NER tools using CRFs are, for instance, ABNER (A Biomedical Named Entity Recognizer) (Settles, 2005) and BANNER (Leaman and Gonzalez, 2008). Hybrid methods combine machine learning methods with dictionary- or rule-based techniques. For instance, ChemSpot (Rocktäschel et al., 2012) integrates results of a CRF model with a dictionary-matching module for chemical NER, and Gimli (Campos et al., 2013) applies post-processing steps like parentheses balancing to the output of the CRF models.
All these methods build upon pre-defined sets of features, whose correlations with other features and the target class of tokens are learned from the gold standard annotations. In contrast, methods based on deep ANNs also consider non-linear combinations of feature values (Hastie et al., 2001). This drastically increases the search space, leading to the fact that ANNs for long were considered impractical for many applications. This situation changed only recently due to the steep increase in the compute power of machines. When combined with specifically trained word embeddings, deep learning with ANNs has been shown to outperform other methods in many areas, such as sentiment analysis (Dai and Le, 2015) or language modeling (Jozefowicz et al., 2016). These methods are also gradually entering the field of biomedical information extraction, yet results so far have been mixed. Segura-Bedmar et al. (2015) used word embeddings as input to a CRF and reported only marginal effects in chemical NER. Liu et al. (2015b) found word embeddings to only marginally improve the performance of a CRF-based method using comprehensive dictionaries as features. Tang et al. (2014) compared several ways of obtaining word embeddings and reported up to 2% increase in recall. The method we use in this paper, LSTM-CRF, was proposed as a general NER algorithm by Lample et al. (2016). A few previous works have applied LSTM-CRF for biomedical NER (e.g. Chalapathy et al., 2016a,b). However, all these evaluations considered at most a handful of corpora. In contrast, our evaluation of biomedial NER methods based on 33 evaluations using 24 corpora to our knowledge is the most comprehensive one ever performed in this field. Larger previous evaluations we are aware of were performed in Campos et al. (2012), who measured the performance of different machine learning-based NER methods on 14 corpora, and Batista-Navarro et al. (2015), who reported on the performance of a CRF-based NER tool using nine different corpora.
5 Conclusion
In summary, our results indicate that LSTM-CRF improves considerably upon current biomedical NER methods. We find this exciting particularly because the method is completely agnostic to entity types; thus, the costly development of specific tools using specific dictionaries could become superfluous. However, further research is necessary to turn this observation into useful applications. In particular, LSTM-CRF only helps with the recognition of entities in texts; the next step in most text-mining applications, which is mapping recognized entities to standard nomenclature (e.g. Entrez-IDs for genes, ChEBI-IDs for chemicals etc.), is not addressed. Thus, LSTM-CRF should be combined with generic NEN tools, as for instance presented in (Leaman et al., 2013). In the future, we plan to study the inclusion of background knowledge into the LSTM-approach, for instance in the form of post-processing rules to deal with long multi-token entities; the hope is to achieve further improvements especially in terms of precision, though the price will be to lose the entity-independence. Nevertheless, our results imply that the advances in statistical NLP and machine learning can help to make biomedical text mining (i) more accurate, (ii) less laborious to develop and (iii) more robust with respect to the specific texts being mined.
Acknowledgements
We are grateful to the Federal Ministry for Economic Affairs and Energy (BMWi) for its financial support through the BioPatent project [KF2205219BZ4], the Federal Ministry for Research and Education (BMBF) for its financial support through PREDICT project (031L0023A), and the German Research Council (DFG) for funding through the Research Training Group SOAMED (GRK1651).
Conflict of Interest: The authors declare that they have no conflict of interests.
Appendix A
We have provided precision, recall and F1-scores of each NER model for each corpus in Table A1.
Table A1.
