SUMMARY
The stereotyped features of neuronal circuits are those most likely to explain the remarkable capacity of the brain to process information and govern behaviors, yet it has not been possible to comprehensively quantify neuronal distributions across animals or genders due to the size and complexity of the mammalian brain. Here we apply our quantitative brain-wide (qBrain) mapping platform to document the stereotyped distributions of mainly inhibitory cell types. We discover an unexpected cortical organizing principle: sensory-motor areas are dominated by output-modulating parvalbumin-positive interneurons, whereas association, including frontal, areas are dominated by input-modulating somatostatin-positive interneurons. Furthermore, we identify local cell type distributions with more cells in the female brain in seven out of eight sexually dimorphic subcortical areas, in contrast to the overall larger brains in males. The qBrain resource can be further mined to link stereotyped aspects of neuronal distributions to known and unknown functions of diverse brain regions.
Keywords: Cell type, brain, cortex, inhibition, sexual dimorphism
INTRODUCTION
Since the first visualization of brain cytoarchitecture by Golgi and Ramón y Cajal, scientists have been captivated by the astonishing morphological diversity of neurons throughout the mammalian brain, which has now been expanded to encompass additional diversity in biophysical and synaptic properties and gene expression profiles. Linking cellular diversity, morphology and function has been a longtime goal in neuroscience, which in less complex systems such as C. elegans has been greatly aided by comprehensive anatomic mapping studies of the whole nervous system (White et al., 1986). However, the size and complexity of the mammalian brain have been barriers to understanding key questions central to mammalian neurobiology, such as how distinct cell type distributions and ratios correlate with diverse processing capabilities of different brain regions and neural circuits, what aspects of brain organization are stereotyped within or between distinct brain regions, or which brain regions vary based on gender or genetic background differences? To address these types of questions, we have developed a rapid and robust method for quantitative mapping and statistical comparison of distributions of fluorescently labeled neural cell types across the entire mouse brain, which has not previously been possible.
To generate a quantitative whole-brain (qBrain) cell type resource for the mouse and provide a proof of principle for its broad utility, we have mapped and quantified the distribution of seven cell type populations of predominantly inhibitory neurons. GABAergic inhibitory neurons comprise a great diversity of cell types that play critical roles in a broad range of brain functions and are strongly implicated in human neurologic disease. The three major cell types we mapped include neurons expressing somatostatin (SST), parvalbumin (PV) and vasoactive intestinal peptide (VIP), which in the cortex comprise the majority of inhibitory interneurons and are thought to control the inputs, outputs and long range modulation of cortical excitatory circuits respectively (Kepecs and Fishell, 2014; Wang et al., 2004). PV+ interneurons are also involved in generating gamma oscillations—a substrate for neuronal coordination in cortical processing (Wang, 2010), while PV+ and SST+ neurons each regulate circuits for fear learning and memory in the amygdala (Li et al., 2013; Wolff et al., 2014). VIP+ neurons are implicated in diverse circuit functions, such as circadian pacemaking in the suprachiasmatic nucleus (Fan et al., 2015). Deficits in these neural subtypes are linked to human brain disorders, such as the perturbation of cortical PV+ and SST+ neurons in schizophrenia (Lewis et al., 2012; Morris et al., 2008). Therefore, quantifying the distributions and ratios of these neuronal cell types across the entire brain will enable a new quantitative basis for understanding their functions in health and disease, for example by allowing computational circuit modeling based on realistic cell densities that could validate existing predictions and/or generate novel hypotheses, or identifying new features or subdivisions of poorly characterized brain regions.
The gold standard for quantifying cell distribution is design-based stereology (Schmitz and Hof, 2005). The laboriousness of both data collection and data scoring, however, prevents the use of stereology for quantitative analysis at the whole-brain level. A high-throughput alternative is the isotropic fractionator based on counting cell nuclei from dissociated tissue by flow cytometry (Herculano-Houzel et al., 2015). However, since the isotropic fractionator requires the brain regions to be manually dissected, this method would be very difficult to use for comprehensive whole-brain analyses: for example, generating a representative map of the mouse brain’s known functionally distinct areas would require microdissection of over 400 unique regions (Dong, 2008). These technical barriers have led to a piecemeal understanding of the basic organization of brain circuits and limited comparisons between genders or animals of different genetic backgrounds or disease states.
To overcome these barriers, we developed a “cell counting and distribution mapping” platform, based on automated imaging by serial two-photon tomography (STPT) and data analysis by machine learning algorithms (Kim et al., 2015; Ragan et al., 2012). Here we apply this platform to generate the first quantitative whole-brain maps for three major cell types—the PV+, SST+, and VIP+ neurons, and two subtypes of the SST+ and VIP+ populations each. These data and our pipeline comprise the qBrain resource, which includes tables with cell counts and cell densities for over 800 anatomical regions delineated by the Allen Mouse Brain Atlas (Dong, 2008) and graphic visualizations of the data in 3D movies, 2D flatmaps, and serial section datasets at our webportal http://mouse.brainarchitecture.org/cellcounts/ost/.
To demonstrate the value of this resource we apply it to discover a novel cortical organizing principle based on the ratios of the output-controlling PV+ interneurons and input-controlling SST+ interneurons. We then use these distributions to build inhibitory circuit models that predict region-specific differences in local cortical computation and long-range cortico-cortical communication. We also identify sexually-dimorphic distributions of the VIP+, SST+ and PV+ cell types in nine brain areas, most of which belong to the structures of the vomeronasal system regulating reproductive and social behaviors. Finally, we highlight other examples of cell type-based biological insights that can be derived from the analysis of the current data.
These findings constitute a proof of principle that the qBrain resource can be mined by the scientific community to uncover additional novel brain features and stimulate new research across a broad range of brain functions. New datasets involving more cell types as well as mouse disease models can be readily analyzed by our pipeline and added to this database, enabling a broad range of future studies with the ultimate goal to generate a comprehensive cell type-based atlas of the mouse brain.
RESULTS
Quantifying and comparing cell type distributions in the mouse brain
The cell type distribution data were collected using our cell counting and distribution mapping pipeline comprising 1) brainwide cell-type labeling, 2) automated brain imaging at a single-cell resolution, and 3) computational analyses to enable reliable cell type detection, 3D volume registration, and anatomical segmentation.
While this method can be applied to any fluorescently labeled cell population, here we took advantage of a well characterized “knock-in” mice with cell type-specific expression of the Cre and Flp recombinases based on the expression of genes that define distinct, mainly inhibitory neuronal populations (He et al., 2016; Taniguchi et al., 2011). PV+, SST+, and VIP+ cells were labeled by crossing the respective gene-IRES-Cre driver mice with reporter mice expressing green fluorescent protein fused to histone 2B (H2B-GFP) upon Cre-mediated recombination (Figure 1A). The SST+ and VIP+ cell types were each further subdivided into cells co-expressing SST and calretinin (SST:CR) and SST and neuronal nitric oxide synthase (SST:nNOS), and cells co-expressing VIP and CR (VIP:CR) and VIP and cholecystokinin (VIP:CCK). This was achieved by crossing cell type-specific IRES-Cre and IRES-Flp driver mice with reporter mice expressing the red fluorescent protein tdTomato upon both Cre and Flp recombination (He et al., 2016) (Figure 1A).
Figure 1. Quantitative cell type mapping in the mouse brain.
(A) Genetic strategy to label brain cell types by Cre and Cre and Flp drivers. Top: the cell type-specific Cre and Flp mouse lines; bottom: the fluorescent protein-based reporter lines.
(B) Reconstructed SST-Cre:H2B-GFP mouse brain imaged by STPT (B1) and the detected SST+ cell distribution (B2). Each brain is registered to a reference STP (RSTP) brain aligned with the common coordinate framework Allen Brain atlas (B3) for anatomical segmentation of the whole-brain cell type distribution.
(C) Cell densities per mm3 of each of the seven cell types analyzed in our study are visualized as whole brain flatmaps (see Figure S1 for a high resolution flatmap; complete cell counts and cell densities are given in Tables S1 and S3).
To achieve reliable and rapid brain imaging, we applied our automated whole-mount microscopy STPT to image the entire brains at XY resolution of 1 µm and Z-spacing of 50 µm, resulting in 270-serial coronal section datasets (Ragan et al., 2012) (Figure 1B; Methods). Automated detection of each cell type in the serial section datasets was achieved using convolutional neural networks (CNs) trained to recognize the nuclear H2B-GFP or the cytoplasmic tdTomato signals (Jain et al., 2010; Kim et al., 2015) (Figure S1). The resulting serial 2D cell counts were converted to 3D cell counts by a stereological conversion factor estimated based on computational and manual cell counts (Figure S1E; Methods). To obtain cell type-specific cell counts per anatomical areas, each imaged brain was warped to a reference STP (RSTP) brain aligned to the Allen Mouse Brain Atlas (Figure 1B; Table S1; Methods) (Dong, 2008; Kim et al., 2015). Finally, region-specific cell densities per cubic mm (mm3) were calculated from the estimated cell counts and the volume of each brain area (Figure S1E; Table S2 and S3; Methods).
To enable broad use of these generated data as a resource for the scientific community, the whole brain cell type datasets that can be interrogated using three different formats. First, we provide a visual overview of each cell type distribution (per mm3) as whole-brain flatmaps adapted from the rat brain atlas (Swanson, 2004) (Figure 1C and S1G) and 3D movies of serial sections (Movie S1 and S2). Second, the total cell counts and the cell densities for all anatomical ROIs are given in Tables S1 and S3, respectively. And third, the full resolution serial section images of cell type distributions are accessible for viewing via our cell type-dedicated repository http://mouse.brainarchitecture.org/cellcounts/ost/.
Analysis of cortical inhibitory interneurons reveals unexpected cell type-specific regional distributions
To demonstrate the value of the qBrain resource we first examined the distributions of the mapped neurons in different regions of the neocortex, where these cell types represent the majority of inhibitory interneurons and are known to play cell type-specific roles in orchestrating cortical functions (Kepecs and Fishell, 2014; Wang et al., 2004). The quantitative data we generated allowed us to address a key unanswered question relating to cortical processing, that is whether the archetypal local inhibitory circuit architecture described in the visual cortex (Pfeffer et al., 2013) is repeated throughout the entire neocortex, or if interneuron distribution shows cortical area-specificity suggesting that distinct circuit architectures may underlie the diverse processing capabilities of different cortical regions.
Visualizing the cell density of inhibitory cell types in flat maps of the isocortex revealed that the PV+ and SST+ cell densities differ considerably among cortical areas. The PV+ cell density appeared lower and the SST+ cell density higher in regions involved in higher cognitive functions, including medial frontal and lateral associational areas (Figure 2; see also Figure S2). To qualitatively validate this data, we examined PV and SST in situ hybridization data from the Allen Brain data portal (Ng et al., 2009), which supported our findings (Figure S3). However, it is important to note that the Allen Brain results typically derive from only one brain and therefore could not be used to independently derive our findings.
Figure 2. Uneven distribution of the three major interneurons in the isocortex.
(A) Unbiased voxel-based quantitative mapping. The first three columns show the PV+, SST+, and VIP+, and the last column the combined signals overlaid on the RSTP brain with anatomical segmentation information (see also Movie S1). The heatmap represents number of cells per voxel (a sphere of 100µm diameter). The A/P bregma position is shown in the first column. (Tables Sl–3 list the full anatomical names).
(B–E) Cell density mapping using a cortical flatmap (B). The heatmap displays of cell density per mm3 for the VIP+ (C), PV+ (D), and SST+ (E) cell populations. Note the low density of PV+ and high density of SST+ in the medial frontal (blue arrow) and lateral association cortices (orange arrow).
(F –G) Cell density plots (F) and relative distribution (G) of the three interneuron cell types across anatomical regions of the isocortex arranged in five subnetworks based on their anatomical connectivity (Zingg et al., 2014) (see also Tables S3 for complete dataset). The values in (F) = mean ± standard deviation.
We also note that similar cortical networks comprising the medial frontal and lateral associational areas were recently inferred based on long-range cortico-cortical connectivity (Zingg et al., 2014). This surprising link is clearly seen when the interneuron cell density data are organized according to the Zingg et al data: the PV+ to SST+ ratio appears balanced in the motor and somatosensory, audio-visual and medial associational areas, while, in contrast, this ratio is strongly skewed in favor of the SST+ interneurons in the lateral associational and medial frontal networks, suggesting the existence of distinct local circuits for the higher-order cortical areas (Figure 2F–G; Figures S2) (see more below).
