Abstract
Pif1 family helicases have multiple roles in maintenance of nuclear and mitochondrial DNA in eukaryotes. S. cerevisiae Pif1 is involved in replication through barriers to replication such as G-quadruplexes and protein blocks and reduces genetic instability at these sites. Another Pif1 family helicase in S. cerevisiae, Rrm3, assists in fork progression through replication fork barriers at the rDNA locus and tRNA genes. ScPif1 also negatively regulates telomerase, facilitates Okazaki Fragment processing, and acts with polymerase δ in break induced repair. Recent crystal structures of bacterial Pif1 helicases and the helicase domain of human PIF1 combined with several biochemical and biological studies on the activities of Pif1 helicases have increased our understanding of the function of these proteins. This review article focuses on these structures and the mechanism(s) proposed for Pif1’s various activities on DNA.
Helicases are a family of molecular motors involved in all phases of nucleic acids metabolism including DNA replication, recombination, repair, transcription, translation, and RNA processing [1–3]. The Pif1 family is a group of DNA helicases that have been identified in all eukaryotes and some prokaryotes and viruses [4]. Although their functions in bacteria and viruses are not well known, the role of Pif1 helicases in eukaryotic genome maintenance is better understood. S. cerevisiae contains two Pif1 family helicases: Pif1 (referred to as ScPif1 here) and Rrm3 [5]. However, most higher eukaryotes encode only one Pif1 helicase [4]. ScPif1 is required for maintenance of mitochondrial DNA [6–8] and has been suggested to be the mitochondrial replicative helicase [9]. In the nucleus, ScPif1 is involved in Okazaki Fragment processing [10,11], regulation of telomerase at telomeres and double strand breaks (DSBs) [12,13], maintenance of the rDNA barrier [14], break induced repair [15,16], and removal of protein barriers to replication [17] and at telomeres [18]. ScPif1 has also been shown to reduce genomic instability at G-quadruplex DNA (G4DNA) motifs in the CEB1 minisattelite [19]. For more information about the function of Pif1 in cells we direct the reader to several recent reviews on the subject [9,20–23].
Structure
Pif1 family helicases are classified as superfamily (SF) 1B helicases based on amino acid sequence motifs and 5′-to-3′ direction of translocation on single-stranded DNA (ssDNA). Pif1 family helicases are composed of a central helicase domain and N-terminal and C-terminal accessory domains. However, their similarity is limited to the core helicase domain [5]. The variable N-terminal domains are not required for helicase activity in vitro [24]; however, the Rrm3 N-terminal domain is required for function in vivo [25]. Although the precise functions of the N- and C-terminal domains are unknown, non-conserved regions are proposed to be involved in protein-protein interactions, oligomerization, and substrate recognition [26]. Indeed, the N-terminal domain of ScPif1 reportedly interacts with the chromatin assembly factor I subunit, Cac1 [27] and is required for increased double stranded DNA (dsDNA) unwinding activity by ScPif1 when the yeast mitochondrial ssDNA binding protein, Rim1, is present [28]. For human PIF1, the N-terminal domain has been proposed to be involved in strand annealing [29]. Deletion of the C-terminal domain decreases the processivity of unwinding for ScPif1 [30]. Additionally, phosphorylation of the C-terminal domain of ScPif1 on T763 and S766 in response to DSBs is required for ScPif1 inhibition of telomerase at DSBs but not at telomeres [31].
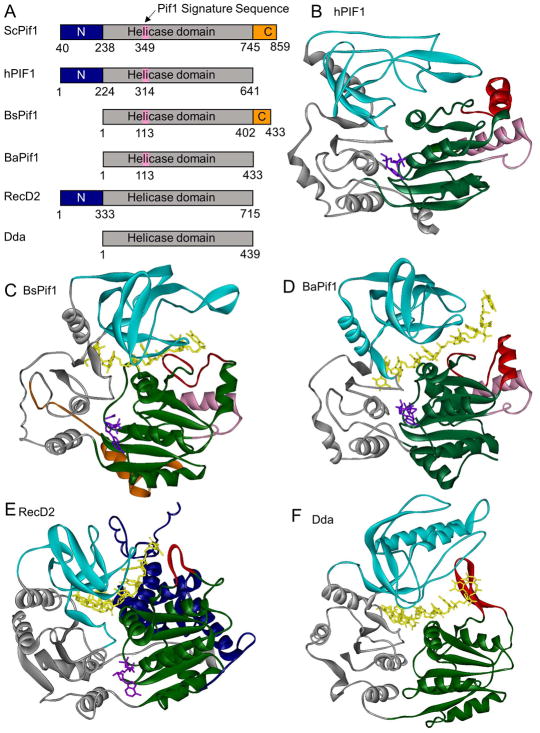
The crystal structures of BsPif1 from Bacteroides spp [32], BaPif1 from Bacteroides sp. 2-1-16 [33], and the human PIF1 helicase domain (hPIF1-HD) [33] have been reported (Figure 1). Pif1 helicases, like other SF1B helicases such as Deinococcus radiodurans RecD2 [34] and bacteriophage T4 Dda [35], contain two RecA-like domains referred to as domains 1A (green) and 2A (gray) in their catalytic core where the helicase motifs are clustered. Domains 1A and 2A are separated by a cleft where ATP (purple) binding and hydrolysis occurs. The 2B domain of Pif1, RecD2, and Dda helicases forms a SH3-like domain (cyan). The 1B (red) domains of RecD2 and Dda are composed of β-hairpins. In BsPif1, the 1B domain forms an ordered loop, and in both BaPif1 and hPif1-HD, the 1B domain is formed by a loop and an α-helix. In all cases, the 1B domain is proposed to form a pin or wedge that splits the incoming duplex with the translocase strand passing into the active site on one side of the pin/wedge and the displaced strand separated and passed along the exterior of the enzyme [34,35].
Figure 1. Structures of superfamily 1B helicases.
(A) Domain maps of SF1B helicases. N-terminal domains are shown in blue, helicase domains in gray, C-terminal domains in orange, and the Pif1 signature sequence in pink. (B) Structure of hPIF1-HD (PDB: 5FHH) [33]. Domain 1A is shown in green, 1B in red, 2A in gray, 2B in cyan, and the Pif1 signature sequence in pink. ADP-AlF4− is shown in purple. (C) Structure of BsPif1 (PDB: 5FTE) [32]. Colors are the same as in B with the C-terminal domain in orange, AMPPNP in purple, and DNA is in yellow. (D) Structure of BaPif1 (PDB: 5FHD) [33]. Colors are as in B with DNA shown in yellow. (E) Structure of D. radiodurans RecD2 (PDB: 3GPL) [34]. Colors are the same as in B with the N-terminal domain in blue, AMPPNP in purple, and DNA in yellow. (F) Structure of Bacteriophage T4 Dda (PDB: 3UPU) [35]. Colors are as in B with DNA in yellow.