Precision, recall and F1-scores obtained for each corpus by the baselines and eight variants of the CRF and the LSTM-CRF methods
| Baseline | CRF | CRF (i) | CRF (ii) | CRF (iii) | LSTM- CRF | LSTM- CRF (i) | LSTM- CRF (ii) | LSTM- CRF (iii) | LSTM-CRF (iii) 5% | LSTM-CRF (iii) 10% | LSTM-CRF (iii) 15% | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a) Precision scores | |||||||||||||
| Chemicals | CHEMDNER patent | 82.56 | 84.52 | 84.29 | 84.42 | 84.29 | 81.09 | 82.25 | 83.35 | 83.33 | 84.66 | 86.14 | 87.44 |
| CHEBI | 71.46 | 80.59 | 78.56 | 78.68 | 79.67 | 71.58 | 71.1 | 72.95 | 69.5 | 71.3 | 72.85 | 74.57 | |
| BioSemantics | 72.56 | 82.17 | 82.1 | 81.93 | 81.96 | 80.64 | 82.07 | 79.72 | 80.88 | 82.4 | 83.81 | 85.11 | |
| CHEMDNER | 83.23 | 90.4 | 90.52 | 90.66 | 90.79 | 87.31 | 87.27 | 88.25 | 87.83 | 89.97 | 91.77 | 93.47 | |
| CDR | 89.19 | 91.04 | 91.47 | 91.81 | 92.37 | 88.19 | 91.12 | 92.18 | 92.57 | 94.61 | 95.98 | 97.16 | |
| Genes/Proteins | CHEMDNER patent | 67.02 | 67.01 | 67.33 | 67.66 | 67.13 | 61.05 | 64.02 | 65.74 | 66.23 | 67.3 | 68.32 | 69.29 |
| BioCreative II GM | 79.92 | 76.45 | 77.07 | 77.27 | 77.22 | 73.79 | 77.22 | 78.99 | 77.5 | 79.6 | 81.5 | 83.81 | |
| JNLPBA | 72.74 | 73.28 | 73.21 | 73.39 | 73.65 | 71.86 | 73.07 | 74.77 | 74.83 | 76.59 | 78.16 | 79.71 | |
| CellFinder | 81.98 | 74.39 | 77.27 | 84.05 | 83.77 | 78.49 | 82.01 | 83.56 | 88.73 | 91.19 | 93.12 | 95.28 | |
| OSIRIS | 83.33 | 77.92 | 76.88 | 81.38 | 78.61 | 74.07 | 76.52 | 76.73 | 78.02 | 80 | 81.73 | 82.74 | |
| DECA | 81.89 | 76.89 | 76.6 | 78.1 | 77.66 | 64.11 | 67.22 | 72.91 | 75.27 | 76.76 | 78.67 | 80.05 | |
| Variome | 90.32 | 90.08 | 89.56 | 89.92 | 90.12 | 87.63 | 87.64 | 86.59 | 87.47 | 89.41 | 90.72 | 91.33 | |
| PennBioIE | 88.12 | 88.14 | 88.32 | 89.5 | 89.58 | 83.31 | 86.51 | 88.02 | 86.97 | 89.34 | 91.21 | 93.11 | |
| FSU-PRGE | 87.79 | 87.04 | 86.71 | 87.33 | 87.2 | 81.69 | 85.09 | 86.03 | 87.26 | 89.7 | 91.51 | 92.88 | |
| IEPA | 90.68 | 88.28 | 87.08 | 90.11 | 88.89 | 91.01 | 84.67 | 86.6 | 88.01 | 89.21 | 89.35 | 90.76 | |
| BioInfer | 94.16 | 91.57 | 91.05 | 92.38 | 92.01 | 87.73 | 90.87 | 91.71 | 92.64 | 94.72 | 95.86 | 96.76 | |
| miRNA | 87.35 | 83.45 | 80.31 | 85.11 | 84.69 | 70.15 | 69.94 | 75 | 75.95 | 78.47 | 80.38 | 80.82 | |
| Diseases | NCBI Disease | 80.11 | 84.81 | 84.25 | 84.87 | 85.18 | 81.7 | 86.16 | 86.43 | 85.31 | 87.49 | 89.1 | 90.4 |
| CDR | 80.61 | 83.54 | 83.26 | 83.36 | 83.72 | 79.24 | 81.06 | 83.3 | 84.19 | 84.63 | 86.4 | 88.17 | |
| Variome | 78.42 | 85.42 | 84.68 | 85.17 | 85.28 | 83.13 | 83.42 | 85.12 | 84.51 | 86.21 | 87.62 | 88.89 | |
| Arizona Disease | 76.11 | 79.54 | 78.41 | 80.91 | 79.89 | 72.64 | 74.52 | 79.15 | 76.64 | 78.62 | 81.17 | 82.89 | |