Data from the intersectional mice revealed that the SST/CR interneurons, a population enriched for Martinotti cells, show similar distribution as the total SST neurons, while the SST/nNOS interneurons, called type I nNOS neurons, have a more uniform distribution across the isocortex (Figure S4). Both VIP/CR interneuron-selective cells and multipolar VIP/CCK small basket cells show similar distribution to the total VIP interneurons (Figure S4).
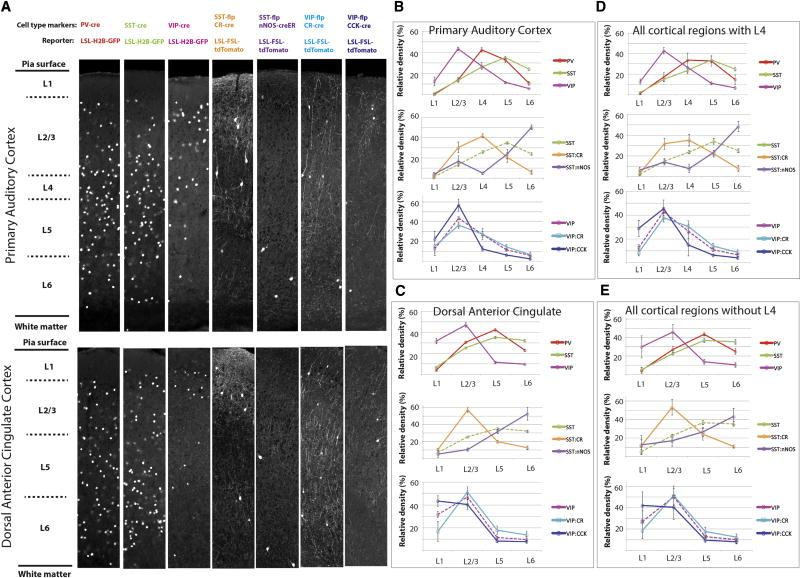
Next, we asked whether relative laminar distribution of interneurons across cortical layers, independent of the cell density-based differences, is conserved between cortical regions. Previous immunohistochemical studies characterized the relative laminar distribution of major interneuron cell types in the somatosensory and visual cortex (Gonchar et al., 2007; Pfeffer et al., 2013; Pronneke et al., 2015; Rudy et al., 2011; Xu et al., 2010), and our data showed that this pattern is indeed repeated across the entire isocortex: the PV+ and SST+ interneurons are most abundant in the layers 4–5 (PV+) and layers 4–6 (SST+), while the VIP+ interneurons show high density in the layers 1–4, with a peak in the layer 2/3 (Figure 3; see also Figure S5 for distribution based on distance from the brain surface).
Figure 3. Cell type-specific laminar cortical distribution.
(A) Examples of the seven cell types in the six-layer primary auditory (top panel) and five-layer dorsal anterior cingulate (bottom panel) cortex (full datasets are deposited at http://mouse.brainarchitecture.org/cellcounts/ost/).
(B–C) Relative cortical layer cell densities from the primary auditory (B) and dorsal anterior cingulate (C) cortices, with each cell type color-coded as shown on the right (the values = mean ± standard deviation).
(D–E) Average relative cortical layer cell densities from all 6-layer (D) and 5-layer (E) cortical areas, shown with the same figure composition as in (B–C) (the values = mean ± standard deviation). Note the stereotypic layer distribution of the interneuron subtypes across the mouse isocortex.
The inhibitory intersectional subpopulations also showed a stereotypical laminar distribution, with some cell type-specific distinctions: The SST/CR neurons show relatively high density in the superficial layers, while the SST/nNOS neurons are relatively more abundant in deep cortical layers (Figure 3). Both the VIP/CR and VIP/CCK interneurons show expression patterns similar to the entire VIP+ population, except that the VIP/CCK cells are relatively more abundant in the layer 1 (Figure 3).
We conclude that, while the total numbers of the main populations of PV+ and SST+ interneurons significantly vary based on cortical regions, the laminar organization of the inhibitory cell types is comparable across the isocortex.
Areal hierarchy in the cortical PV+ to SST+ cell density ratios
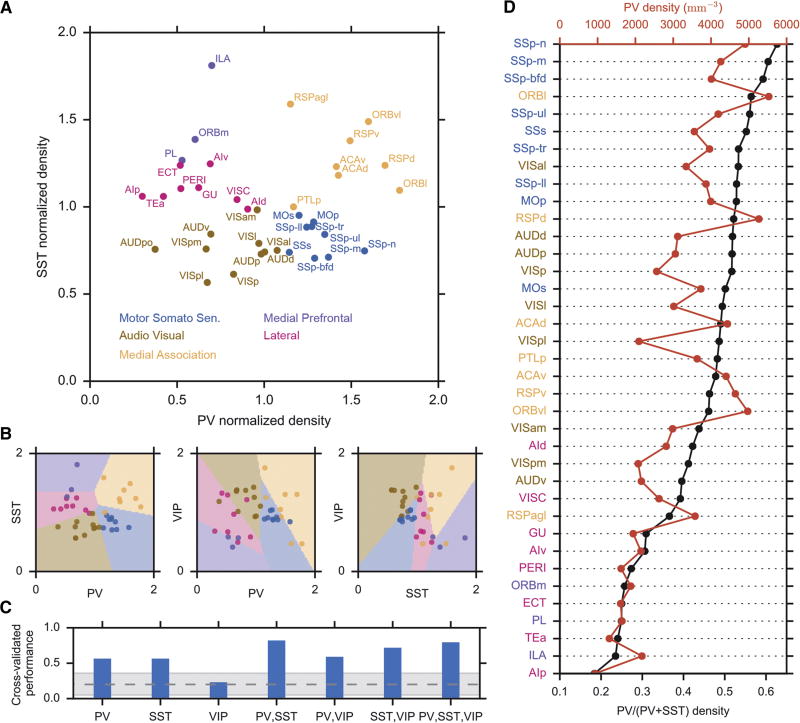
To understand the cortical interneuron differences more quantitatively, we next plotted each layer 2/3 area within a two-dimensional PV+ to SST+ cell density plane, with the cell-type specific density normalized by its average across areas, which confirmed that the local PV+/SST+ cell densities are indeed most similar within the Zingg et al. (2014) defined cortical networks (Figure 4A). Similar analysis of the VIP+/PV+ and VIP+/SST+ interneuron distribution revealed lesser separation (Figure 4B–C), suggesting that the link between local circuits and cortical networks is more specific to the balance between the output-controlling PV+ interneurons and input-controlling SST+ interneurons (see also Figure S6 for cortical layer 5 analysis). Finally, the notion of a cortical area hierarchy related to the PV+/SST+ cell densities is confirmed when the cortical areas are sorted according to their PV+/SST+ ratio (Felleman and Van Essen, 1991): areas belonging to the motor-somatosensory subnetwork tend to have the highest PV+/SST+ density ratios, corresponding to lowest levels in the hierarchy, while the areas from the lateral and the medial frontal subnetworks occupy the highest levels in the hierarchy, with the lowest PV+/SST+ density ratios (Figure 4D).
Figure 4. Cortical area hierarchy in L2/3 PV+ to SST+ density ratios.
(A) Cortical areas are segregated in space of L2/3 PV+ and SST+ density according to their cortical subnetworks (color coded).
(B) Decision boundaries of linear discriminant analysis classifiers using, from left to right, PV+/SST+, PV+/VIP+, or SST+/VIP+ cell densities.
(C) Cross-validated classifier performances on left-one-out data, when different combinations of density information are used. Gray area indicates the 95% confidence interval of classifier performances on shuffled data.
(D) Cortical areas ranked by their PV+/SST+ cell density ratios.
Modeling the consequences of the distinct PV+ and SST+ cell densities on local cortical circuits
What may be the functional consequences of the measured cortical interneuron densities? A straightforward prediction is that an increase in the density will correspondingly strengthen the cell type’s role in controlling cortical excitatory neurons.
To test this prediction, we studied a simple linear rate-based circuit model consisting of excitatory (E), PV+, SST+, and VIP+ cell populations connected according to a circuit diagram measured in the visual cortex (Figure 5A) (Pfeffer et al., 2013), with the cell density modeled as a scaling factor to all output projection weights of that population. PV+, SST+, and VIP+ neurons all receive long-range inputs from other cortical and subcortical areas that serve a range of important functions, including feed-forward inhibition and feedback disinhibition (Kepecs and Fishell, 2014; Wall et al., 2016). Therefore, we focused on circuit responses to external inputs driving the three interneuron cell types, starting with inputs targeting the PV+ populations.
Figure 5. Modeling response properties of local circuits while varying cell densities.
(A) Cortical circuit model with an excitatory (E) and three inhibitory populations (PV, SST, and VIP) (Pfeffer et al., 2013). Output weights of an inhibitory population are proportional to the density of that population.
(B,C) Comparing cell densities (B) and local circuit responses (C) in areas SSp-bfd and ILA. (C) From top to bottom: E population activity, PV-to-E, SST-to-E, VIP-to-SST, and E-to-E currents in response to external inputs driving the PV population. Spontaneous activities are kept the same across areas.
(D,E) Same comparisons as in (B,C), for areas AUDpo and RSPv.
(F–H) Responses of E population activity (F) and PV-to-E current (G) depend on both PV+ and SST+ cell densities. (H) Maps of circuit responses overlaid with the distribution of cortical areas in the PV+/SST+ density plane.
(I) Increasing SST+ density strengthens the PV-SST-PV effective disinhibitory loop, leading to a stronger PV-to-E current response.
We first compared responses in models of the somatosensory barrel field (SSp-bfd) (low SST to PV L2/3 ratio) and the infralimbic (ILA) cortex (high SST to PV L2/3 ratio) (Figure 4A and 5B). As expected, in response to the inputs targeting the PV+ population, the steady-state response of the E population is more suppressed in the SSp-bfd, which has higher PV+ cell density than the ILA area (Figure 5C, top trace). The circuit model also suggests that the larger suppression of E activity in the SSp-bfd is not due to stronger responses of inhibitory currents (Figure 5C, middle traces) but, instead, due to reduced steady-state excitatory recurrent currents reflecting the reduced steadystate E activity (Figure 5C, bottom trace).
Next, we compared responses in the posterior auditory (AUDpo) and the ventral restrosplenial (RSPv) areas, which have approximately even SST+ to PV+ ratio, but with densities of both cell types higher in the RSPv (Figure 4A and 5D). Here, we find that the steady-state response of E population activity is similar in the two areas (Figure 5E, top trace), though the density difference is reflected in stronger responses of the PV-to-E and SST-to-E currents in the RSPv (Figure 5E).
Moving beyond the selected example areas, we examined the responses to inputs targeting the PV+ population while systematically varying the PV+ and SST+ cell densities. The steady-state E activity indeed becomes more suppressed with higher PV+ cell density, but it also becomes more disinhibited with higher SST+ cell density (Figure 5F). This suggests that both PV+ and SST+ densities are effective at altering the circuit responses when external input targets the PV+ cell population. Under the same conditions, the PV-to-E current response strengthens (becomes more negative) with both higher PV+ and higher SST+ cell density (Figure 5G), and the SST-to-E current response also strengthens (becomes more positive/disinhibited) with higher PV+ and SST+ density (Figure S7A).
These findings are not trivial outcomes of the inhibitory connectivity or the circuit diagram (Figure S7B), as they require sufficiently strong recurrent excitatory connections (see STAR method). Furthermore, these findings remain valid after addition of other putative connections in the cortical circuit, including PV-to-SST, VIP-to-PV, and VIP-to-E connections (Pi et al., 2013). Finally, these results were also reproduced in a neural circuit model of 5,000 realistic spiking neurons and data-constrained synaptic weights (Figure S7C–E) (Litwin-Kumar et al., 2016; Wang et al., 2004).