Superfamily 1B helicases contain 12 conserved helicase motifs in their RecA-like domains which are involved in ATP binding and hydrolysis, DNA binding, and coordination between the ATP and DNA binding sites [36]. Residues in motifs I, III, IV, and VI contact the ATP in the BsPif1 and BaPif1 structures [32,33]. In BsPif1, Q145 from motif IV (and the analogous Q499 in RecD2) have been proposed to act as sensors to detect the presence or absence of the γ-phosphate [32,34]. The DNA (yellow) passes between the RecA-like domains and the 2B domain. In both the BsPif1 and BaPif1 structures, the proteins contact the 6 nucleotides visible in the structures through stacking interactions with the bases and hydrogen bonding and electrostatic interactions with the backbone. F75 and P74 in the 1B domain (pin-loop or wedge domain) of both BaPif1 and BsPif1 stack with bases of the DNA [32,33]. Similar stacking interactions were observed between F98 in the 1B (pin) domain of Dda and the DNA [35]. Characterization of several Pif1 variants with single amino acid substitutions in the ATPase and DNA binding sites [32,33] confirms the roles previously established for these motifs based on studies of other helicases [37]. The DNA in the structure of BaPif1 with bound DNA and nucleotide makes a 90° turn as it enters the DNA binding site [33]. Similar turns have been observed for the SF1A helicases PcrA [38] and UvrD [39]. As has been observed with other helicases, binding of ATP and ssDNA results in large conformational changes in both BsPif1 and BaPif1 with the 2A and 2B domains rotating and shifting relative to the 1A domain. Introduction of a disulfide bond that prevents this rotation in BsPif1 or introduction of proline residues in the hinge between the 2A and 2B domains of BaPif1 abolishes dsDNA unwinding activity [32,33].
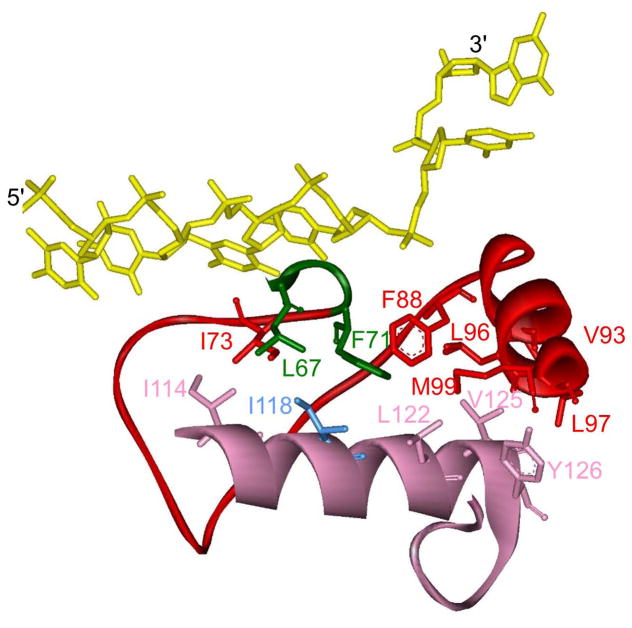
Pif1 family helicases also contain a 21 amino acid Pif1 signature sequence located between helicase motifs II and III [40]. Structurally, the Pif1 signature sequence (Figures 1 and 2; pink) is composed of an α-helix and a turn in all three reported Pif1 structures [32,33] and is located at the entrance to the DNA binding site, opposite the strand separation pin/wedge. Hydrophobic interactions between the Pif1 signature sequence and domains 1A and 1B maintain F71 in domain 1A, which is essential for enzymatic activity, in the appropriate position to separate the DNA strands [32]. L319 in hPIF1 (I118 in BaPif1 in Figure 2, blue), within the Pif1 signature sequence, is conserved as Leu or Ile among Pif1 family helicases, and an L319P variant has been linked with an increased risk of breast cancer [41]. This variant lacks DNA unwinding activity, likely due to destabilization of interactions of the Pif1 signature sequence with domains 1A and 1B [33]. This indicates that the function of the Pif1 signature sequence may be to maintain the appropriate arrangement of residues in the DNA binding site for DNA unwinding. The localization of the Pif1 signature sequence at the entrance to the DNA binding site also suggests this sequence may be involved in substrate specificity although there is no evidence for this.
Figure 2. Structure of the Pif1 signature sequence.
Hydrophobic amino acids in the BaPif1 (PDB: 5FHD) [33] signature sequence (pink) interact with amino acids in domains 1A (green) and 1B (red) stabilizing this region of the protein. Ile118 which is essential for helicase activity [33] and corresponds to L319 in hPIF1 is shown in blue. DNA is shown in yellow.
Quaternary structure
An important feature of helicase enzymology is the quaternary structure of the enzyme. Although monomeric forms of some helicases can function, [30,42–48] multimeric forms can provide multiple DNA binding sites which typically increases processivity and can provide additional avenues for regulation of activity. Dimeric helicases have been described in which the conformations of each respective monomer can activate or inhibit the enzyme [48,49]. Helicases that are part of the main replication machinery are often hexameric, with a central channel through which DNA can pass [50]. Another mode of action available to some helicases in SF1 and SF2 involves multiple helicase monomers aligning on the same strand of nucleic acid to enhance activity (so-called helicase trains) [44,51–54]. Interactions of monomers with other proteins can also result in enhanced activity [28,55,56].
Pif1’s quaternary structure has been investigated carefully by the Galletto lab. Initial reports indicated that upon binding to ssDNA, ScPif1 formed a dimer [57]. Additional, extensive characterization followed and clearly showed that monomeric ScPif1 can indeed function to translocate on ssDNA [58] and unwind duplex DNA [30]. One revealing experiment involved attachment of ScPif1 to a magnetic bead, followed by extensive washing to ensure only monomeric ScPif1 remained on the bead. The resulting measures of DNA unwinding showed that ScPif1 could function as a monomer. Kinetic analysis using a novel FRET-based assay illustrated that the activity is modulated by the composition of the 3′-ssDNA tail of a forked substrate [30], suggesting that ScPif1 contains a second DNA binding site in addition to the translocation site [59,60]. Although some reports have suggested that a monomer of ScPif1 is unable to unwind DNA:DNA duplexes but can unwind DNA:RNA duplexes [61], the Galletto lab has shown that ScPif1 monomers can unwind forked dsDNA substrates [30].