| SCAI | 76.99 | 82.85 | 81.47 | 81.51 | 81.3 | 67.4 | 74.97 | 77.21 | 78.46 | 80.1 | 81.61 | 83.49 | |
| miRNA | 79.01 | 81.56 | 77.9 | 80.27 | 79.54 | 80.39 | 77.44 | 80.84 | 80.86 | 82.81 | 83.92 | 85.59 | |
| Species | S800 | 77.66 | 77.33 | 76.55 | 76.8 | 76.8 | 69.67 | 73.39 | 72.67 | 74.55 | 75.95 | 77.27 | 78.99 |
| CellFinder | 82 | 93.18 | 89.9 | 90.22 | 91.3 | 81.98 | 83.76 | 81.67 | 79.03 | 80.51 | 83.04 | 83.02 | |
| Variome | 32.17 | 82.98 | 81.25 | 83.33 | 83.33 | 50.46 | 66.27 | 62.92 | 59 | 61.05 | 62.22 | 64.71 | |
| LocText | 84.78 | 95.45 | 92.5 | 95.65 | 94.37 | 94.74 | 93.48 | 90.36 | 91.01 | 92.94 | 92.59 | 93.42 | |
| Linneaus | 87.4 | 98.26 | 96.21 | 97.73 | 97.63 | 93.23 | 91.54 | 92.49 | 93.57 | 95.27 | 96.38 | 96.99 | |
| miRNA | 72.54 | 92.67 | 85.06 | 94.19 | 94.16 | 91.67 | 89.29 | 86.13 | 87.88 | 92.95 | 94.59 | 94.29 | |
| Cell lines | CLL | 83.33 | 71.88 | 73.97 | 77.78 | 81.97 | 76.47 | 82.43 | 77.63 | 86.84 | 87.67 | 91.3 | 90.77 |
| Gellus | 93.18 | 93.6 | 86.25 | 93.55 | 93.42 | 70.89 | 85.64 | 90.23 | 89.53 | 91.46 | 91.61 | 94.56 | |
| CellFinder | 98.72 | 94.12 | 95.45 | 97.01 | 96.67 | 89.47 | 87.34 | 97.06 | 92.52 | 93.14 | 95.88 | 95.6 | |
| JNLPBA | 65.34 | 64.8 | 64.19 | 64.11 | 64.11 | 51.48 | 54.2 | 59.83 | 61.73 | 63.54 | 64.75 | 66.95 | |
| b) Recall scores | |||||||||||||
| Chemicals | CHEMDNER patent | 84.53 | 83.09 | 83.99 | 84.14 | 84.14 | 86.23 | 90.41 | 87.72 | 87.53 | 85.28 | 82.19 | 78.8 |
| CHEBI | 66.87 | 60.61 | 64.81 | 64.06 | 65.58 | 71.44 | 80.27 | 78.52 | 79 | 76.99 | 74.52 | 72.05 | |
| BioSemantics | 78.37 | 78.12 | 78.29 | 78.38 | 78.34 | 79.29 | 82.56 | 84.42 | 83.14 | 80.47 | 77.54 | 74.37 | |
| CHEMDNER | 85.08 | 78.27 | 80.51 | 81.58 | 81.8 | 79.8 | 84.96 | 83.17 | 85.45 | 83.36 | 80.54 | 77.48 | |
| CDR | 88.83 | 81.25 | 84.63 | 86.03 | 86.46 | 85.58 | 89.86 | 89.94 | 88.77 | 86.18 | 82.84 | 79.18 | |
| Genes/Proteins | CHEMDNER patent | 69.85 | 64.19 | 67.69 | 68.88 | 68.17 | 69.9 | 77.76 | 76.74 | 74.34 | 72.84 | 70.04 | 67.1 |
| BioCreative II GM | 75.21 | 67.97 | 69.69 | 70.83 | 70.89 | 72.46 | 77.72 | 78.16 | 78.13 | 76.19 | 73.9 | 71.78 | |
| JNLPBA | 72.77 | 73.31 | 73.93 | 74.96 | 75.14 | 75.32 | 79.11 | 79.22 | 79.82 | 77.62 | 75.04 | 72.27 | |
| CellFinder | 63.17 | 56.53 | 63.47 | 67.47 | 68.8 | 36 | 63.2 | 65.07 | 65.07 | 63.47 | 61.33 | 59.2 | |
| OSIRIS | 63.49 | 48.58 | 57.89 | 61.94 | 63.97 | 56.68 | 76.52 | 76.11 | 73.28 | 71.26 | 68.83 | 65.99 | |
| DECA | 58.24 | 55.12 | 57.7 | 61.25 | 61.09 | 67.68 | 73.45 | 66.82 | 66.16 | 64.08 | 62.21 | 59.78 | |
| Variome | 93.51 | 90.2 | 92.24 | 93.47 | 93.06 | 91.56 | 94.56 | 96.6 | 96.87 | 94.15 | 90.48 | 85.99 | |