Based on these results we can interpret the cell-density plots in Figure 4 in the following way. The largest areal difference defined by the PV+/SST+ ratio is an anti-correlated change in PV+ and SST+ densities, which is equivalent to traveling along the anti-diagonal direction in the PV+/SST+ density plane. Such a change strongly affects the excitatory population response, as the E activity is more suppressed in areas with higher PV+/SST+ ratio (Figure 5H). Regarding the underlying changes in the local circuit currents, the reduced E activity is reflected in reduced recurrent excitatory currents, but without changes in steady state responses of inhibitory currents. On the other hand, audio-visual and medial association subnetworks are separated along the diagonal direction in the PV+/SST+ density plane, which reflects comparable PV+/SST+ cell ratios and such a change does not affect the E steady-state activity, though it does lead to differences in steady-state responses of inhibitory currents (Figure 5H).
How can we explain the surprising outcome for the regions lying along the diagonal and anti-diagonal directions of the PV+/SST+ plot? Based on the circuit model, inputs onto the PV+ population will suppress the E activity, which, in turn, leads to a suppression of recurrent excitation on the SST+ cells. The loss of SST+ activity then disinhibits the PV+ population and this disinhibitory loop is stronger in areas with higher SST+ density, resulting in a stronger PV-to-E current response (Figure 5I; see STAR method for more rigorous derivations).
We have also carried out similar analysis of the L2/3 circuit properties in response to external inputs targeting the other cell populations (Figure S7A). In response to inputs targeting the SST+ population, the steady-state E activity becomes more suppressed with higher SST+ cell density, but more disinhibited with higher PV+ cell density. In response to external inputs targeting the VIP+ population, the steady-state E activity becomes more disinhibited with higher SST+ cell density, and less disinhibited with higher PV+ cell density. Finally, in response to inputs onto the excitatory population, the steady-state E activity as well as the E-to-E current is reduced with both higher PV+ and SST+ cell density.
In summary, the functional impact of the empirical cell densities needs to be interpreted beyond the immediate effect of each cell type, as for example the SST+ density can strongly influence how PV+ interneurons inhibit excitatory neurons. These results provide a rationale for further studies of other circuit parameters (e.g. connection probability and synaptic weights) in the lateral associational and frontal cortical areas in comparison to the sensorimotor areas.
Subcortical maps of the PV+, SST+ and VIP+ cells
Subcortical neurons expressing PV, SST and VIP comprise a broad variety of cell types, including long-range projection inhibitory neurons, for example, in the basal forebrain (Henny and Jones, 2008), and even projection excitatory neurons (Shang et al., 2015; Wallace et al., 2017). Since the functional significance of many of these neurons is only beginning to be understood, analyses of their ratios and distributions within diverse subcortical brain regions are likely to provide potential anatomical bases for known functions and inspire new functional and developmental studies to understand the origin and consequence of these stereotyped features of brain circuits. Here, we highlight several observations that suggest novel cell type-based hypotheses about organizations of key subcortical regions and circuits. More detailed anatomical descriptions are given in the STAR method section.
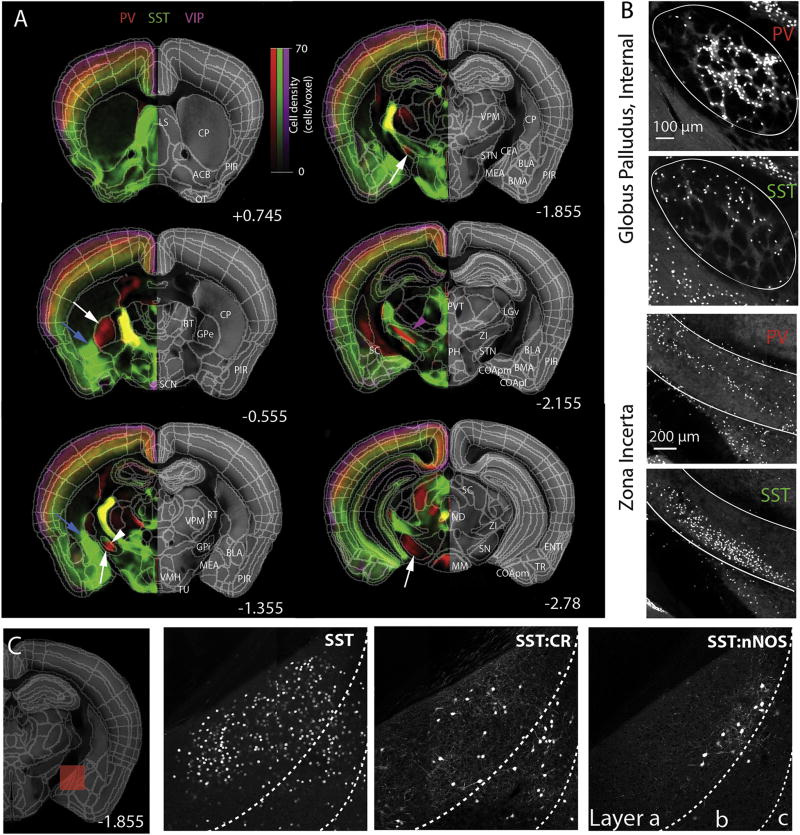
SST+ neurons are most abundant in the lateral septum, bed nuclei stria terminalis, and the amygdala (range ~14,000 to 24,000 cells per mm3) (Table S3). We identified a novel high-density band of the SST+ field extending rostrally from the CEA and MEA across the caudal half caudoputamen (CP) (light blue arrows in Figure 6A), which is reminiscent of the rat “amygdalo-striatal transition zone”. We therefore propose that this region is organized in an analogous way in the mouse brain and should be distinguished from the larger CP proper (see STAR methods).
Figure 6. Subcortical distribution patterns of seven cell types.
(A) Virtual density overlay reveals distinct anatomical distribution of each cell type. For example, enriched expression of SST neurons in the central amygdala is extended to more anterior ventral part of caudate putamen (light blue arrows). Some area is exclusively occupied by one cell type (PV in GPe, STN, SN, white arrow). PV and SST neurons showed dorsal and ventral expression in the GPi (white arrow head) and in the ZI (purple arrow head). PV neurons are highly expressed in basal ganglia (white arrows). Numbers represent A/P bregma. See also Movie S1.
(B) High resolution images to show topographical separation of PV and SST neurons expression in GPi and ZI.
(C) Genetic intersection approach reveals specific expression from SST subtypes in medial amygdala. Numbers represent A/P bregma (see also Movie S2).
PV+ cells are abundant in the cerebellum, inferior colliculus, reticular thalamus, dorsal pallidum and substantia nigra (range ~4,000 to 47,000 cells per mm3) (Figure 6A). We note a marked spatial separation between SST+ and PV+ cells within the basal ganglia globus pallidus internal segment (GPi; also called entopeduncular nucleus) (white arrowhead in Figure 6A and 6B), suggesting distinct input/output circuits in the GPi. This hypothesis is indeed in agreement with a recent study identifying three GPi projection neuron classes—SST+ cells co-releasing glutamate and GABA and PV+ cells releasing either glutamate or GABA (Wallace et al., 2017). We also note a similar within-region spatial segregation between PV+ and SST+ cell populations in the subthalamic zona incerta (purple arrowhead in Figure 6A and 6B). Finally, we note that the SST:nNOS++ neurons are spatially clustered in the posterior MEA, suggesting distinct circuit function and/or connectivity for this cell subtype (Figure 6C).
Sex dimorphism in SST+ and VIP+ cell type distribution
Gender differences in brain anatomy have been of great interest for their capacity to underlie sexually dimorphic behaviors and to offer neural substrates for male-female differences in disease susceptibility or responses to drugs (Bayless and Shah, 2016; Simerly, 2002). Mining the qBrain resource enabled us to quantitatively contrast the distributions of the major PV+, SST+ and VIP+ cell types in male and female mice, leading to several new discoveries.
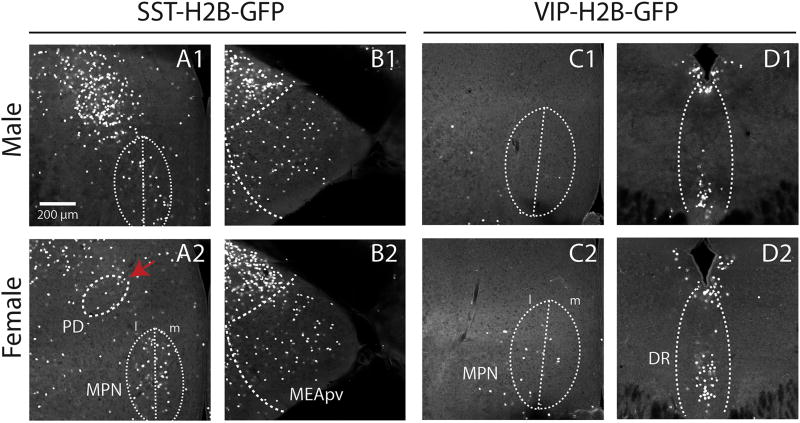
In agreement with a magnetic resonance imaging (MRI) study (Spring et al., 2007), our data revealed a bias toward larger brain volumes in the male mice, with 192 anatomical ROIs (of the total 818) found to be larger by more than 3% in the male brain (Table S2). Surprisingly and in contrast to the brain volume measures, analysis of sex-specific cell-type distribution revealed significantly higher cell numbers of the SST+ and VIP+ cells in several brain structures in the female brain, suggesting novel female-specific circuit mechanisms for regulating reproductive and social behaviors. We observed female specific cell enrichment in regions linked to pheromonal processing, including SST+ cells in the medial preoptic nucleus (MPNc and MPNl), medial amygdala (MEApv), and paraventricular nucleus of the thalamus (PVT), and more VIP+ cells in the accessory olfactory bulb (AOBgl and AOBmi), cortical amygdala (COApm1), and medial preoptic nucleus (MPNm) (Figure 7 and Table S4). In addition, female brains contain more VIP+ cells in the inferior colliculus (central nucleus, ICc) of the auditory pathway and the neuromodulatory dorsal and central linear nuclei raphe (DR and CLI) (Table S4).
Figure 7. Sexually dimorphic expression of SST+ and VIP+ neurons.
(A–D) Examples of sexually dimorphic regions (A–B) SST showed higher number of cells in females than males in medial preoptic nucleus (MPN) and posteroventral medial amygdala (MEApv), and lower number in posterodorsal preoptic nucleus (PD, red arrow). (C–D) VIP showed higher number of cells in the female than males in MPN and dorsal raphe (DR). See Table S4 for the complete list including the cell counting.
Unexpectedly, the only example of male-specific enrichment was an increased density of SST+ cells in the posterodorsal preoptic nucleus (PD), a structure previously linked to male ejaculation (red arrows in Figure 7B and Table S4) (Heeb and Yahr, 1996; Heeb and Yahr, 2000). PV+ cells, in contrast, did not show any sexually dimorphic distributions.
DISCUSSION
Comprehensive cell type resource
Here we describe the first brainwide maps of cell type distribution in the mouse brain and we present the data as an easily accessible resource comprising whole-brain flatmaps (Figure 1), supplementary movies and tables, and serial section datasets at http://mouse.brainarchitecture.org/ost/. To illustrate the biological insights that can be derived from these datasets, we describe: 1) cortical organizations based on the distribution of PV+ and SST+ interneurons, 2) spatial organizations of PV+ and SST+ cell types in several brain regions, and 3) several regions with cell type-based sexual dimorphism, revealing a surprising bias towards increased circuitry in the female brain. While these findings are discussed further below, we anticipate that the use of these datasets by the scientific community will generate many more insights and hypotheses, as the functional roles of these cell types especially in subcortical areas are only beginning to be understood.
The use of STPT and cell type-specific Cre/Flp driver mouse lines to estimate cell type distribution
We previously demonstrated the use of our STPT pipeline for automated analysis of behavior-driven c-fos induction or anatomical anterograde or retrograde tracing (Hou et al., 2016; Jeong et al., 2016; Kim et al., 2016; Kim et al., 2015; Ragan et al., 2012). The use of this platform for quantitative brainwide mapping of cell type distribution represents a dramatic methodological advancement compared to laborious stereological methods (Schmitz and Hof, 2005) or the isotropic fractionator that can only analyze larger brain structures and does not preserve the spatial information (Herculano-Houzel et al., 2015).