Biochemical Activity
ScPif1 preferentially unwinds forked dsDNA substrates relative to substrates lacking a 3′-ssDNA overhang [59,60]. The rates of unwinding forked and non-forked duplexes are similar [60]; however, the processivity for unwinding is enhanced with forked duplexes [59,60]. ScPif1 has also been shown to preferentially unwind an DNA:RNA hybrid duplex where the translocase strand is DNA and the displaced strand is RNA [61,62]. It has been shown recently that this preference is due to enhanced processivity on the DNA:RNA hybrid substrates [63].
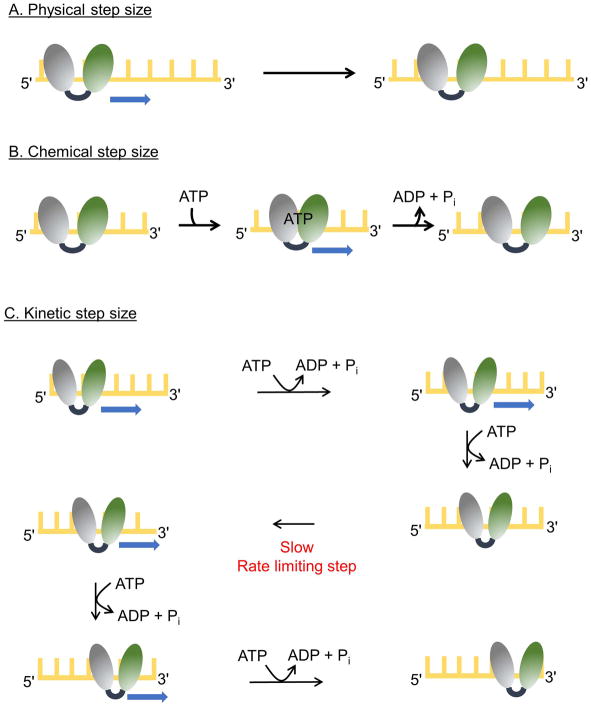
Pif1 helicases unwind duplex DNA in the 5′-to-3′ direction using the energy of ATP hydrolysis [58,59]. This directional unwinding is likely due to unidirectional translocation of ScPif1 on ssDNA [58,60]. This movement occurs in discrete steps [38] referred to as the physical step size of the helicase (Figure 3A). A physical step size of 1 nucleotide translocated can be inferred from structural studies [32,33]. In the simplest case, each single nucleotide step is driven by the energy of hydrolysis of one ATP molecule (chemical step size) (Figure 3B). Indeed, chemical step sizes of one ATP hydrolyzed per nucleotide translocated have been measured for many helicases, including ScPif1 [60], B. stearothermophilus PcrA [64], E. coli DnaB [65], E. coli UvrD [66], and Hepatitis C virus NS3-4A [67]. However, for some helicases, such as Hepatitis C Virus NS3h, coupling of ATP hydrolysis to translocation is inefficient, resulting in a chemical step size of less than one nucleotide [67]. A third measurement, the kinetic step size (Figure 3C), is the number of base pairs separated between rate limiting kinetic steps. The kinetic step size for ScPif1 for translocation on ssDNA and dsDNA unwinding have both been determined to be one nucleotide [60,61,68]. For ScPif1, all three step sizes are one [60] but larger kinetic step sizes have been observed for many helicases [46,69–72].
Figure 3. Step sizes of for helicases.
The two RecA-like domains of a helicase are shown (gray and green) and the yellow lattice represents DNA. (A) A helicase with a physical step size of one nucleotide is depicted. The enzyme moves by one nucleotide in each step. (B) A helicase with chemical and physical step sizes of one nucleotide is shown. Binding of ATP results in movement of the rear domain forward by one nucleotide closing the cleft between the two RecA-like domains. Release of ADP and Pi results in movement of the leading domain forward by one nucleotide as the cleft opens. (C) The scheme illustrates movement by a helicase with physical and chemical step sizes of one nucleotide and a kinetic step size of 2 nucleotides. The enzyme moves in single nucleotide steps, each powered by the hydrolysis of a single ATP molecule. After two forward steps, a slow kinetic step occurs which limits the overall rate, resulting in an observed step size of 2 nucleotides. After the slow kinetic step, ATP hydrolysis and forward movement resume.
ScPif1 unwinds duplex DNA at a rate of about 75–100 bp/s under excess enzyme conditions [30,60,63,68] and as a monomer [30]. However, according to another report a monomer of ScPif1 was unable to unwind and excess ScPif1 unwound a non-forked DNA duplex at a much slower rate [61]. This discrepancy remains unresolved. ScPif1 unwinds 10 base pairs of a forked DNA duplex on average before dissociation but translocates 25 nucleotides on ssDNA before dissociation [60].
Role of Pif1 in replication through barriers
Genomic DNA is known to be wrapped around histones or bound to other proteins such as the shelterin complex at telomeres. Removal of proteins bound to DNA is a prerequisite for replication, repair, transcription, and essentially all metabolic events associated with DNA. Helicases are known to perform this function [73,74]. ScPif1 can displace telomerase from telomeric DNA thereby regulating telomere length [12,13]. Further, ScPif1 removes telomerase from the ends of DSBs [12,75], which reduces genomic instability by preventing telomerase from adding telomeric DNA to these sites. Recent work from the Galletto lab supports the ability of monomeric ScPif1 to remove protein blocks from dsDNA such as the Rap1 protein [17]. ScPif1 also increases replication through a telomeric protein barrier in vivo [18]. Rrm3, and in its absence ScPif1, displace transcription complexes and resolve R-loops at tRNA genes [76,77]. Rrm3 also displaces Fob1, allowing replication through the rDNA replication fork barrier [14,78] while ScPif1 maintains the replication fork barrier [14]. The specific mechanism for protein displacement is unknown, but clearly the ATPase-driven translocase activity is required [17]. As a model system, streptavidin attached to biotinylated DNA can serve as a protein block [79]. ScPif1 has been shown to readily displace streptavidin [58,60].