| PennBioIE | 82.61 | 80.54 | 81.82 | 82.35 | 82.76 | 82.98 | 87.09 | 85.44 | 86.54 | 84.47 | 81.7 | 78.77 | |
| FSU-PRGE | 84.14 | 81.25 | 82.27 | 83.42 | 83.44 | 84.25 | 87.55 | 88.23 | 87.24 | 85.79 | 82.91 | 79.48 | |
| IEPA | 85.18 | 75.33 | 78.67 | 79 | 80 | 81 | 84.67 | 88.33 | 85.67 | 82.67 | 78.33 | 75.33 | |
| BioInfer | 89.44 | 88.42 | 89.98 | 90.06 | 90.14 | 90.06 | 92.72 | 91.78 | 89.59 | 87.01 | 83.41 | 79.5 | |
| miRNA | 72.93 | 42.76 | 54.77 | 56.54 | 58.66 | 49.82 | 80.57 | 79.51 | 78.09 | 75.97 | 73.85 | 69.96 | |
| Diseases | NCBI Disease | 79.69 | 76.62 | 79.74 | 80.69 | 81.59 | 75.86 | 83.06 | 82.92 | 83.58 | 81.45 | 78.56 | 75.3 |
| CDR | 78.36 | 73.21 | 76.47 | 79.15 | 79.17 | 76.98 | 83.15 | 83.38 | 82.79 | 81.54 | 78.87 | 76.01 | |
| Variome | 74.66 | 81.2 | 81.47 | 81.64 | 81.82 | 84.38 | 84.82 | 86.32 | 87.64 | 85 | 81.82 | 78.38 | |
| Arizona Disease | 59.3 | 57.64 | 62.63 | 64.75 | 64.3 | 65.05 | 70.8 | 70.65 | 72.47 | 70.65 | 69.14 | 66.72 | |
| SCAI | 70.6 | 48.45 | 59.6 | 61.02 | 62.01 | 56.07 | 76.55 | 70.34 | 70.48 | 68.22 | 65.82 | 63.56 | |
| miRNA | 79.41 | 65.05 | 73.18 | 71.11 | 71.97 | 70.93 | 78.37 | 76.64 | 75.26 | 73.36 | 70.42 | 67.82 | |
| Species | S800 | 69.08 | 55.79 | 60.09 | 59.72 | 60.93 | 62.9 | 64.95 | 69.35 | 69.81 | 67.57 | 65.14 | 62.9 |
| CellFinder | 87.85 | 77.36 | 83.96 | 78.3 | 79.25 | 85.85 | 92.45 | 92.45 | 92.45 | 89.62 | 87.74 | 83.02 | |
| Variome | 98.48 | 59.09 | 59.09 | 60.61 | 60.61 | 83.33 | 83.33 | 84.85 | 89.39 | 87.88 | 84.85 | 83.33 | |
| LocText | 86.66 | 68.48 | 80.43 | 71.74 | 72.83 | 78.26 | 93.48 | 81.52 | 88.04 | 85.87 | 81.52 | 77.17 | |
| Linneaus | 64.56 | 79.37 | 84.64 | 90.78 | 90.34 | 76.21 | 94.03 | 87.53 | 93.24 | 90.25 | 86.39 | 82.09 | |
| miRNA | 71.15 | 89.1 | 94.87 | 93.59 | 92.95 | 91.67 | 96.15 | 95.51 | 92.95 | 92.95 | 89.74 | 84.62 | |
| Cell lines | CLL | 77.92 | 59.74 | 70.13 | 63.64 | 64.94 | 67.53 | 79.22 | 76.62 | 85.71 | 83.12 | 81.82 | 76.62 |
| Gellus | 49.8 | 47.37 | 55.87 | 58.7 | 57.49 | 61.13 | 67.61 | 63.56 | 62.35 | 60.73 | 57.49 | 56.28 | |
| CellFinder | 60.63 | 25.81 | 33.87 | 52.42 | 46.77 | 41.13 | 55.65 | 79.84 | 79.84 | 76.61 | 75 | 70.16 | |
| JNLPBA | 56.95 | 55.31 | 57.13 | 59.14 | 58.47 | 63.44 | 67.94 | 66.7 | 64.98 | 63.54 | 61.34 | 59.9 | |
| c) F1-scores | |||||||||||||
| Chemicals | CHEMDNER patent | 83.53 | 83.8 | 84.14 | 84.28 | 84.22 | 83.58 | 86.14 | 85.48 | 85.38 | 84.97 | 84.12 | 82.9 |
| CHEBI | 69.09 | 69.18 | 71.03 | 70.63 | 71.94 | 71.51 | 75.41 | 75.63 | 73.95 | 74.03 | 73.67 | 73.29 | |
| BioSemantics | 75.35 | 80.1 | 80.15 | 80.12 | 80.11 | 79.96 | 82.32 | 82.01 | 81.99 | 81.42 | 80.55 | 79.27 | |