The reporter mice are based on “knock-in” Cre and Flp drivers that express the recombinases constitutively from an IRES (internal ribosomal entry site) cassette inserted in the 3’ untranslated region (UTR) of the cell type-specific marker genes (Madisen et al., 2010; Taniguchi et al., 2011), which can lead to labeling of cell populations that express the protein marker only transiently during development and not in the adult brain. This does not seem to be the case in the adult isocortex, where the Cre-driven labeling of the PV+, SST+, and VIP+ cells colocalizes in >95% cells with the expression of the respective reporter proteins (Pfeffer et al., 2013; Pronneke et al., 2015; Taniguchi et al., 2011). Similarly, subcortical PV-Cre and VIP-Cre labeling agrees well with available data for the respective mRNA and protein marker expression in the adult brain, while several thalamic and hypothalamic SST-Cre-positive areas do not appear to express SST in the adult brain, suggesting that these cell populations are indeed marked due to a transient developmental SST expression (see Anatomical Description in the STAR method).
Region-specific distribution of cortical interneurons: modeling of their impact on local inhibitory circuits
The isocortex is thought to consist of repeated stereotypical circuits that represent local computational units (Douglas and Martin, 2007). At the same time, it has been argued that modification of the numbers or ratios of interneurons could allow different cortical areas to perform qualitatively distinct computations or functions: for example, the frontal cortex is believed to require sufficiently strong recurrent excitation to generate persistent activity that could underlie its function in working memory as well as to benefit from more dendrite-targeting inhibitory neurons than primary sensory areas in order to gate inputs and filter out distractors (Wang, 2006). Our brainwide analysis clearly supports the hypothesis that distinct local circuits may enable area-specific functions, as high order association areas (the medial frontal and lateral association networks) fall into high SST+ / low PV+ category, while sensory areas (auditory-visual and somatosensory network) show balanced to low SST+ / high PV+ interneuron ratio distribution. This hierarchical cortical organization based on the PV+/SST+ ratio parallels network classification based on cortical functions and connectivity (Zingg et al., 2014), suggesting a link between local circuits and cortico-cortical communication. Finally, to account for the striking differences in cortical cell densities, especially with respect to the PV+ interneurons, our data point to a novel developmental regulation of cortical region-specific interneuron cell migration and/or pruning.
To understand the computational significance of our data, we examined a L2/3 circuit model built with the area-corresponding cell densities. These models predict that anti-correlated change of the PV+ and SST+ cell ratio will have a major effect on the activity of the excitatory neurons. For example, excitatory neuronal activity is more suppressed by PV inputs in areas with high PV+/SST+ ratios than with lower ratios. Alternatively, a correlated change of the PV+ and SST+ cell densities alters the PV+- and SST+-mediated currents without affecting excitatory activity.
Another major difference between the PV+ and SST+ interneurons concerns their long-range inputs. Long-range feedforward inputs strongly target PV+ neurons, while long-range feedback inputs tend to target VIP+ neurons, which in turn inhibit SST+ neurons (Pfeffer et al., 2013; Pi et al., 2013; Wall et al., 2016). As a result of a high PV+/SST+ ratio, excitatory neurons in the motor somatosensory areas will have a stronger response to feedforward inhibition. Because of the low PV+/ SST+ ratio, excitatory neurons in the medial frontal and lateral areas are predicted to respond more strongly to feedback inhibition or disinhibition.
Finally, since SST+ interneurons target the dendrites of excitatory neurons, it was suggested that SST+ neurons may gate the inputs onto excitatory neurons (Lee et al., 2017; Wang et al., 2004), and computational models suggest that SST+ interneurons can selectively gate inputs that originate from different areas or pathways (Yang et al., 2016). Therefore, higher SST+ interneuron density in the medial associational and lateral areas can also allow for a larger repertoire of selective neurons.
Of note, our models assume that local circuit parameters, namely the connection probability and synaptic weights, remain approximately constant across cortical areas. While this assumption is supported by some experimental measurements from frontal and sensory cortical areas (Packer and Yuste, 2011; Pi et al., 2013), our data provide a strong rationale for determining these parameters systematically at least across the areas with the largest cell density differences, such as the frontal infralimbic cortex and the somatosensory barrel cortex.
Sexual dimorphism
The majority of previous studies of brain specific gender dimorphisms have identified male specific increases in cell numbers or gene expression levels, but here we observe the opposite (Bayless and Shah, 2016; Simerly, 2002). We identified higher SST+ and VIP+ cell numbers in all but one sex dimorphic regions in the female brain, of which several were linked to regions involved in pheromone perception and its translation into innate behaviors. For example, the AOB, which transmits pheromonal signals from the vomeronasal organ to the brain was reported to be larger in the male rat (Segovia and Guillamon, 1993), and we observe a similar ~5% volume increase in male mice. However, female mice have an increased number of AOB VIP+ neurons, which in the main olfactory bulb (MOB) were shown to regulate circadian augmenting of olfactory sensitivity (Gracia-Llanes et al., 2003). Therefore, one testable hypothesis suggested by these data is that female mice may exhibit augmented sensitivity in pheromonal processing, or perhaps be more sensitive to circadian rhythms than males.
The MEA, COApm, and MPN lie downstream of the AOB signaling and were also reported to be larger and comprise more cells in male rats (Bayless and Shah, 2016; Simerly, 2002), in agreement with our data of ~4% increased volume in male mice. Our finding of larger numbers of SST+ cells in the MEApv and MPN, and VIP+ cells in the COApm and MPN in the female brain, in addition to the above AOB data, implies a novel form of sex dimorphic regulation of reproductive and social behaviors in female mice and offers genetically targetable cell populations to tease out the functional consequences of this anatomic dimorphism.
Other regions with female specific enrichment include the VIP+ cells in the DR and CLI. This is of interest because these neurons are thought to play a role in anxiety, especially with a social context, which may be differentially regulated in males and females (Lee et al., 2008).
Rather surprisingly, given previous work, we identified only a single region with enrichment in these cell types in male mice. This three-fold increase in the density of SST+ neurons in the PD region of the hypothalamic medial preoptic area has not been reported and the region has not received wide attention. Nevertheless, several studies of the region link it to ejaculation (Coolen et al., 1996; Heeb and Yahr, 2000), which is a function quite consistent with a requirement for male-specific circuitry, and one that could represent a novel target for pharmacologic intervention if present in humans.
In summary, our study provides the first comprehensive and quantitative qBrain resource of cell type distribution in the mammalian brain. The unexpected findings of cortical hierarchy, areal subdivisions, and sex based anatomic dimorphism demonstrate the power of unbiased cell type mapping in uncovering novel structural insights and hypotheses about brain functions. Computational modeling revealed an organization of the density space of input-controlling SST+ interneurons and output-controlling PV+ interneurons into two orthogonal subspaces, illustrating how the new dataset can be used as a resource for discoveries from a modeling perspective. Further, because these cell populations are defined by genetic reporter mouse strains that are compatible with Cre/Flp inducible viruses, the functional consequence of these newly defined circuit features can be readily interrogated using optogenetic or chemogenetic tools or studied developmentally. Finally, the resource we provide can be easily expanded to include maps of the growing number of genetically or immunologically defined neural subtypes emerging from functional and transcriptional studies of neuronal diversity.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Pavel Osten: osten@cshl.edu.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Animal procedures were approved by the Cold Spring Harbor Laboratory Institutional Animal Care and Use Committee (IACUC). All animals were housed under constant temperature and light conditions (12-hour cycle lights ON: 0600, lights OFF: 1800) and given food and water ad libitum. For major GABA neurons, we crossed Cre drivers (Parvalbumin-IRES-Cre, Somatostatin-IRES-Cre, and VIP-IRES-Cre) were crossed with reporter mice (CAG-LoxP-STOP-LoxP-H2B-GFP). For intersectional approach, we crossed intersectional driver (SST-flp:CR-cre, SST-flp:nNOS-creER, VIP-flp:CR-cre, or VIP-flp:CCK-cre) with Ai65 reporter mice (CAG-Frt-STOP-Frt-LoxP-STOP-LoxPtdTomato). To induce nNOS-CreER, tamoxifen was prepared by dissolving in corn oil at room temperature overnight with constant rotation and the same dose of 20mg/mL was administrated by intraperitoneal injection three times, once every other day, beginning after P14. 8- to 10-week old mice from both female and male were used for the study. For major GABA neurons, 5 males and 5 females were used per kind. For the intersectional lines, 3 – 4 male and females were used per kind. All brain samples were acquired as single groups without replication. Detailed information of each mouse including its sex can be found in the online database http://mouse.brainarchitecture.org/cellcounts/ost/. Parvalbumin-IRES-Cre was purchased from the Jackson laboratory and all other cell type specific driver and reporters were directly obtained from Dr. Z Josh Huang. All Cre and Flp drivers, and Ai65 reporter mice used in this study are also available from Jackson Laboratory as indicated in the KEY RESOURCES TABLE.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Allen in situ data | Allan R Jones (Lein, 2007 #4147) | http://mouse.brain-map.org/ |
| Cell type specific whole brain map data | This paper | http://mouse.brainarchitecture.org/ost |
| Experimental Models: Organisms/Strains | ||
| Mouse: PV-IRES-Cre, (B6;129P2-Pvalbtm1(cre)Arbr/J) | Jackson Laboratory | Cat#008069 |
| Mouse: SST-IRES-Cre (Ssttm2.1(cre)Zjh/J) | Z. Josh Huang, Jackson Laboratory | Cat#013044 |
| Mouse: VIP-IRES-Cre (Viptm1(cre)Zjh/J) | Z. Josh Huang, Jackson Laboratory | Cat#010908 |
| Mouse: CR-IRES-Cre (B6(Cg)-Calb2tm1(cre)Zjh/J) | Z. Josh Huang, Jackson Laboratory | Cat#010774 |
| Mouse: nNOS-CreER-KI (B6;129S-Nos1tm1.1(cre/ERT2)Zjh/J) | Z. Josh Huang, Jackson Laboratory | Cat#014541 |
| Mouse: CCK-IRES-Cre (Ccktm1.1(cre)Zjh/J) | Z. Josh Huang, Jackson Laboratory | Cat#012706 |
| Mouse: Ai65 (B6;129S-Gt(ROSA)26Sortm65.1(CAG-tdTomato)Hze/J) | Z. Josh Huang, Jackson Laboratory | Cat#021875 |
| Mouse: SST-IRES-Flp (Ssttm3.1(flpo)Zjh/J) | Z. Josh Huang, Jackson Laboratory | Cat#028579 |
| Mouse: VIP-IRES-Flp (Viptm2.1(flpo)Zjh/J) | Z. Josh Huang, Jackson Laboratory | Cat#028578 |
| Mouse: CAG-LoxP-STOP-LoxP-H2B-GFP | Z. Josh Huang, (Taniguchi, 2011 #2169) | N/A |
| Software and Algorithms | ||
| STPT imaging stitching algorithm | Osten Laboratory | http://mouse.brainarchitecture.org/ost/ |
| Convolutional Neural network for cell counting | Osten Laboratory | http://mouse.brainarchitecture.org/ost/ |
| Elastix for image registration | Elastix | http://elastix.isi.uu.nl/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Volocity | Perkin Elmer | http://cellularimaging.perkinelmer.com/downloads/detail.php?id=14 |
| R for statistical computing | R project | https://www.r-project.org/ |
| Statistical algorithm | Osten Laboratory | http://mouse.brainarchitecture.org/ost/ |
| Whole brain flatmap | Larry Swanson | http://larrywswanson.com/?page_id=1415 |
| Python algorithms for digital flatmap | This paper | http://mouse.brainarchitecture.org/ost/ |
| Linear Discriminant Analysis | Python package Scikit-learn | http://scikit-learn.org/stable/ |
| Rate and spiking network simulation and analysis | This paper | https://github.com/xjwanglab |
| Illustrator | Adobe | http://www.adobe.com/products/illustrator.html |
METHOD DETAILS
Brain sample preparation and STPT imaging
Transgenic mice were anesthetized with ketamine/xylazine mix and killed by transcardiac perfusion with isotonic saline followed by 4% paraformaldehyde (PFA) in 0.1M phosphate buffer (PB, pH 7.4). The brains were post-fixed overnight at 4°C, and stored at 0.05M PB until imaging. The fixed brains were embedded and cross-linked with oxidized 4% agarose for precise vibratome cutting. To make oxidized 4% agarose, 2g of agarose (Fisher Scientific, cat.no. BP1356) and 0.21 g sodium periodate (NaIO4)(Sigma, cat.no. S1878) were added in 100ml of 0.05M PB and the solution was gently stirred for 2–3 hours at room temperature under fume hood while protected from light. Then, the solution was washed three times in distilled water followed by one time washing with 0.05M PB using a funnel with filter paper and vacuum suction. The agarose was then re-suspended in 50 ml of 0.05M PB for the brain embedding (this can be stored at 4°C for up to 14 days). For the crossing-linking, sodium borate buffer solution was made by adding 19g Borax (Sodium tetraborate decahydrate, Sigma, cat.no. S9640) and Boric acid (Sigma, cat.no. B0394). Then, 0.2g of sodium borohydrate (NaBH4, Sigma, cat.no. 452882) was added in 100ml of the sodium borate buffer to make sodium borohydride solution, which can be used up to 7 days. For the brain embedding, the oxidized agarose was heated in the microwave. Once the solution became clear and cooled down to 60°C, it was poured into the custom-built mold to embed the brain in consistent 3D angles. When the agarose was solidified, the agarose block was trimmed and placed in 50ml Falcon tube with the sodium borohydride solution for cross-linking at 4°C overnight or at room temperature for 2 – 4 hrs. For the STPT imaging, cross-linked agarose block was glued to a glass slide with magnetic bars underneath, which was then placed in a chamber filled with 0.05M PB. The chamber was placed in the motorized stage in Tissuecyte 1000 (Tissuevision) for the STPT imaging. The block was initially sectioned with 300 µm thickness until the tip of the olfactory bulb was cut, followed by 3 additional cuts with 200 µm and 2 additional cuts with 50 µm. Two-photon excitation focus was set at 60 µm from the sample surface. Once the stable cutting was confirmed, a series of images was acquired in 12 × 16 XY tiles (700 × 700 pixels field of view, 1 × 1 µm resolution) with 270 z section in every 50µm (Kim et al., 2015; Ragan et al., 2012). Badly prepared samples due to poor perfusion and/or dissection errors, and poorly imaged brains were excluded from further analysis.