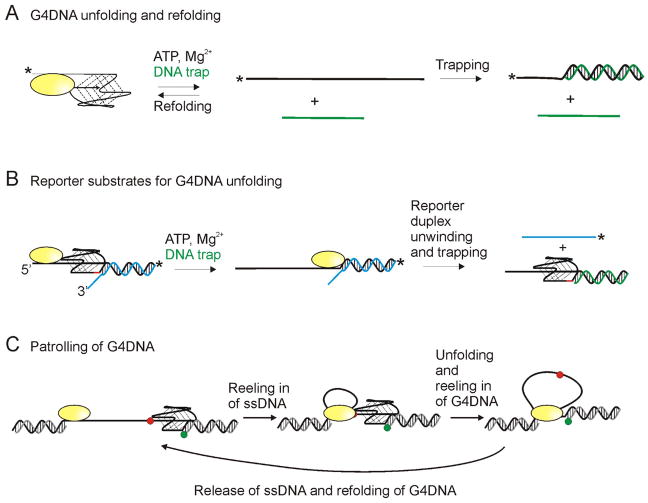
Alternative DNA structures such as G4DNA can also be barriers to replication and ScPif1 has been shown to localize to and reduce genome instability at these sites [19,80,81]. However, the absence of ScPif1 was later shown to have no effect on fork progression past a G4DNA motif [18]. ScPif1 has been shown to localize to and unfold G4DNA structures late in S phase, after replication of the G4DNA sequence, suggesting that it may be involved in resolving G4DNA structures in preparation for mitosis but not for replication [80] which may explain this apparent discrepancy. ScPif1 has also been shown to unfold G4DNA in vitro [19,61,68,81–83]. Some have reported very fast unfolding of G4DNA structures by ScPif1 (within a few seconds) and others report much slower G4DNA unfolding (within a few minutes). These differences are possibly the result of rapid refolding of the G4DNA thereby decreasing product formation in ensemble experiments (Figure 4A). It is also possible that in some smFRET experiments, partial melting of G4DNA structures is observed, which may explain a major discrepancy in folding rates of G4DNA. Measurements of G4DNA unfolding are affected by the rate of G4DNA folding. The consensus is that folding of G4DNA structures proceeds slowly to the thermodynamic equilibrium; however, fast folding to various intermediate occurs [84,85]. These reported folding rates differ from those reported in smFRET studies, which indicate very rapid folding [61,86]. It is possible that that the rapid conformational changes observed in smFRET are between unfolded and intermediate states or the fully folded and intermediates states as opposed to fully folded and fully unfolded states. It should be noted that many of the intermediates observed during folding are G4DNA structures; therefore, transitions between unfolded DNA and an intermediate could be conversions of ssDNA to G4DNA even if it is not the most thermodynamically stable G4DNA structure. It is also possible the presence of the helicase leads to much faster folding than observed with G4DNA alone, which could explain the rapid transitions observed in the smFRET. Due to these complications, the relative rates of melting dsDNA and G4DNA are not clear; however, the ATP hydrolysis rate on G4DNA is slower than the rate of hydrolysis on ssDNA [82] and slower than the rates of translocation on ssDNA and unwinding of dsDNA [60,82].
Figure 4. Substrates utilized for G4DNA unfolding studies.
(A) Some experiments for measuring G4DNA unfolding rely on trapping the unfolded duplex as dsDNA to prevent refolding of the G4DNA structure. Experiments typically start with the enzyme prebound to the G4DNA substrate and unfolding is initiated by addition of ATP, Mg2+, and a DNA trap (green) complementary to the G4DNA. G4DNA substrate and dsDNA product can be separated by electrophoresis. * indicates either a fluorescent or radiolabel. Since refolding is a unimolecular process and trapping is a bimolecular process, trapping can be inefficient. (B) Alternatively, G4DNA unfolding can be monitored by measuring the unwinding of a reporter duplex following the G4DNA. Appearance of ssDNA provides a “report” on the unfolding of the G4DNA. A gap of 2 nucleotides (red) between the G4DNA and reporter duplex is necessary for folding of both the G4DNA and dsDNA to occur [83]. (C) Based on smFRET studies, some helicases have been reported to remain anchored at a ssDNA/dsDNA junction while repetitively reeling in the ssDNA and unfolded G4DNA. Green and red dots represent Cy3 and Cy5.
Several unresolved observations related to G4DNA unfolding by Pif1 remain. Several groups have utilized substrates containing G4DNA followed by dsDNA so that unwinding of the duplex can serve as a reporter for unfolding of the quadruplex (Figure 4B) [68,82,83,87]. The Xi lab reported that G4DNA stimulates dsDNA unwinding [68,87]. However, no other groups observed similar stimulation despite a variety of methods and conditions [61,82,83]. Of note, the experiments from the Xi lab were preformed exclusively in Na+, resulting in a less stable quadruplex which may give the appearance of faster activity. Another possible difference is the substrates from the Xi lab contained no space between the G4DNA and the dsDNA (Figure 4B) which results in destabilization of the G4DNA. A gap of 2 nucleotides between the G4DNA and dsDNA reporter is needed in order for both structures to fold properly [83]. Hence, it remains to be determined whether ScPif1 is actually stimulated by G4DNA.
Some researchers have attempted to compare G4DNA unfolding to dsDNA unwinding leading to statements that G4DNA is unfolded more efficiently than dsDNA by ScPif1. Enzyme efficiency is typically measured as kcat/KM. However, the two substrates (dsDNA and G4DNA) are extremely different making this comparison difficult. Duplexes can vary in length and GC content which affect the stability; G4DNA can vary in both structure and stability, and both can have different numbers of nucleotides that must be traversed by the enzyme before the structures spontaneously melt. A thorough examination of the free energy for duplex stability vs helicase catalyzed unwinding rate was reported by the Patel lab for NS3 and T7 helicases, and they found that the unwinding rates increased as the stability of the duplex decreased [88]. However, Dda, which is in the same family as Pif1, was observed to show little correspondence between unwinding rate and GC content of the duplex [89], suggesting that the effect of duplex stability on the unwinding rate varies between helicases. Comparisons of dsDNA unwinding and G4DNA unfolding using structures with similar free energies of melting would be a useful comparison; however, without knowledge of how many nucleotides must be removed from each structure for spontaneous melting to occur, the comparison of duplex and quadruplex melting will still be difficult. Also, comparisons of G4DNA unfolding relative to dsDNA unwinding can be complicated by refolding of G4DNA and re-annealing of the ssDNA products of dsDNA unwinding [30,90]. Due to these complications, it is premature at this point to conclude that G4DNA is melted more efficiently than dsDNA by ScPif1.