| CHEMDNER | 84.15 | 83.9 | 85.22 | 85.88 | 86.06 | 83.38 | 86.1 | 85.63 | 86.62 | 86.54 | 85.79 | 84.73 | |
| CDR | 89.01 | 85.87 | 87.92 | 88.83 | 89.31 | 86.87 | 90.48 | 91.05 | 90.63 | 90.2 | 88.93 | 87.25 | |
| Genes/Proteins | CHEMDNER patent | 68.4 | 65.57 | 67.51 | 68.26 | 67.65 | 65.17 | 70.22 | 70.81 | 70.05 | 69.96 | 69.17 | 68.18 |
| BioCreative II GM | 77.49 | 71.96 | 73.2 | 73.91 | 73.92 | 73.12 | 77.47 | 78.57 | 77.82 | 77.86 | 77.52 | 77.33 | |
| JNLPBA | 72.75 | 73.3 | 73.57 | 74.17 | 74.39 | 73.55 | 75.97 | 76.93 | 77.25 | 77.1 | 76.57 | 75.81 | |
| CellFinder | 71.35 | 64.24 | 69.69 | 74.85 | 75.55 | 49.36 | 71.39 | 73.16 | 75.08 | 74.84 | 73.95 | 73.03 | |
| OSIRIS | 72.06 | 59.85 | 66.05 | 70.34 | 70.54 | 64.22 | 76.52 | 76.42 | 75.57 | 75.37 | 74.73 | 73.42 | |
| DECA | 68.06 | 64.21 | 65.82 | 68.65 | 68.39 | 65.85 | 70.2 | 69.73 | 70.42 | 69.85 | 69.48 | 68.45 | |
| Variome | 91.88 | 90.14 | 90.88 | 91.66 | 91.57 | 89.55 | 90.97 | 91.32 | 91.93 | 91.72 | 90.6 | 88.58 | |
| PennBioIE | 85.27 | 84.17 | 84.94 | 85.78 | 86.04 | 83.14 | 86.8 | 86.71 | 86.75 | 86.84 | 86.19 | 85.34 | |
| FSU-PRGE | 85.92 | 84.05 | 84.43 | 85.33 | 85.27 | 82.95 | 86.3 | 87.12 | 87.25 | 87.7 | 87 | 85.66 | |
| IEPA | 87.84 | 81.29 | 82.66 | 84.19 | 84.21 | 85.71 | 84.67 | 87.46 | 86.82 | 85.81 | 83.48 | 82.33 | |
| BioInfer | 91.73 | 89.97 | 90.52 | 91.2 | 91.07 | 88.88 | 91.79 | 91.75 | 91.09 | 90.7 | 89.21 | 87.29 | |
| miRNA | 79.49 | 56.54 | 65.13 | 67.94 | 69.31 | 58.26 | 74.88 | 77.19 | 77 | 77.2 | 76.98 | 75 | |
| Diseases | NCBI Disease | 79.89 | 80.51 | 81.94 | 82.73 | 83.35 | 78.67 | 84.58 | 84.64 | 84.44 | 84.36 | 83.5 | 82.16 |
| CDR | 79.46 | 78.03 | 79.72 | 81.2 | 81.38 | 78.1 | 82.09 | 83.34 | 83.49 | 83.06 | 82.46 | 81.64 | |
| Variome | 76.49 | 83.26 | 83.04 | 83.37 | 83.51 | 83.75 | 84.11 | 85.71 | 86.05 | 85.6 | 84.62 | 83.3 | |
| Arizona Disease | 66.66 | 66.84 | 69.64 | 71.93 | 71.25 | 68.64 | 72.61 | 74.66 | 74.49 | 74.42 | 74.67 | 73.93 | |
| SCAI | 73.65 | 61.14 | 68.84 | 69.79 | 70.35 | 61.22 | 75.75 | 73.61 | 74.26 | 73.68 | 72.87 | 72.17 | |
| miRNA | 79.2 | 72.38 | 75.47 | 75.41 | 75.57 | 75.37 | 77.9 | 78.69 | 77.96 | 77.8 | 76.58 | 75.68 | |
| Species | S800 | 73.11 | 64.82 | 67.33 | 67.19 | 67.95 | 66.11 | 68.91 | 70.97 | 72.1 | 71.51 | 70.69 | 70.03 |
| CellFinder | 84.82 | 84.54 | 86.83 | 83.84 | 84.85 | 83.87 | 87.89 | 86.73 | 85.22 | 84.82 | 85.32 | 83.02 | |
| Variome | 48.49 | 69.03 | 68.42 | 70.18 | 70.18 | 62.86 | 73.83 | 72.26 | 71.08 | 72.05 | 71.79 | 72.85 | |
| LocText | 85.7 | 79.75 | 86.05 | 81.99 | 82.21 | 85.71 | 93.48 | 85.71 | 89.5 | 89.27 | 86.71 | 84.52 | |
| Linneaus | 74.26 | 87.81 | 90.05 | 94.13 | 93.84 | 83.86 | 92.77 | 89.94 | 93.4 | 92.7 | 91.11 | 88.92 | |