STPT data processing
Image files were reassembled in 2D and 3D using custom built software (Kim et al., 2015; Ragan et al., 2012). For cell counting, we used CNs that were trained based on nuclear c-fos-GFP signaling (Kim et al., 2015) for H2B-GFP signaling or cytoplasmic signal for tdTomato signal from intersectional approaches. For H2B-GFP signal, we used 6803 cells in 28 randomly chosen image tiles (800×800 XY pixels) (N = 3 mice). For tdTomato cytoplasmic signal, we used 710 cells from 34 randomly chosen image tiles (800×800 XY pixels) (N = 4 mice). Two human experts independently marked up positive signals from the selected image tiles while blind to another person’s and computer’s assessment. Marked cells by at least one expert are used as ground truth data to calculate CNs performance. To enhance signal to noise ratio of tdTomato signal, we used “Subtract Background” function with rolling ball radius = 10 in Fiji (ImageJ) before CNs cell counting. The CNs performance was evaluated using the F-score measure, which represents the harmonic mean of the precision (index for false positive) and recall (index for false negative), where 1 is the best and 0 is the worst.
For the image registration, autofluorescent background is used to register the reference STP (RSTP) brain by the warping algorithm Elastix (3D affine transformation with 4 resolution level, followed by a 3D B-spline transformation with 6 resolution level). Mattes Mutual information was used to calculate the similarity between the moving images (brain samples) and the target reference brain as described previously (Kim et al., 2015; Ragan et al., 2012). Centroids of CNs detected signal in each brain were registered to the RSTP brain based on the registration parameters of each brain.
Automatic cell counting in anatomically defined areas was achieved by computing number of registered signals in each anatomical label using custom built codes (Kim et al., 2015). Cell counting in evenly spaced and partially overlapping 3D sphere voxels (100µm diameter, 20µm apart) was used to digitize and visualize detected signal distribution in the entire brain in an unbiased way using custom built codes (Figure 2, Movie S1 and S2) (Kim et al., 2015). Virtual overlay was done in FIJI (ImageJ) and 3D rendering was done in Volocity (Perkin Elmer).
Cortical and whole brain flatmap
For the cortical flatmap, we adapted the previously used method (Watabe-Uchida, 2012 #273). First, isocortex area of the RSTP brain was binarized with every 200um z interval from bregma anterior/posterior axis +3.045 to −4.855. Second, zero line was placed along the most dorsal medial corner of the cortex and evenly spaced bins were generated in every 600um in the middle guideline along the medial and lateral axis of the cortex. Centroid of cells detected by automated CNs counting was registered to the RSTP brain with the cortical bins, and number of cells in each bin was computed by a custom-built code. For the whole brain flatmap, we used a rat brain flatmap generated by Larry Swanson (version 4.0 beta3, 2015) downloaded from http://larrywswanson.com/?page_id=1415. We used custom-built Python code to map our density measurement onto the flatmap for 2D visualization.
Interneuronal circuit model
We studied linear dynamics of a four-population rate model.
The weight matrix W has the form
The connection from λ to X, WXY, is scaled by the normalized density of the projecting population, . The connectivity value when ρE = ρP = ρS = ρV = 1 follows (Litwin-Kumar et al., 2016), therefore
ρE is omitted since they are set to equal to 1. The normalized density ρX for each area is the absolute density normalized by the absolute density averaged across all areas. For the main text, we used cell densities from L2/3. We set
These time constant parameters have no effect on the steady-state responses, however, they could change the stability of the fixed point.
In this model, the steady state of the system, given it is stable, is
1 is the identity matrix. The linear response of the steady-state activity to external inputs is
The current from population X to Y is IYX = WYXrX. The current response to external inputs targeting population Z, , is defined as .
Spiking neural circuit model
Each population X (= E, P, S, V) is modeled with NX adaptive exponential integrate-and-fire neurons, where the H-th neuron is described by:
is the external input, decomposed into a mean and a noise term. ηi(t) is a white noise Gaussian process. The adaptation current follows
Whenever a neuron spikes, is increased by an amount bX. The synaptic conductance from population Y to the i-th neuron in population X follows
tj,k is the time of the k-th spike from neuron j in population Y.
is the connection weight from the j-th neuron of population Y to the i-th neuron of population X. Ideally, we would like to set with probability , and otherwise. However, this will introduce additional variability when we compare simulations with different parameters (cell density). In order to reduce this variability, the connections from population Y to X, if exist, are all-to-all. The connection weight is then the original connection weight GXY multiplied by the would-be probability .
Parameters are mainly taken from (Litwin-Kumar et al., 2016). We used 4000 excitatory neurons, 500 PV neurons, 250 SST neurons, and 250 VIP neurons. The membrane capacitances are and . The leak conductances are , and . The membrane time constants are τE = 28.8ms, τP= 8ms, and τS = τV = 16ms. The resting potential is −60mV for all neurons. The threshold voltages are and . The EIF slope parametr ΔT is 0.25 for PV neurons and 1 for other neurons. The reset potential Vre is −60mV for all neurons. The refractory period τref is 2ms for all neurons. The subthreshold adaptation a is 4nS for all populations except for PV neurons that do not adapt (a = 0nS). Similarly, the spike-triggered adaptation b = 8pA for all neurons except for PV neurons (bP = 0pA). The adaptation time constant τw is always 150ms. The standard deviation for external input σext = 3.5mV for all neurons. The reversal potential Esyn is 0mV for excitatory synapses while it is −67mV for inhibitory synapses. The synaptic time constants are , and .
For consistency with Litwin-Kumar et al., 2016, we assume that neurons are tuned to orientation. This assumption has little effect on our results. Neurons' preferred orientations span (−π/2, π/2) uniformly. And the connection probability is given by
θi is the preferred orientation of the i-th neuron from population X. The connection weight matrix, and the would-be connection probability matrices are given below.
In Figure S7E, we varied the density of PV and SST neurons. If we keep the external background inputs to the system the same while varying cell density, then the spontaneous activities of the network would be very different, which in turn affects how neurons respond to inputs. So a critical step is to ensure the spontaneous activities of the network is more or less the same when varying cell density. In order to achieve this uniform spontaneous activity, we used an optimization algorithm to find the proper mean background inputs , X = E, P, S, V.
We compared the spontaneous activity condition with the PV-activated condition, in which the mean level of external input to the PV population was increased by 0.5 mV. This is a relatively modest increase aimed at probing the linear response of the system. We computed the change in the average currents from PV, SST, and E neurons to E neurons.
Gradients of circuit responses with respect to cell densities
We studied a simplified linear circuit model
In component form,
where X, Y ∈ {E, P, S, V}. The output connection weights of a population are scaled by the cell density of that population, .
Rate responses
In this model, the steady state of the system, given it is stable, is
where 1 is the identity matrix. We denote
the linear response matrix. An entry of this matrix,
represents the change in X population activity when unit-level external inputs target the Y population. MXY takes into account not only the direct connection WXY (if exists), but also all the indirect connections through other types of neurons. For the linear system, we have
We are interested in how the response depends on cell densities. Therefore we now calculate the derivative:
First, we calculate this derivative
Notice that
Then we have
Therefore when X ≠ Z, we have simply
Current responses
We next study how the current responses depend on cell densities. An unit-level external input targeting a Y population uY will change the X population steady-state activity by MXY, which then changes the steady-state X-to-V current by WVX MXY.Mathematically speaking, the current response
We can calculate the derivative of this current response with respect to the density of population Z, ρZ:
We can understand the signs of all rate and current responses presented in the main text with these expressions. For example, increasing SST+ density strengthens the PV-to-E current response when external inputs target the PV population. Mathematically, this means
The above equation holds true because
and MPS < 0, MSP < 0. MPSMSP describes an effective disinhibitory loop which is strengthened with higher SST+ density.
Conditions for findings
We observe several interesting and non-trivial phenomena in the rate and current response maps (Figure 5, Figure S7). The E rate response maps have similar contours when external inputs target the PV or the SST population (Figure S7A, top row). When the external input targets the PV population, the PV-to-E current response map has similar contours as the SST-to-E current response map (Figure S7A, second column). Similar finding is seen when the external input targets the SST population (Figure S7A, third column). More specifically, the PV-to-E current response and the SST-to-E current response usually have opposite signs but similar magnitudes, regardless of the cell densities.
Below we show that all these phenomena are closely related, and they all hold true when the recurrent E-to-E excitation roughly balances the own leakage of the E population.
Rate responses
The E rate response is described by MEP when external inputs target the PV population, and MES when external inputs target the SST population. For these responses to have similar contours in the PV+/SST+ density plane, their gradients with respect to the cell densities
and
should point to the exact opposite directions. Therefore we need
| (1) |
as well as
| (2) |
Because
So equation (1) becomes:
Below we will compute MPP MSS − MPS MSP.
The inverse of a matrix A can be written as its adjugate adj(A) divided by its determinant det(A),
The VIP population plays minimal role when external inputs do not directly target them. So for simplicity, here we ignore the VIP population, and consider only excitatory, PV, and SST populations. Then the connectivity becomes
The response matrix also becomes 3-by-3,
So we have
Since M = (1 − W)−1 is invertible, we have det(M) ≠ 0. So for equation (1) to hold exactly we need
Next we derive conditions for the signs to be correct as in equation (2). For the reduced 3-dimensional system, we can explicitly calculate the determinant and the adjugate of the matrix 1 − W. The determinant is
And the adjugate matrix is
When WEE = 1 we have
Because
we have
Also
Therefore the signs of and are fixed. On the other hand,
Because
and
in order to satisfy equation (2), we need [adj(1 − W)]ES < 0 or equivalently
which means that the SST population needs to be overall net inhibitory to the E population. The left-hand-side represents the strength of the recurrent E-SST-E inhibitory loop, while the right-hand-side represents the strength of the E-SST-PV-E recurrent disinhibition loop. Here we see a direct tradeoff between SST's roles in inhibition and disinhibition (through PV). In our model, the parameter choice made SST overall inhibitory, satisfying equation (2).
Current responses
Now we show that when WEE = 1 the PV-to-E current response and the SST-to-E current response will always cancel out each other perfectly, leading to a net-zero change of inhibitory currents in response to external inputs onto inhibitory neurons.
Because
we have
This is simply stating that the rate response is the result of currents from PV, SST, and E populations.
When WEE = 1 we have
and
Therefore
and
Similarly,
and
These equations mean that the total inhibitory current onto the excitatory population does not change in response to external inputs onto the inhibitory populations.