ScPif1 has also been described as having patrolling activity in which it binds to a 3′-ssDNA tailed dsDNA junction and repetitively reels in the tail in single nucleotide steps [61,86] (Figure 4C). Because the helicase remains anchored at the 3′-ssDNA junction, a loop of ssDNA is created as the helicase reels in the tail as it translocates toward the 3′-end of the DNA. Similar repetitive activity has been observed previously for E. coli Rep [91], E. coli UvrD [92], B. stearothermophilus PcrA [93], S. cerevisiae Srs2 [94], and human FANCJ [95], RHAU [96], WRN [96], and BLM [96,97] helicases. Such an activity could have biological significance in clearing DNA of proteins and secondary structures. This action is proposed to occur repetitively (about 200 times to unwind a 31 base pair DNA:RNA hybrid duplex [61]). Multiple groups found that G4DNA was unfolded in each patrolling cycle, but dsDNA downstream of the G4DNA was not melted which suggested a mechanism whereby ScPif1 repetitively unfolds the G4DNA which refolds rapidly but leaves adjacent dsDNA intact [61,86]. However, other groups have reported that ScPif1 is able to unwind a duplex following a quadruplex [68,82,83]. The differences are likely due to the substrate design and experimental conditions because some reports used non-forked duplexes [61,68,83,86] which are known to be non-preferred substrates for ScPif1 while another used both forked and non-forked duplexes [82]. Some experiments were performed under single-cycle conditions [61,68,82,86] and others under multiple turnover conditions [82,83]. Some used excess enzyme [68,82] and others used excess substrate [61,83,86]. It is likely that the differences in observed results are due to these differences in experimental conditions. Whether ScPif1 unwinds a dsDNA duplex downstream of G4DNA and/or patrols the DNA in a cell remains to be determined.
Standardization of experimental conditions would allow for easier comparisons of results from different laboratories. We suggest that since K+ is the predominant monovalent cation in cells and G4DNA stability is affected by the identity of the monovalent cation, experiments should be performed in K+ (≥ 50 mM) as opposed Na+. The variability of results when studying different G4DNA structures combined with the ability of naturally occurring G4DNA sequences to fold into different structures suggests that experiments should be performed using multiple quadruplex sequences when possible. It is well established that Pif1 prefers forked duplexes so duplex substrates should contain these structures. Since both ScPif1 [98] and hPIF1 [99] are low abundance proteins, excess substrate conditions would seem to be more biologically relevant. However low abundance is no guarantee of monomeric protein in vivo so both excess enzyme and excess substrate conditions could be relevant. In general, it would appear that the best method to monitor G4DNA unfolding is smFRET. However, placement of the probes in multiple positions on the substrate is needed to confirm the interpretation of the results. Furthermore, it would be useful to supplement smFRET with gel-based experiments.
Regulation of Pif1 activity
Although ScPif1 serves an important role in genome maintenance, overexpression of ScPif1 is toxic [100] so its activity must be regulated. DNA damage signaling in response to DSBs activates a series of kinases including Rad53 in yeast which results in cell cycle arrest and DNA repair [101]. De novo telomere synthesis at DSBs by telomerase can lead to chromosomal deletions [102] but ScPif1 has been shown to inhibit this telomere addition [12,13]. The Blackburn lab has shown that DSBs, but not stalled replication forks, result in Rad53 mediated phosphorylation of the C-terminus of ScPif1 at T763 and S766 which inhibits telomerase specifically at DSBs [31]. Both ScPif1 and Rrm3 are also phosphorylated in response to replication fork stalling in the N-terminal domain by Rad53 [103]. These phosphorylations inhibit ScPif1 and Rrm3 which prevents fork reversal and genome instability.
Unanswered questions
Recent data has provided much insight into the activities of Pif1 helicases; however, several questions remain. Surprisingly, despite the well documented specificity of ScPif1 for G4DNA [61,81,82], little structural information exists on where and how the enzyme interacts with G4DNA. The helicase domain of ScPif1 has similar affinity for G4DNA as the full length protein, indicating that the specificity for G4DNA comes from the helicase domain itself [82]. Investigators had hopes that the structures would reveal some unique structural feature; however, no obvious G4DNA interaction motif was found in the Pif1 structures [32,33]. For some G4 interacting helicases detailed information about the interaction of the protein with the G4DNA is known. The 13 amino acid RSM motif of DHX36 interacts with G4 structures [104] and a solution structure shows a DHX36 peptide including the RSM interacting with the exposed face of a tetrad in a parallel G4DNA [105]. Residues in the RecQ-C-terminal domain of WRN helicase involved in interaction with G4DNA have also been identified [106], and G4DNA docks into the same position as dsDNA with good complementarity in a structure of RecQ [107].
The stepwise mechanism for dsDNA unwinding has been well characterized [60]. Several recent papers have investigated G4DNA unfolding by ScPif1. One report concluded that unfolding occurs in three steps [61], with each step corresponding to melting of one column of a quadruplex structure. Another group reported that unfolding is a two-step process [86]. The basic ATP-driven motor of ScPif1 has been shown to operate in one base steps, with each base being translocated by one ATP hydrolysis event [60]. The manner in which this stepwise mechanism might be utilized for G4DNA unfolding and information on the coupling of ATP hydrolysis to G4DNA unfolding remains to be clarified.
ScPif1’s patrolling activity could provide a means to clear DNA of proteins and structural barriers to replication. This mechanism has been suggested to be conserved among many helicases [96]. However, it is not known whether patrolling happens in vivo or how patrolling occurs. One possibility is that the enzyme, which normally translocates unidirectionally in the 5′-to-3′ direction in an ATP dependent manner slides backwards along the DNA (3′-to-5′) as suggested recently for WRN helicase [108]. Alternatively, the enzyme could release the substrate from its DNA binding site but a second binding site which interacts with the 3′-ssDNA-dsDNA junction could remain bound causing the enzyme to snap back to the junction. Hence, additional work is needed to address the mechanism of patrolling.
Multiple ScPif1 molecules have been suggested to function cooperatively resulting in increased protein displacement [60]. Additionally, with many substrates, ScPif1 appears to be more active under excess enzyme conditions when the DNA substrate would be expected to be coated with ScPif1 molecules compared to conditions where only a monomer would be expected to be bound, suggesting that ‘trains’ of ScPif1 may be more active. However, whether these helicase trains occur in vivo is unknown.