| miRNA | 71.83 | 90.85 | 89.7 | 93.89 | 93.55 | 91.67 | 92.59 | 90.58 | 90.34 | 92.95 | 92.11 | 89.19 | |
| Cell lines | CLL | 80.54 | 65.25 | 72 | 70 | 72.46 | 71.72 | 80.79 | 77.12 | 86.27 | 85.33 | 86.3 | 83.1 |
| Gellus | 64.91 | 62.9 | 67.81 | 72.14 | 71.18 | 65.65 | 75.57 | 74.58 | 73.51 | 72.99 | 70.65 | 70.56 | |
| CellFinder | 75.12 | 40.51 | 50 | 68.06 | 63.04 | 56.35 | 67.98 | 87.61 | 85.71 | 84.07 | 84.16 | 80.93 | |
| JNLPBA | 60.86 | 59.68 | 60.46 | 61.52 | 61.16 | 56.84 | 60.3 | 63.08 | 63.31 | 63.54 | 63 | 63.23 | |
CRF and LSTM-CRF stand for the CRF and the LSTM-CRF methods without word embedding features. The other ones represented by numbers are using one of the three word embeddings: (i) Patent, (ii) PubMed-PMC and (iii) Wiki-PubMed-PMC. The ones represented by percentage numbers indicate the performance of the method after removing a percentage of predicted entities with lower confidence values.
References
- Aerts S. et al. (2006) Gene prioritization through genomic data fusion. Nat. Biotechnol., 24, 537–544. [DOI] [PubMed] [Google Scholar]
- Akhondi S.A. et al. (2014) Annotated chemical patent corpus: a gold standard for text mining. PloS One, 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagewadi S. et al. (2014) Detecting miRNA mentions and relations in biomedical literature. F1000Research, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Navarro R. et al. (2015) Optimising chemical named entity recognition with pre-processing analytics, knowledge-rich features and heuristics. J. Cheminform., 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos D. et al. (2012) Theory and Applications for Advanced Text Mining, Chapter Biomedical Named Entity Recognition: A Survey of Machine-Learning Tools. INTECH Open Access Publisher, Rijeka, Croatia, pp. 175–195. [Google Scholar]
- Campos D. et al. (2013) Gimli: open source and high-performance biomedical name recognition. BMC Bioinformatics, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalapathy R. et al. (2016. a) Bidirectional LSTM-CRF for clinical concept extraction. In Proceedings of the Clinical Natural Language Processing Workshop, Osaka, Japan, pp. 7–12.
- Chalapathy R. et al. (2016b) An investigation of recurrent neural architectures for drug name recognition. In Proceedings of the Seventh International Workshop on Health Text Mining and Information Analysis, Austin, TX.
- Culotta A., McCallum A. (2004) Confidence estimation for information extraction. In Proceedings of NAACL-HLT, Boston, MA, pp. 109–112.
- Dai A.M., Le Q.V. (2015) Semi-supervised sequence learning. In Advances in Neural Information Processing Systems, Montreal, Canada, pp. 3079–3087.