Anatomical description of PV+, SST+ and VIP+ neuronal distributions in the major divisions of the mouse brain
We provide detailed description of the three major GABA types to facilitate anatomical understanding of their subcortical expression pattern. We also encourage readers to examine high resolution raw data from each cell type in our web site (http://mouse.brainarchitecture.org/cellcounts/ost/).
Basal nuclei
Overall, the basal nuclei are mostly labeled by SST+ neurons. The basal nuclei SST+ neurons (~300,000) are about one order of magnitude more numerous than the PV+ (31,914), and two orders of magnitude than the VIP+ neurons (2,969). PV is consistently present only in a few regions of basal ganglia, and VIP+ neurons are mostly in the order of tens per nucleus. Even though the PV+ neurons are the most numerous, they are not uniformly distributed, and tend to form specific patterns.
Rostral-most, the nucleus accumbens (ACB) is intensely labeled by SST around the anterior commissure (ac). The rostral lateral septum (LSr) and substantia innominata (SI) are also heavily labeled around this level (+1.64 mm from bregma). Proceeding caudally, the SST field in the ACB extends dorsally and labels the lateral wall of the rostral caudoputamen (CP). The remaining SST label in ACB is restricted to its dorsal part and around ac. Further caudally, the SST label of ACB is reduced to a band just below the ac, which separates in two segments. The dorsomedial segment tends to migrate toward the lateral septum, while the lateral part of this SST field migrates toward the piriform cortex (PIR). The ACB becomes again intensely labeled by the SST in its caudalmost part.
The lateral SST field in the rostralmost CP becomes thinner more caudally, merges with the SST field in the lateroventral ACB, and gradually extends horizontally, to form the ventral border of CP at about 0 mm from bregma. This SST field continues to label the ventral CP to its remaining rostrocaudal extent (see below).
The fundus of the striatum (FS) is labeled by the SST almost entirely across its rostrocaudal extent. In its rostral part, FS is labeled mostly dorsally, close to the border with CP, while caudally SST labels it entirely and strongly.
The SI is diffusely labeled by SST in its more caudal part. However, it includes an ovoid and restricted PV field just below the rostral BSTal. Proceeding caudally, this field becomes elongated in the dorsal SI, and it will migrate in the ventral sector of the rostralmost part of the external globus pallidus (GPe). The PV field of SI does no intermingle with the surrounding SST.
The LSr is very intensely SST-labeled with the exception of a lateral zone long of about 1 mm where the label is moderate at best, after which it becomes entirely labeled. The caudal and ventral parts of the lateral septum (LSc; LSv) are also intensely labeled by SST. However, their neighbors, the septofimbrial nucleus (SF) and the triangular nucleus of the septum (TRS) are weakly labeled by SST, and only rostrally and laterally.
The rostral parts of the medial septum (MS) and the nucleus of the diagonal band (NDB) are diffusely labeled by SST. More caudally, the NDB includes a restricted PV field, which does not intermingle with the SST+ neurons. Further caudally, the SST label in MS becomes weak, and strong in NDB. This field also labels the magnocellular nucleus (MA) at least partially.
The olfactory tubercle (OT) is void of any label rostrally. However, PV+ neurons are found in layer II, and SST is expressed across all its three layers more caudally.
The anterior bed nuclei of the stria terminalis (BST) are intensely labeled by SST, but only those dorsal from the ac. Ventral from the ac, the SST label is strong only in the ventral part of the anterolateral BST (BSTal), moderate in the anteromedial nucleus (BSTam), and in the fusiform nucleus (BSTfu). The nuclei of the posterior division, however, show different SST labeling patterns. Thus, the principal nucleus of the BST (BSTpr) is labeled very strongly on its rostrocaudal extent, as well as the dorsal part of the interfascicular nucleus (BSTif), and the rostral segment of the transverse BST (BSTtr).
The amygdalar nuclei associated with the striatum are intensely labeled by the SST, with the exception of the ventral part of the anterior amygdala (AAA), the basal amygdala (BA), and the ventral tips of the anteroventral and anterodorsal nuclei of the medial amygdala (MEAav; MEAad), respectively. The capsular, lateral and medial nuclei of the central amygdala (CEAc, CEAl, CEAm) are exceptionally labeled by SST, and form a continuous field with the ventral sector of the caudal CP.
Finally, the GPe is labeled by PV rostrocaudally, and the internal segment of the globus pallidus (GPi) includes both PV+ and SST+ neuronal populations. These populations tend to be segregated spatially (see Figs. 7a and 7b).
Thalamus
The nuclei of the thalamus are labeled almost exclusively by SST and PV, though a number of the SST-Cre:H2BGFP+ nuclei do not appear to express SST mRNA or protein in the adult brain based on the available data from the Allen Mouse Brain in situ hybridization Atlas and/or previous literature {Fitzpatrick-McElligott, 1988 #4028;Johansson, 1984 #4030;Sunkin, 2013 #2069;Priestley, 1991 #4031}. These nuclei, which most likely express SST transiently during the development, include the central medial nucleus (CM), intermediodorsal nucleus (IMD), anteromedial nucleus (AM), anteroventral nucleus (AV), mediodorsal nucleus (MD), interanterodorsal nucleus (IAD), interanteromedial nucleus (IAM), paracentral nucleus (PCN), and parataenial nucleus (PT). The SST+ cells are, however, prominent in the reticular nucleus (RT), even in the adult brain.
Large PV+ neurons are found also in the RT and in the caudal thalamus in the ventral posteromedial nucleus (VPM), in the VPL, and in the paracentral nucleus (PCN). Finally, both lateral (LH) and medial (MH) are labeled by SST. Hence, an extended and intense ventrodorsal SST field is observed in the caudalmost thalamus, covering SPA, IMD, PVT, LH, and MH.
Hypothalamus
The SST+ cells in the hypothalamus are more abundant than PV+ neurons (13,000), and the latter more numerous than the VIP+ neurons, though in four hypothalamic structures (the anterodorsal and anteroventral preoptic nucleus, vascular organ of the lamina terminalis, and lateral mammillary nucleus) SST-Cre:H2BGFP labeling is likely due to developmental SST+ expression that is not maintained in the adult brain, similarly to the thalamic data described above {Fitzpatrick-McElligott, 1988 #4028;Johansson, 1984 #4030;Priestley, 1991 #4031;Sunkin, 2013 #2069}. Most of the VIP+ hypothalamic neurons are concentrated in the suprachiasmatic nucleus (SCH).
In the rostral part of the hypothalamus, SST+ neurons are distributed along the 3rd ventricle (V3), in the preoptic part of the periventricular nucleus (PVpo). A second group of SST+ neurons are located in the ventral part of the medial preoptic area (MPO), lateral from the SCH. This field has a roughly ovoid form and extends caudally into the retrochiasmatic area (RCH). The dorsal part of MPO and medial preoptic nucleus (MPN) are diffusely SST labeled, as the ventral extension of the BSTpr field.
More caudally, SST+ neurons form a lens-like field around a circular PV+ field, in the anterior hypothalamic nucleus. This PV+ “lens”-like field migrates laterally, in the lateral hypothalamic area (LHA), and surrounds the fornix (fx) dorsally. At this level (approx. −1 mm from bregma), the zona incerta (ZI) is very intensely labeled by SST. The ZI SST field is continuous with the one in VM, which curves dorsally around the mammilothalamic tract (mmt).
This is also the rostralmost level of a very conspicuous SST field that covers the ventral floor of the hypothalamus for more than 1.2 mm rostrocaudally. This field initially follows the dorsal borders of the arcuate (ARH) and tuberal (TU) nuclei. More caudally, it labels the ventrolateral part of the ventromedial nucleus (VMHvl), the ventral part of the ventral premammillary nucleus (PMv), the posterior periventricular nucleus (PVp), and the dorsal and ventral tuberomammillary nuclei (TMd; TMv).
Dorsally, a second conspicuous SST field forms at about the same level with the ventral one. This field includes the ventrolateral ZI, the ventral VM, and the SMT SST+ neurons, which initially cover the mtt ventrally and completely surround it more caudally. This dorsal hypothalamic field extends caudally in the posterior hypothalamus (PH), and in the anterior and posterior parts of the dorsomedial hypothalamic nucleus (DMHa; DMHp). The ZI includes an additional PV+ population of neurons, besides the SST+ neurons that are preferentially located more rostrally and ventrally. This PV field starts in the subthalamic nucleus (STN), and then migrates into the ventral ZI where it extends for about 600 microns. Different of the SST field, the PV+ neurons are spatially grouped in a bar-shaped field positioned in the middle of ZI, and surrounded by SST+ neurons. The PV+ neurons tend to be spatially segregated from the SST+ except at the periphery of the bar-shaped field, and in its caudal end. In the STN, the PV field is replaced by SST+ neurons.
The caudal hypothalamus is largely void of labeled neurons, except for a mediolateral PV field that starts as two circular subfields in the rostral supramammillary (SUM) and lateral mammillary (LM) nuclei, respectively. Further caudally, the medial aspect of this PV field develops a hook-like shape, in the mediodorsal half of the mammillary bodies (MM). This field includes about 25% of the PV+ hypothalamic neurons.
Midbrain
As in the basal nuclei, thalamus and hypothalamus, the midbrain SST+ neurons are the most numerous (365,000) of the three classes. The PV+ neurons are about 1/3 of the SST+ (135,000), while the VIP+ neurons do not exceed 8000 (see Table S1). The majority of the labeled neurons are in the sensory-related midbrain nuclei: 59% of the PV+ neurons, 62% of the SST+, and 81% of the VIP-expressing cells. The motor-related midbrain nuclei include 33% of the PV+, 35% of the SST+ and 13% of the VIP neurons. Hence, only 4% to 6% of each of the three classes are located in the behavioral staterelated midbrain subdivision.
In the superior colliculus (SC), the SST-expressing neurons make the majority of the three classes in most of its layers. One exception is the most superficial layer of SC, the zonal layer (SCzo) where the PV+ and the SST+ neurons are in almost equal numbers. The SST+ neurons are evenly distributed in these layers, while the PV-expressing neurons tend to occupy the most superficial (dorsal) part of it. The second exception is found in the b sublayer of the intermediate gray layer (SCig-b) where the PV+ neurons form a latero-ventral “wing”, rostrally.
The inferior colliculus (IC) includes about half of the PV+ and SST+ and most of the VIP+ neurons identified in the sensory-related division of the midbrain. Rostralmost, the external IC (ICe) is labeled strongly and uniformly by the SST. The PV+ neurons are concentrated in a circular field located in the ventral ICe. Proceeding caudally, the SST labels very strongly and relatively uniformly all IC subdivisions with the exception of a tear-shaped region, ventrally. The PV+ neurons tend to be distributed mostly in the ICe and in the dorsal subdivision (ICc). Topographically, the PV+ neurons extend into the IC subdivisions along a ventro-dorsal gradient. PV+ and SST+ neurons tend to be intermingled across IC, except in the ventral tear-shaped region mentioned above. The VIP+ neurons are preferentially located in the more caudal IC, where they form a dorsomedial wing-shaped field. Finally, the VIP+ neurons occupy the entire caudalmost IC.
In the midbrain motor-related subdivision, the midbrain reticular nucleus (MRN) does not include high numbers of any of the three neuronal classes, but a small and distinct SST field can be observed in the rostral MRN, just above the fx. A specific feature of motor-related subdivision may be that of predominance of PV+ neurons relative to the SST+. Thus, the reticular part of the substantia nigra (SNr), the anterior pretectal nucleus (APN), the anterior tegmental nucleus (ATN), the oculomotor nucleus (III), the red nucleus (RN) and the ventral tegmental nucleus (VTN) include PV fields that are restricted within their respective boundaries.
The PV+ and SST+ neurons intermediate and deep layers of SC are partially segregated topographically. Thus, the PV+ neurons tend to be located in the central parts of the motor-related SC, while the SST+ neurons tend to form laterodorsal “wings” around the PV field, with the highest density laterally.
The rostral periacqueductal gray (PAG) is moderately labeled by the SST, and it includes few PV+ neurons. However, the nucleus of Darkschewitsch (ND) and the interstitial nucleus of Cajal (INC), which are PAG’s ventrolateral neighbors, express high levels of both SST and PV. More caudally, the PAG SST+ neuronal populations tend to be organized in topographical quadrants, akin to the subdivisions identified in other species (Swanson, 1998; Swanson, 2004). The PAG also includes VIP+ neurons, which are about 67% from the total of VIP+ cells identified in the midbrain motor division. These neurons form a strong field in the caudal PAG, centrally, along the midline.