In addition to interacting with itself, ScPif1 has been shown to interact with other proteins. ScPif1 interacts physically with Rim1, the yeast mitochondrial ssDNA binding protein, and this interaction increases the product formation for dsDNA unwinding [28]. ScPif1 also interacts with Sub1 [109], a co-transcriptional activator that reduces genome instability at G4DNA sequences in yeast [109]. Sub1 binds tightly to G4DNA but does not unfold G4DNA structures [110]. This suggests that Sub1 may instead recruit a G4DNA resolving protein to the G4DNA site. Indeed, Sub1 and ScPif1 interact physically and genetic experiments confirmed that Sub1 and ScPif1 act in the same pathway to suppress G4DNA associated genome instability [109]. ScPif1 has also been shown to interact functionally with Polδ-PCNA through a physical interaction with PCNA [15]. This interaction results in displacement of the newly synthesized strand in break induced repair [15]. The Paeschke lab has shown there is a functional interaction between ScPif1 and Mms1, a component of the E3 ubiquitin ligase complex, with Mms1 recruiting ScPif1 to G4DNA sites [111]. It remains to be determined whether there is a direct interaction between Mms1 and ScPif1. The biochemical and in vivo role of Pif1 interactions with these and other proteins remains to be determined.
Pif1 has proven to be a most versatile enzyme in terms of the variety of biological processes in which it participates. It is likely to continue to be an intensely investigated protein in light of its breadth of functions. S. cerevisiae Pif1 appears to be an excellent system in particular for long-term studies in which the biochemical mechanism can be related to biological function.
Acknowledgments
Funding information
Our work on Pif1 is supported by National Institutes of Health Grants R01 GM098922 and R35 GM122601 (to K. D. R.).
Abbreviations
- ScPif1
Saccharomyces cerevisiae Pif1
- hPIF1
human PIF1
- DSB
double strand breaks
- G4DNA
G-quadruplex DNA
- ssDNA
single-stranded DNA
- SF
superfamily
- dsDNA
double-stranded DNA
- BsPif1
Bacteroides spp Pif1
- BaPif1
Bacteroides sp. 2-1-16 Pif1
- hPIF1-HD
human PIF1 helicase domain
- smFRET
single molecule Förster resonance energy transfer
Footnotes
Declarations of interest
The authors declare that conflict of interest associated with this manuscript.
Author contributions
All authors wrote and edited the manuscript.
References
- 1.Crouch JD, Brosh RM., Jr Mechanistic and biological considerations of oxidatively damaged DNA for helicase-dependent pathways of nucleic acid metabolism. Free Radic Biol Med. 2017;107:245–257. doi: 10.1016/j.freeradbiomed.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrd AK, Raney KD. Superfamily 2 helicases. Front Biosci. 2012;17:2070–2088. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raney KD, Byrd AK, Aarattuthodiyil S. Structure and Mechanisms of SF1 DNA Helicases. Adv Exp Med Biol. 2013;767:17–46. doi: 10.1007/978-1-4614-5037-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochman ML, Judge CP, Zakian VA. The Pif1 family in prokaryotes: what are our helicases doing in your bacteria? Mol Biol Cell. 2011;22:1955–1959. doi: 10.1091/mbc.E11-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessler JB, Torredagger JZ, Zakian VA. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol. 2001;11:60–65. doi: 10.1016/s0962-8924(00)01877-8. [DOI] [PubMed] [Google Scholar]
- 6.Foury F, Lahaye A. Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J. 1987;6:1441–1449. doi: 10.1002/j.1460-2075.1987.tb02385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion. 2007;7:211–222. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Cheng X, Qin Y, Ivessa AS. Loss of mitochondrial DNA under genotoxic stress conditions in the absence of the yeast DNA helicase Pif1p occurs independently of the DNA helicase Rrm3p. Mol Genet Genomics. 2009;281:635–645. doi: 10.1007/s00438-009-0438-6. [DOI] [PubMed] [Google Scholar]
- 9.Bochman ML. Roles of DNA helicases in the maintenance of genome integrity. Mol Cell Oncol. 2014;1:e963429. doi: 10.4161/23723548.2014.963429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi ML, Pike JE, Wang W, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J Biol Chem. 2008;283:27483–27493. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz VP, Zakian VA. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 13.Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 14.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 15.Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, Ira G. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koc KN, Singh SP, Stodola JL, Burgers PM, Galletto R. Pif1 removes a Rap1-dependent barrier to the strand displacement activity of DNA polymerase delta. Nucleic Acids Res. 2016;44:3811–3819. doi: 10.1093/nar/gkw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand RP, Shah KA, Niu H, Sung P, Mirkin SM, Freudenreich CH. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 2012;40:1091–1105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabouri N. The functions of the multi-tasking Pfh1Pif1 helicase. Curr Genet. 2017;63:621–626. doi: 10.1007/s00294-016-0675-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geronimo CL, Zakian VA. Getting it done at the ends: Pif1 family DNA helicases and telomeres. DNA Repair (Amst) 2016;44:151–158. doi: 10.1016/j.dnarep.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendoza O, Bourdoncle A, Boule JB, Brosh RM, Jr, Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung WH. To Peep into Pif1 Helicase: Multifaceted All the Way from Genome Stability to Repair-Associated DNA Synthesis. J Microbiol. 2014;52:89–98. doi: 10.1007/s12275-014-3524-3. [DOI] [PubMed] [Google Scholar]
- 24.Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bessler JB, Zakian VA. The amino terminus of the Saccharomyces cerevisiae DNA helicase Rrm3p modulates protein function altering replication and checkpoint activity. Genetics. 2004;168:1205–1218. doi: 10.1534/genetics.104.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall MC, Matson SW. Helicase motifs: the engine that powers DNA unwinding. Mol Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 27.Monson EK, de BD, Zakian VA. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci U S A. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramanagoudr-Bhojappa R, Blair LP, Tackett AJ, Raney KD. Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 2013;41:1029–1046. doi: 10.1093/nar/gks1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y, Masuda Y, Kamiya K. Biochemical analysis of human PIF1 helicase and functions of its N-terminal domain. Nucleic Acids Res. 2008;36:6295–6308. doi: 10.1093/nar/gkn609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh SP, Koc KN, Stodola JL, Galletto R. A Monomer of Pif1 Unwinds Double-Stranded DNA and It Is Regulated by the Nature of the Non-Translocating Strand at the 3′-End. J Mol Biol. 2016;428:1053–1067. doi: 10.1016/j.jmb.