- Ding J. et al. (2002) Mining MEDLINE: abstracts, sentences, or phrases. In Proceedings of the Pacific Symposium on Biocomputing, Lihue, Hawaii, USA, pp. 326–337. [DOI] [PubMed]
- Doğan R.I. et al. (2014) NCBI disease corpus: a resource for disease name recognition and concept normalization. J. Biomed. Inform., 47, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltyeb S., Salim N. (2014) Chemical named entities recognition: a review on approaches and applications. J. Cheminform., 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong L.I. et al. (2008) OSIRISv1. 2: a named entity recognition system for sequence variants of genes in biomedical literature. BMC Bioinformatics, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner M. et al. (2010) LINNAEUS: a species name identification system for biomedical literature. BMC Bioinformatics, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg T. et al. (2015) Linked annotations: a middle ground for manual curation of biomedical databases and text corpora. BMC Proc., 9, 1–3. [Google Scholar]
- Graves A., Schmidhuber J. (2005) Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw., 18, 602–610. [DOI] [PubMed] [Google Scholar]
- Gurulingappa H. et al. (2010). An empirical evaluation of resources for the identification of diseases and adverse effects in biomedical literature. In Proceedings of the 2nd Workshop on Building and evaluating resources for biomedical text mining, Valletta, Malta, pp. 15–22.
- Habibi M. et al. (2016) Recognizing chemicals in patents - a comparative analysis. J. Cheminform., 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn U. et al. (2010). A proposal for a configurable silver standard. In Proceedings of the 4th Linguistic Annotation Workshop at ACL, Uppsala, Sweden, pp. 235–242.
- Hastie T. et al. (2001) The Elements of Statistical Learning, Vol. 1 Springer series in statistics Springer, Berlin. [Google Scholar]
- Hettne K.M. et al. (2009) A dictionary to identify small molecules and drugs in free text. Bioinformatics, 25, 2983–2991. [DOI] [PubMed] [Google Scholar]
- Hochreiter S., Schmidhuber J. (1997) Long short-term memory. Neural Comput., 9, 1735–1780. [DOI] [PubMed] [Google Scholar]
- Jozefowicz R. et al. (2016). Exploring the limits of language modeling. arXiv preprint arXiv:1602.02410v2.
- Kaewphan S. et al. (2016) Cell line name recognition in support of the identification of synthetic lethality in cancer from text. Bioinformatics, 32, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-D. et al. (2004). Introduction to the bio-entity recognition task at JNLPBA. In Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications, Geneva, Switzerland, pp. 70–75.
- Krallinger M. et al. (2013) Overview of the chemical compound and drug name recognition (CHEDNER) task. In BioCreative Challenge Evaluation Workshop, Washington, DC, vol. 2, pp. 2–33.
- Krallinger M. et al. (2015a) The CHEMDNER corpus of chemicals and drugs and its annotation principles. J. Cheminform., 7, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M. et al. (2015b) Overview of the CHEMDNER patents task. In Proceedings of the 5th BioCreative Challenge Evaluation Workshop, Sevilla, Spain, pp. 63–75.
- Kulick S. et al. (2004) Integrated annotation for biomedical information extraction. In Proceedings of NAACL-HLT, Boston, MA, pp. 61–68.
- Lafferty J. et al. (2001) Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In Proceedings of the Eighteenth International Conference on Machine Learning, ICML, pp. 282–289.
- Lample G. et al. (2016) Neural architectures for named entity recognition. In Proceedings of NAACL-HLT, San Diego, CA, pp. 260–270.
- Leaman R., Gonzalez G. (2008) BANNER: an executable survey of advances in biomedical named entity recognition. In Proceedings of the Pacific Symposium on Biocomputing, Big Island, Hawaii, USA, pp. 652–663. [PubMed]
- Leaman R., Lu Z. (2016) TaggerOne: Joint named entity recognition and normalization with Semi-Markov models. Bioinformatics, 2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman R. et al. (2009). Enabling recognition of diseases in biomedical text with machine learning: corpus and benchmark. In Proceedings of the Symposium on Languages in Biology and Medicine, Seogwipo-si, Jeju Island, South Korea, pp. 82–89.