The nuclei of the behavioral state-related subdivision mostly include SST+ neurons, which are uniformly distributed within their borders. The pedunculopontine nucleus (PPN), the interpedunculopontine nucleus (IPN), and the dorsal raphé (DRN) contain the highest numbers of the SST+ neurons. DRN also includes most of the VIP+ neurons of this midbrain subdivision.
Hindbrain
The general theme of the PV+, SST+ and VIP+ distributions in the mouse subcortical main divisions is also true in the hindbrain and its sudivisons: SST+ neurons are more numerous than PV+, and the VIP+ neurons are only in the order of thousands in an entire basic neuroanatomical division. However, the functionally related subdivions of both pons and medulla have specific ratios of PV+ and SST+ neurons.
Pons
In the sensory-related pons, the nucleus of the lateral lemniscus (NLL), the nuclei of the superior olivary complex (SOC), and the principal nucleus of the trigeminal (PSV) include more PV+ than SST+ neurons. Specifically, in the dorsal and the ventral NLL, the PSV, and in the periolivary region of SOC (POR) the PV+ neurons are from about. 1.5 to about 3 times more than the SST+. These two populations of neurons tend to partially overlap. The PV+ neurons tend to be concentrated in the middle of POR and NLLv, while SST+ neurons form an exterior ring in POR and border medially in the NLLv, respectively. In the PSV, the PV+ and SST+ neurons are completely intermingled. The nuclei of the parabrachial nucleus (PB) include more SST+ neurons than PV+. The VIP+ neurons are few in this pons subdivision (342), and almost half of them (149) can be found in SOC. The VIP+ neurons in SOC tend to group in two distinct fields that are roughly circular, and placed caudalmost.
In the motor-related pons, the PV+ and SST+ neurons of the pontine central gray (PCG) and of the tegmental reticular nucleus (TRN) are specifically organized. Rostralmost, the PCG SST+ neurons form a ring with the highest density in the ventrolateral corner, while the PV+ neurons assume the shape of the nucleus with the highest density centrally. More caudally, the PV+ neurons form a lens-shaped field, overlapping with the SST+ field. Further caudally, at about −4.15 mm from bregma, both neuronal populations occupy the entire PCG and TRN, each field having two zones of maximal density, respectively. Finally, at the levels where only TRN remains present, the maximal densities of the PV+ and SST+ fields are somewhat spatially complementary: the SST+ neurons are densest in the dorsomedial corner of the nucleus, while the maximal density of the PV+ field is located centrally. Two pontine motor nuclei include more PV+ than SST+ neurons: the motor nucleus of the trigeminal (V), and the supratrigeminal nucleus (SUT). Both rostral and caudal divisions of the pontine reticular nucleus (PRNr; PRNc), the first included in the motor-related and the second in the behavioral state-related pons, have the SST+ neurons more or less uniformly distributed in a diffuse field. The VIP+ neurons identified in this subdivision of pons tend to group in three nuclei: PRNc, SUT, and V. The VIP+ neurons in V and SUT form a ring-like field across them.
In the behavioral state-related pontine nuclei, the ratios between SST+ neurons and the PV+ can be as high as an order of magnitude. Consequently, these nuclei appear very highly labeled by SST. Topographically, they tend to group either along the midline, as the raphé nuclei and the nucleus incertus (NI), or dorsally.
Medulla
As in the sensory division of pons, the PV+ neurons tend to be more numerous than the SST+ in many nuclei of the sensory-related medulla. These include the cochlear nuclei (CO), the cuneate (CU) and external cuneate (ECU) nuclei, nucleus gracilis (GR), and the nucleus of the trapezoidal body (NTB). Rostrocaudally, CO, CU, ECU and GR are neighbors in the laterodorsal part of the medulla. Furthermore, the lateral and the superior salivatory nuclei (LAV; SUV) of the motor-related medullar nuclei also have larger PV+ than SST+ neuronal populations. Thus, many if not most of the PV+ neurons are distributed latero-dorsally in the medulla. Both PV+ and SST+ neurons are heavily present in the interpolar part of the spinal nucleus of the trigeminal overlap (SPIV), where they overlap. A second PV+ field is observed in the lateral part of the perigigantocellular nucleus (PGRNl), where is segregated from the SST+ field.
The regions of medulla where SST+ neurons dominate tend to be more medial than the nuclei with high PV+ populations. The former includes the nucleus of the solitary tract (NTS) and its parts, which form a rostrocaudal dense SST field around the fx, the gigantocellular (GRN) and the intermediate reticular (IRN) nuclei, and the area postrema (AP).
The VIP+ neurons are few, if any, in the nuclei of the medulla. However, few nuclei include these neurons as distinct fields. VIP+ neurons are first observed as two fields in the facial motor nucleus (VII), where they migrated from the caudal SOC. A second VIP+ population, organized as a round field near the midline, is observed in the caudal part of the nucleus raphé obscurus (RO). Finally, the hypoglossal nucleus (XII) includes a small population of VIP+ neurons, organized as an elongated field near the midline, at about the same level where RO field ends.
Patterns
As described above, the SST+ neurons are more numerous than the PV+ in the basic subcortical divisions of the mouse brain. In the sensory-related pons and medulla, however, the PV+ neurons predominate over those that express SST. The VIP+ are the least numerous, in the order of thousands in each basic subcortical division, at best (the entire thalamus includes only 75).
The SST+ neurons tend to be organized in fields that cross neighboring nuclei, rather than spatially restricted regions within neuroanatomical nuclei or regions. This is the case of the very strong SST field that covers the ventral corner of the caudal half CP and the subdivisions of the CEA, MEA, as well as AAA and COAa. The CP portion of this SST field, especially its most caudal aspect, corresponds at least partially to the rat “amygdalo-striatal transition zone” (AStr; deOlmos et al., 2004). The AStr receives a specific set of cortical inputs: visual and auditory associative (Shi & Cassell, 1997; McDonald, 1998), polymodal from the ectorhinal cortex (ECT; Shi & Cassell, 1998), and from the anterior insular areas (Shi & Cassell, 1998). Another example of SST field crossing neighboring regions is in the caudal thalamus, where it labels all midline nuclei in ventrodorsally. In the hypothalamus, such field is present for more than 1 mm rostrocaudally, in its ventral part.
A second feature of SST-labeled neurons is that of organizing around major fiber tracts, surrounding them partially or completely, as seen in frontal sections. Thus, SST+ neuronal populations can be identified in the close proximity or around the ac from the rostralmost ACB to 0 mm from bregma. Further caudally, the internal capsule (int) is bordered by an SST field, to RT (−0.4 mm from bregma). A second SST forms around the temporal limb of the anterior commissure (act) at about the same bregma levels. In the ventral thalamus and in the hypothalamus the same can be observed around the mmt and the fx, respectively. Further caudally, the vertical SST field in the caudalmost thalamus that was mentioned above, makes the medial border of the fx and tends to curve around it. Further caudally, in the midbrain, the SST+ neurons of the central linear (CLI) and dorsal (DR) raphé nuclei are in close vicinity of the medial longitudinal fasciculus (mlf). Thus, several of the mouse brain major tracts, such as the ac, fx or mft, may be seen as rostrocaudally “embedded” along some of their sectors by SST-expressing neurons. Hence, one may hypothesize that a generic function of the subcortical SST-expressing neurons may be that of modulation of the input that comes via these major tracts.
Different of the SST+, the PV+ and VIP+ neurons tend to group in specific nuclei and regions. The PV+ and VIP+ neurons also tend to be located more in the midbrain and hindbrain than in the more rostrally located subcortical divisions. The majority of PV+ neurons identified in the midbrain can be found in the sensory-related nuclei, and most of those in the IC. However, as described above, the motor-related midbrain subdivision includes several nuclei that have high ratios of PV+ relative to SST+ neurons. Interestingly, these nuclei have very few VIP+ neurons, if any. In the hindbrain, however, the PV+ neurons are the majority in the sensory subdivisions of both pons and medulla. Topographically, the PV+ neuronal populations in the hindbrain tend to concentrate in a “ring” of nuclei, which starts rostrally ventrally, continues laterally, and it is closed dorsally at the caudal-most levels (see movie S1).
QUANTIFICATION AND STATISTICAL ANALYSIS
3D cell counting and density measurement
To convert CNs 2D cell counting in the 3D counting estimates, sample brains for H2B-GFP and tdTomato were imaged at xyz resolution 1 × 1 × 5 µm to achieve complete 3D imaging without sub-sampling. Then, H2B-GFP+ or tdTomato+ cells were manually counted respectively in 3D in 60 image tiles (200 × 200 × 50 µm in xyz) or 31 image tiles (600 × 600 × 50 µm, xyz) randomly selected from the whole brain, respectively. The 2D-to-3D conversion was then derived by dividing the manual 3D counts in 3D boxes (H2B-GFP: 365 cell, tdTomato: 250 cells) by CNs based 2D counts (H2B-GFP: 246 cell, tdTomato: 173 cells) from the middle section in each 3D box (Figure S1E). Lastly, the obtained conversion factors of 1.5 for H2B-GFP and 1.4 for tdTomato were used to multiply the 2D region of interest (ROI) counts in order to estimate the total numbers of cells in each anatomical volume (Table S1).
To measure volume of anatomical ROIs, the average RSTP brain synchronized with the Allen reference atlas (ARA) segmentation was reverse-registered onto each individual brain using the Elastix warping procedure described above. Then, the ARA segmentation was registered onto each individual brain based on the registration parameters. Lastly, number of voxels that belong to each ROI in the transformed ARA segmentation were counted, multiplied by 20 × 20 × 50 µm3 (3D volume of anatomical voxel unit), and used as volumes of each anatomical ROI (Table S2). The 3D estimates of cell numbers in each ROI were divided by corresponding ROI volume to generate density measurement per mm3 in each anatomical ROI (Table S3).
Statistical analysis
Number, volume, and density of different cell types in different anatomical regions were presented as mean ± standard deviation (Figure 2D, Figure 3B–E, Figure S4A, Figure S5B–C, Table S1, Table S2, Table S3, Table S4). Mean values were used for the flatmap display and relative density (Figure 1C, Figure 2A–E and G, Figure 4D, Figure S2, Figure S4B, and Figure S6D).
Linear Discriminant Analysis (Figure 4B,C, Figure S6B,C) is implemented with the Python package scikit-learn using the default parameters. Other linear classification methods, including linear support vector machine (SVM), yield similar results. The classifier performance is computed using leave-one-out cross-validation. The classifier performance is also tested on 400 instances of shuffled subnetwork labels to obtain the 95% confidence interval for classifier performance on random labeling.
To identify sexually dimorphic brain regions, we performed statistical comparisons between males and females in the three major GABA neurons based on anatomical ROIs using open source statistical package “R”.
Sample size estimate. We estimated our sample size using the power analysis as described in (Kim et al., 2015). We used PV cell counting data in different ROIs from male brains (n = 5) and performed statistical simulation using variable effect sizes (ranging from 0.1 to 2.45) at selected significance level (α < 0.05). We calculated number of ROIs that has over 80% power as the sample size was changed in the reasonable range (3 to 14). We observed “elbow” at N = 5 sample size with the given effect size (0.85), represent the sample size that addition power gained by adding sample starts to decrease (Kim et al., 2015). Our simulation data estimated that 88% ROIs (695 out of 799 regions) reach sufficient power with N = 5 samples per group.
Statistical analysis between groups. We assumed the cell counts at a given anatomical area, Y, follow a negative binomial distribution whose mean is linearly related to an experimental condition (sex difference), X: E[Y]=α+βX as described in (Kim et al., 2015). We found the maximum coefficients α and β through iterative reweighted least squares, obtaining estimates for sample standard deviations in the process and the significance of the β coefficient. A significant β infers the group status is related to the cell count intensity at the specified anatomical location. The z-values correspond to this β coefficient normalized by its sample standard deviation, which under the null hypothesis of no group effect, has an asymptotic standard normal distribution. The p-values represent the probability of obtaining a β coefficient as extreme as the one observed by chance assuming this null hypothesis is true. To account for multiple comparisons across all ROI locations, we thresholded the p-values and reported false discovery rates with the Benjamini-Hochberg procedure. In contrast to correcting for type I error rates, this method controls the number of false positives among the tests that have been deemed significant.