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11:1383–1386. doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen WF, Dai YX, Duan XL, Liu NN, Shi W, Li N, Li M, Dou SX, Dong YH, Rety S, Xi XG. Crystal structures of the BsPif1 helicase reveal that a major movement of the 2B SH3 domain is required for DNA unwinding. Nucleic Acids Res. 2016;44:2949–2961. doi: 10.1093/nar/gkw033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Ren W, Bharath SR, Tang X, He Y, Chen C, Liu Z, Li D, Song H. Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1. Cell Rep. 2016;14:2030–2039. doi: 10.1016/j.celrep.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 35.He X, Byrd AK, Yun MK, Pemble CW, Harrison D, Yeruva L, Dahl C, Kreuzer KN, Raney KD, White SW. The T4 Phage SF1B Helicase Dda Is Structurally Optimized to Perform DNA Strand Separation. Structure. 2012;20:1189–1200. doi: 10.1016/j.str.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dillingham MS, Soultanas P, Wiley P, Webb MR, Wigley DB. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc Natl Acad Sci U S A. 2001;98:8381–8387. doi: 10.1073/pnas.131009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 39.Lee JY, Yang W. UvrD helicase unwinds DNA bone base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bochman ML, Sabouri, Zakian VA. Unwinding the functions of the Pif1family helicases. DNA Repair. 2010;9:237–249. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chisholm KM, Aubert SD, Freese KP, Zakian VA, King MC, Welcsh PL. A genomewide screen for suppressors of Alu-mediated rearrangements reveals a role for PIF1. PLoS One. 2012;7:e30748. doi: 10.1371/journal.pone.0030748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris PD, Tackett AJ, Babb K, Nanduri B, Chick C, Scott J, Raney KD. Evidence for a functional monomeric form of the bacteriophage T4 DdA helicase. Dda does not form stable oligomeric structures. J Biol Chem. 2001;276:19691–19698. doi: 10.1074/jbc.M010928200. [DOI] [PubMed] [Google Scholar]
- 43.Nanduri B, Byrd AK, Eoff RL, Tackett AJ, Raney KD. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc Natl Acad Sci U S A. 2002;99:14722–14727. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levin MK, Wang YH, Patel SS. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J Biol Chem. 2004;279:26005–26012. doi: 10.1074/jbc.M403257200. [DOI] [PubMed] [Google Scholar]
- 45.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikora B, Eoff RL, Matson SW, Raney KD. DNA unwinding by Escherichia coli DNA helicase I (TraI) provides evidence for a processive monomeric molecular motor. J Biol Chem. 2006;281:36110–36116. doi: 10.1074/jbc.M604412200. [DOI] [PubMed] [Google Scholar]
- 47.Jennings TA, Mackintosh SG, Harrison MK, Sikora D, Sikora B, Dave B, Tackett AJ, Cameron CE, Raney KD. NS3 helicase from the hepatitis C virus can function as a monomer or oligomer depending on enzyme and substrate concentrations. J Biol Chem. 2009;284:4806–4814. doi: 10.1074/jbc.M805540200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comstock MJ, Whitley KD, Jia H, Sokoloski J, Lohman TM, Ha T, Chemla YR. Protein structure. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science. 2015;348:352–354. doi: 10.1126/science.aaa0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maluf NK, Fischer CJ, Lohman TM. A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 50.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat Struct Mol Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 52.Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- 53.Byrd AK, Raney KD. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rad B, Forget AL, Baskin RJ, Kowalczykowski SC. Single-molecule visualization of RecQ helicase reveals DNA melting, nucleation, and assembly are required for processive DNA unwinding. Proc Natl Acad Sci U S A. 2015;112:E6852–E6861. doi: 10.1073/pnas.1518028112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chisty LT, Toseland CP, Fili N, Mashanov GI, Dillingham MS, Molloy JE, Webb MR. Monomeric PcrA helicase processively unwinds plasmid lengths of DNA in the presence of the initiator protein RepD. Nucleic Acids Res. 2013;41:5010–5023. doi: 10.1093/nar/gkt194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J Biol Chem. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- 57.Barranco-Medina S, Galletto R. DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry. 2010;49:8445–8454. doi: 10.1021/bi100984j. [DOI] [PubMed] [Google Scholar]
- 58.Galletto R, Tomko EJ. Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res. 2013;41:4613–4627. doi: 10.1093/nar/gkt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lahaye A, Leterme S, Foury F. PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J Biol Chem. 1993;268:26155–26161. [PubMed] [Google Scholar]
- 60.Ramanagoudr-Bhojappa R, Chib S, Byrd AK, Aarattuthodiyil S, Pandey M, Patel SS, Raney KD. Yeast Pif1 helicase exhibits a one-base-pair stepping mechanism for unwinding duplex DNA. J Biol Chem. 2013;288:16185–16195. doi: 10.1074/jbc.M113.470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chib S, Byrd AK, Raney KD. Yeast Helicase Pif1 Unwinds RNA:DNA Hybrids with Higher Processivity than DNA:DNA Duplexes. J Biol Chem. 2016;291:5889–5901. doi: 10.1074/jbc.M115.688648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 65.Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 66.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajagopal V, Gurjar M, Levin MK, Patel SS. The protease domain increases the translocation stepping efficiency of the hepatitis C virus NS3-4A helicase. J Biol Chem. 2010;285:17821–17832. doi: 10.1074/jbc.M110.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duan XL, Liu NN, Yang YT, Li HH, Li M, Dou SX, Xi XG. G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding. J Biol Chem. 2015;290:7722–7735. doi: 10.1074/jbc.M114.628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ali JA, Lohman TM. Kinetic Measurement of the Step Size of DNA Unwinding by Escherichia coli UvrD Helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 70.Lucius AL, Vindigni A, Gregorian R, Ali JA, Taylor AF, Smith GR, Lohman TM. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J Mol Biol. 2002;324:409–428. doi: 10.1016/s0022-2836(02)01067-7. [DOI] [PubMed] [Google Scholar]