- Leaman R. et al. (2013) DNorm: disease name normalization with pairwise learning to rank. Bioinformatics, 29, 2909–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman R. et al. (2015) tmChem: a high performance approach for chemical named entity recognition and normalization. J. Cheminform., 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser U., Hakenberg J. (2005) What makes a gene name? Named entity recognition in the biomedical literature. Brief. Bioinform., 6, 357–369. [DOI] [PubMed] [Google Scholar]
- Li G. et al. (2015) miRTex: A text mining system for miRNA-gene relation extraction. PLoS Comput. Biol., 11, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. (2016) BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. (2015a) Drug name recognition: approaches and resources. Information, 6, 790–810. [Google Scholar]
- Liu S. et al. (2015b) Effects of semantic features on machine learning-based drug name recognition systems: word embeddings vs. manually constructed dictionaries. Information, 6, 848–865. [Google Scholar]
- Mackin R. (1978) On collocations: Words shall be known by the company they keep. In Honour of as Hornby, pp. 149–165.
- Narayanaswamy M. et al. (2003) A biological named entity recognizer. In Proceedings of the Pacific Symposium on Biocomputing, Lihue, Hawaii, USA, pp. 427–438. [DOI] [PubMed]
- Neves M. et al. (2012) Annotating and evaluating text for stem cell research. In Proceedings of the 3rd Workshop on Building and Evaluation Resources for Biomedical Text Mining at Language Resources and Evaluation, Istanbul, Turkey, pp. 16–23.
- Okazaki N. (2007) CRFsuite: a fast implementation of conditional random fields (CRFs).
- Pafilis E. et al. (2013) The SPECIES and ORGANISMS resources for fast and accurate identification of taxonomic names in text. PLoS One, 8, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascanu R. et al. (2014) How to construct deep recurrent neural networks. In Proceedings of the 2nd International Conference on Learning Representations, Banff, Canada.
- Pyysalo S. et al. (2007) BioInfer: a corpus for information extraction in the biomedical domain. BMC Bioinformatics, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyysalo S. et al. (2013) Distributional semantics resources for biomedical text processing. In Proceedings of the 5th International Symposium on Languages in Biology and Medicine, Tokyo, Japan.
- Rabiner L.R. (1989) A tutorial on hidden markov models and selected applications in speech recognition. Proceedings of the IEEE, Vol. 77, pp. 257–286. [Google Scholar]
- Rocktäschel T. et al. (2012) ChemSpot: a hybrid system for chemical named entity recognition. Bioinformatics, 28, 1633–1640. [DOI] [PubMed] [Google Scholar]
- Segura-Bedmar I. et al. (2015). Combining conditional random fields and word embeddings for the CHEMDNER-patents task. In Proceedings of the 5th BioCreative challenge evaluation workshop, Sevilla, Spain, pp. 90–93.
- Settles B. (2005) ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics, 21, 3191–3192. [DOI] [PubMed] [Google Scholar]
- Smith L. et al. (2008) Overview of BioCreative II gene mention recognition. Genome Biol., 9, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenetorp P. et al. (2012). Size (and domain) matters: Evaluating semantic word space representations for biomedical text. In Proceedings of the 5th International Symposium on Semantic Mining in Biomedicine, Zurich, Switzerland, pp. 42–49.
- Tang B. et al. (2014) Evaluating word representation features in biomedical named entity recognition tasks. BioMed Res. Int., 2014, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole U. et al. (1979) On the suitability of minimum and product operators for the intersection of fuzzy sets. Fuzzy Sets Syst., 2, 167–180. [Google Scholar]
- Tjong Kim Sang E.F., De Meulder F. (2003). Introduction to the CoNLL-2003 shared task: Language-independent named entity recognition. In Proceedings of the 7th conference on Natural language learning at HLT-NAACL, Edmonton, Canada, pp. 142–147.
- Uzuner Ö. et al. (2011) 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Inform. Assoc., 18, 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspoor K. et al. (2013) Annotating the biomedical literature for the human variome. Database 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. (2010) Disambiguating the species of biomedical named entities using natural language parsers. Bioinformatics, 26, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-Y., Zhang H.-Y. (2013) Rational drug repositioning by medical genetics. Nat. Biotechnol., 31, 1080–1082. [DOI] [PubMed] [Google Scholar]
- Wei C.-H. et al. (2015) Overview of the BioCreative V chemical disease relation (CDR) task. In Proceedings of the 5th BioCreative challenge evaluation workshop, Sevilla, Spain, pp. 154–166.
- Zhou X. et al. (2014) Human symptoms-disease network. Nat. Commun., 5. [DOI] [PubMed] [Google Scholar]