- 71.Eoff RL, Raney KD. Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat Struct Mol Biol. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- 72.Serebrov V, Beran RK, Pyle AM. Establishing a mechanistic basis for the large kinetic steps of the NS3 helicase. J Biol Chem. 2009;284:2512–2521. doi: 10.1074/jbc.M805460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 74.Bruning JG, Howard JL, McGlynn P. Accessory replicative helicases and the replication of protein-bound DNA. J Mol Biol. 2014;426:3917–3928. doi: 10.1016/j.jmb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 75.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Osmundson JS, Kumar J, Yeung R, Smith DJ. Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat Struct Mol Biol. 2017;24:162–170. doi: 10.1038/nsmb.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tran PLT, Pohl TJ, Chen CF, Chan A, Pott S, Zakian VA. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat Commun. 2017;8:15025. doi: 10.1038/ncomms15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohanty BK, Bairwa NK, Bastia D. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morris PD, Raney KD. DNA Helicases Displace Streptavidin from Biotin-Labeled Oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 80.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Byrd AK, Raney KD. A parallel quadruplex DNA is bound tightly but unfolded slowly by pif1 helicase. J Biol Chem. 2015;290:6482–6494. doi: 10.1074/jbc.M114.630749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mendoza O, Gueddouda NM, Boule JB, Bourdoncle A, Mergny JL. A fluorescence-based helicase assay: application to the screening of G-quadruplex ligands. Nucleic Acids Res. 2015;43:e71. doi: 10.1093/nar/gkv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gray RD, Trent JO, Chaires JB. Folding and unfolding pathways of the human telomeric G-quadruplex. J Mol Biol. 2014;426:1629–1650. doi: 10.1016/j.jmb.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aznauryan M, Sondergaard S, Noer SL, Schiott B, Birkedal V. A direct view of the complex multi-pathway folding of telomeric G-quadruplexes. Nucleic Acids Res. 2016;44:11024–11032. doi: 10.1093/nar/gkw1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hou XM, Wu WQ, Duan XL, Liu NN, Li HH, Fu J, Dou SX, Li M, Xi XG. Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem J. 2015;466:189–199. doi: 10.1042/BJ20140997. [DOI] [PubMed] [Google Scholar]
- 87.Zhang B, Wu WQ, Liu NN, Duan XL, Li M, Dou SX, Hou XM, Xi XG. G-quadruplex and G-rich sequence stimulate Pif1p-catalyzed downstream duplex DNA unwinding through reducing waiting time at ss/dsDNA junction. Nucleic Acids Res. 2016;44:8385–8394. doi: 10.1093/nar/gkw669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donmez I, Rajagopal V, Jeong YJ, Patel SS. Nucleic Acid Unwinding by Hepatitis C Virus and Bacteriophage T7 Helicases Is Sensitive to Base Pair Stability. J Biol Chem. 2007;282:21116–21123. doi: 10.1074/jbc.M702136200. [DOI] [PubMed] [Google Scholar]
- 89.Byrd AK, Matlock DL, Bagchi D, Aarattuthodiyil S, Harrison D, Croquette V, Raney KD. Dda helicase tightly couples translocation on single-stranded DNA to unwinding of duplex DNA: Dda is an optimally active helicase. J Mol Biol. 2012;420:141–154. doi: 10.1016/j.jmb.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramanagoudr-Bhojappa R, Byrd AK, Dahl C, Raney KD. Yeast Pif1 Accelerates Annealing of Complementary DNA Strands. Biochemistry. 2014 doi: 10.1021/bi500746v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 92.Tomko EJ, Ja H, Park J, Maluf NK, Ha T, Lohman TM. 5′-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocases. EMBO J. 2010;29:3826–3839. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qiu Y, Antony E, Doganay S, Koh HR, Lohman TM, Myong S. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat Commun. 2013;4:2281. doi: 10.1038/ncomms3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu CG, Spies M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016;44:8742–8753. doi: 10.1093/nar/gkw574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tippana R, Hwang H, Opresko PL, Bohr VA, Myong S. Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc Natl Acad Sci U S A. 2016;113:8448–8453. doi: 10.1073/pnas.1603724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009;28:405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vega LR, Phillips JA, Thornton BR, Benanti JA, Onigbanjo MT, Toczyski DP, Zakian VA. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet. 2007;3:e105. doi: 10.1371/journal.pgen.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mateyak MK, Zakian VA. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle. 2006;5:2796–2804. doi: 10.4161/cc.5.23.3524. [DOI] [PubMed] [Google Scholar]
- 100.Chang M, Luke B, Kraft C, Li Z, Peter M, Lingner J, Rothstein R. Telomerase is essential to alleviate pif1-induced replication stress at telomeres. Genetics. 2009;183:779–791. doi: 10.1534/genetics.109.107631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair (Amst) 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 102.Wilkie AO, Lamb J, Harris PC, Finney RD, Higgs DR. A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature. 1990;346:868–871. doi: 10.1038/346868a0. [DOI] [PubMed] [Google Scholar]
- 103.Rossi SE, Ajazi A, Carotenuto W, Foiani M, Giannattasio M. Rad53-Mediated Regulation of Rrm3 and Pif1 DNA Helicases Contributes to Prevention of Aberrant Fork Transitions under Replication Stress. Cell Rep. 2015;13:80–92. doi: 10.1016/j.celrep.2015.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lattmann S, Giri B, Vaughn JP, Akman SA, Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heddi B, Cheong VV, Martadinata H, Phan AT. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide-quadruplex complex. Proc Natl Acad Sci U S A. 2015;112:9608–9613. doi: 10.1073/pnas.1422605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ketkar A, Voehler M, Mukiza T, Eoff RL. Residues in the RecQ C-terminal Domain of the Human Werner Syndrome Helicase Are Involved in Unwinding G-quadruplex DNA. J Biol Chem. 2017;292:3154–3163. doi: 10.1074/jbc.M116.767699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Manthei KA, Hill MC, Burke JE, Butcher SE, Keck JL. Structural mechanisms of DNA binding and unwinding in bacterial RecQ helicases. Proc Natl Acad Sci U S A. 2015;112:4292–4297. doi: 10.1073/pnas.1416746112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu WQ, Hou XM, Zhang B, Fosse P, Rene B, Mauffret O, Li M, Dou SX, Xi XG. Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci Rep. 2017;7:43954. doi: 10.1038/srep43954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lopez CR, Singh S, Hambarde S, Griffin WC, Gao J, Chib S, Yu Y, Ira G, Raney KD, Kim N. Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res. 2017;45:5850–5862. doi: 10.1093/nar/gkx201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao J, Zybailov BL, Byrd AK, Griffin WC, Chib S, Mackintosh SG, Tackett AJ, Raney KD. Yeast transcription co-activator Sub1 and its human homolog PC4 preferentially bind to G-quadruplex DNA. Chem Commun (Camb ) 2015;51:7242–7244. doi: 10.1039/c5cc00742a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wanzek K, Schwindt E, Capra JA, Paeschke K. Mms1 binds to G-rich regions in Saccharomyces cerevisiae and influences replication and genome stability. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx467. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]