DATA AND SOFTWARE AVAILABILITY
Full resolution images of samples are freely available from http://mouse.brainarchitecture.org/cellcounts/ost/
Cell counting data in each cell type and comparison between males and female brains are available in Table S1
Brain region volume measurement from male and female brain are available in Table S2
Density data in each cell type are available in Table S3.
Custom built codes for the STPT imaging and data processing are available in http://mouse.brainarchitecture.org/ost/ under “Cell Type Analysis Toolbox” link
ADDITIONAL RESOURCES
High resolution images of the cell type specific dataset is easily searchable in http://mouse.brainarchitecture.org/ost/
Allen reference brain with anatomical abbreviation is accessible in http://atlas.brain-map.org/
Supplementary Material
Figure S1. Automated CN-based cell counting with a 3D conversation and 3D volume measurements, Related to Figure 1.
(A–D) An example from SST-Cre:H2B-GFP brain (A), scale bar = 1mm. (B) High magnification image from yellow inset box in (A), scale bar = 50µm. (C) CN based cell detection. (D) Examples of watershed algorithm based cell separation, shown by green and red. CNs automated cell detection reached F score = 0.92 (precision = 0.97 and recall = 0.87) for H2B-GFP-based cell counting, and F score = 0.91 (precision = 0.86 and recall = 0.96) for tdTomato cytoplasmic-based cell counting (see the Methods).
(E) Manual 3D cell counts were obtained by a stereological principle: cells touching 3 sides highlighted in black lines of 3D space were included and the ones touching the other 3 sides highlighted in red lines were excluded in the counting (In the schema, the cells are elongated in the Z dimension, representing the point spread function effect of two-photon imaging. The 3D manual counts were used to derive the 2D to 3D conversion factor for generating cell densities per mm3 (see Methods).
(F) 3D volume measurement of anatomical structures. RSTP brain (F1) was registered to each brain (F2) to match its 3D shape (F3). 2D examples of before registration (F4) and after registration (F7) to SST-Cre:H2B-GFP brain (F6). RSTP label (F5) was transformed (F8) based on 3D image registration (F1–F3). Number of areas occupied each transformed label (F8) can then be calculated as volume.
(G) Whole brain flatmap, modified from the original version in http://larrywswanson.com/?page_id=1415
Figure S6. Cortical areas in L5 cell density spaces, Related to Figure 4
(A–D) Same analyses as in Figure 4, but with L5 density data. Segregation of cortical subnetworks is still present but is weaker in L5 data in comparison to L2/3 data.
Figure S7. Complete current response maps and spiking network simulation, Related to Figure 5
(A) E-E, PV-E, and SST-E current response maps (top to bottom) when external inputs target the E, PV, SST, or VIP population (left to right). The excitatory population rate response is proportional to the E-E current response.
(B) Excitatory population rate responses to external inputs targeting PV (left) or SST (right) population in the intact local circuit (top row), after removing all recurrent excitatory connections (middle row), or after weakening all connections by 70% (bottom row).
(C) Spiking neural network model consisting of 5,000 neurons. (D) Raster plot of all neurons at the spontaneous state. (E) From left to right: the PV-to-E, SST-to-E, and E-to-E current responses to external input currents driving PV neurons.
Table S1. Number of cells from the seven different subtypes across the entire brain, Related to Figure 1, 2, 3, S5, and Table S4
Table S1a tab includes cell counting (average ± standard deviation) data from the seven cell types.
Table S1b tab includes separate male and female cell counting (average ± standard deviation) data from the PV, SST, and VIP cell types with multiple comparison corrected p-value (called “q-value”) in each cell type.
Table S2. Brain volume measurement in the entire brain areas in male and female brain, Related to Figure 7 and S1.
Mean and standard deviation of brain volumes from male and female brain (N = 15 samples in each sex) is list across the entire brain areas.
Table S3. Density of cells from the seven different subtypes across the entire brain, Related to Figure 1, 2, 3, 4, S1, S5, and S6.
Table S4. Sexually dimorphic brain areas in SST and VIP neurons, Related to Figure 7
Cell counting (average ± standard deviation) of males and females (N = 5 mice per sex) and false discovery rate (FDR) adjusted statistical significance (q value) for multiple comparison correction in sexually dimorphic brain regions.
Movie S1. Quantitative display of the three major GABA subtypes and their overlay in evenly spaced voxel map, Related to Figure 1 and 2.
Movie S2. Quantitative display of the SST and VIP subtypes and their overlay in evenly spaced voxel map, Related to Figure 1 and 2.
Figure S2. Deep (5 and 6) and superficial (2/3) layer density of PV, SST, and VIP neurons, Related to Figure 2.
(A–B) Deep layer density distribution.
(A) Relative 3D density of PV+, SST+, and VIP+ neurons in the isocortex areas.
(B) Density mapping of PV+, SST+, and VIP+ neurons in the cortical flatmap (see also Table S3 for cell density in each anatomical area).
(C–D) Superficial layer density distribution.
(C) Relative 3D density of PV+, SST+, and VIP+ neurons in the isocortex areas.
(D) Density mapping of PV+, SST+, and VIP+ neurons in the cortical flatmap (see also Table S3 for cell density in each anatomical area).
Figure S3. STPT data in comparison to Allen Brain in situ hybridization data, Related to Figure 1.
Left two columns of panels: note the low density of PV+ cells in the infralimbic (ILA, row 1) and posterior agranular insular cortex (AIp, row 4) (top and bottom panels) compared to the motor (row 2) and somatosensory cortex (row 3) detected by both STPT imaging of the PV-Cre-H2BGFP+ cells (first column of panels) and Allen in situ hybridization (second column of panels).
Right two columns of panels: In contrast, a comparable SST+ cell density is detected by both methods in all four regions, resulting in high SST-to-PV ratio in the ILA and AIp compared to the motor and somatosensory areas.
Allen in situ data for the PV and the SST are from http://mouse.brain-map.org/experiment/show/79556738 and http://mouse.brain-map.org/experiment/show/1001, respectively.
Figure S4. Cortical density of SST and VIP subtypes, Related to Figure 1 and 2.
(A) Absolute 3D density of SST co-expressing either CR (SST:CR) or nNOS (SST:nNOS), and VIP co-expressing either CR (VIP:CR) or CCK (VIP:CCK). Also see Table S3 for the full dataset
(B) Density mapping of SST and VIP subtypes in the cortical flatmap
Figure S5. Stereotypic laminar distribution of interneuron subtypes, Related to Figure 3.
(A) Cell type distribution of 8 different bins equally spaced between pia surface and corpus callosum. (B–C) Cell density (mean ± standard deviation) of each cell types in auditory cortex (B) and the entire isocortex (C). The result was comparable to cortical layer based analysis in Figure 3.
Highlights.
Quantitative maps reveal cell type-based structural organization of the brain
PV+ and SST+ interneuron distribution defines cortical hierarchies
Circuit architectures differ in frontal/ associational and sensorimotor cortices
SST+ and VIP+ distribution identifies sexually dimorphic subcortical circuits
Acknowledgments
We thank Kristin Baldwin for discussions and critical reading and editing the manuscript, Adam Kepecs, Colin Barnstable and Brett Mensh for discussions and critical reading the manuscript. This work was supported by NIH grants U01MH105971 (P.O.) and R01MH062349 (X.J.W.), Simons Foundation Autism Research Initiative (P.O. and P. M.), STCSM grants 14JC1404900 and 15JC1400104 (X.J.W), the Pennsylvania Department of Health Tobacco CURE Funds (Y.K.), Samuel J. and Joan B. Williamson Fellowship (G.R.Y.), Mathers Charitable Foundation H N Mahabala Chair Professorship (P.M.), and IIT Madras (K.R. and P.M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS:
Conceptualization, Y.K. and P.O.; Computational modeling, G.R.Y, L.C.G.M, and X.J.W; Sample preparation and Data collection, Y.K., G.F., and J.L.; Software and statistical analysis tools, K.P. and K.U.V; Transgenic mice, M.H., and Z.J.H.; Web visualization, K.R. and P.M; Anatomical expression analysis, M.B; Manuscript preparation, Y.K., G.R.Y., X.J.W., and P.O. with the assistance of other authors.
References
- Bayless DW, Shah NM. Genetic dissection of neural circuits underlying sexually dimorphic social behaviours. Philosophical transactions of the Royal Society of London. 2016;371:20150109. doi: 10.1098/rstb.2015.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen LM, Peters HJ, Veening JG. Fos immunoreactivity in the rat brain following consummatory elements of sexual behavior: a sex comparison. Brain research. 1996;738:67–82. doi: 10.1016/0006-8993(96)00763-9. [DOI] [PubMed] [Google Scholar]
- Dong HW. The Allen reference atlas: A digital color brain atlas of the C57Bl/6J male mouse. John Wiley & Sons Inc; 2008. [Google Scholar]
- Douglas RJ, Martin KA. Mapping the matrix: the ways of neocortex. Neuron. 2007;56:226–238. doi: 10.1016/j.neuron.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Fan J, Zeng H, Olson DP, Huber KM, Gibson JR, Takahashi JS. Vasoactive intestinal polypeptide (VIP)-expressing neurons in the suprachiasmatic nucleus provide sparse GABAergic outputs to local neurons with circadian regulation occurring distal to the opening of postsynaptic GABAA ionotropic receptors. J Neurosci. 2015;35:1905–1920. doi: 10.1523/JNEUROSCI.2661-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cerebral cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Gonchar Y, Wang Q, Burkhalter A. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat. 2007;1:3. doi: 10.3389/neuro.05.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracia-Llanes FJ, Crespo C, Blasco-Ibanez JM, Marques-Mari AI, Martinez-Guijarro FJ. VIP-containing deep short-axon cells of the olfactory bulb innervate interneurons different from granule cells. The European journal of neuroscience. 2003;18:1751–1763. doi: 10.1046/j.1460-9568.2003.02895.x. [DOI] [PubMed] [Google Scholar]
- He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Y, et al. Strategies and Tools for Combinatorial Targeting of GABAergic Neurons in Mouse Cerebral Cortex. Neuron. 2016;91:1228–1243. doi: 10.1016/j.neuron.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb MM, Yahr P. c-Fos immunoreactivity in the sexually dimorphic area of the hypothalamus and related brain regions of male gerbils after exposure to sex-related stimuli or performance of specific sexual behaviors. Neuroscience. 1996;72:1049–1071. doi: 10.1016/0306-4522(95)00602-8. [DOI] [PubMed] [Google Scholar]
- Heeb MM, Yahr P. Cell-body lesions of the posterodorsal preoptic nucleus or posterodorsal medial amygdala, but not the parvicellular subparafascicular thalamus, disrupt mating in male gerbils. Physiology & behavior. 2000;68:317–331. doi: 10.1016/s0031-9384(99)00182-1. [DOI] [PubMed] [Google Scholar]
- Henny P, Jones BE. Projections from basal forebrain to prefrontal cortex comprise cholinergic, GABAergic and glutamatergic inputs to pyramidal cells or interneurons. The European journal of neuroscience. 2008;27:654–670. doi: 10.1111/j.1460-9568.2008.06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, von Bartheld CS, Miller DJ, Kaas JH. How to count cells: the advantages and disadvantages of the isotropic fractionator compared with stereology. Cell and tissue research. 2015;360:29–42. doi: 10.1007/s00441-015-2127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XH, Hyun M, Taranda J, Huang KW, Todd E, Feng D, Atwater E, Croney D, Zeidel ML, Osten P, et al. Central Control Circuit for Context-Dependent Micturition. Cell. 2016;167:73–86. e12. doi: 10.1016/j.cell.2016.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Bollmann B, Richardson M, Berger D, Helmstaedter M, Briggman K, Denk W, Bowden J, Mendenhall J, Abraham W, et al. Boundary Learning by Optimization with Topological Constraints. Paper presented at: CVPR 2010 [Google Scholar]
- Jeong M, Kim Y, Kim J, Ferrante DD, Mitra PP, Osten P, Kim D. Comparative three-dimensional connectome map of motor cortical projections in the mouse brain. Scientific reports. 2016;6:20072. doi: 10.1038/srep20072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Perova Z, Mirrione MM, Pradhan K, Henn FA, Shea S, Osten P, Li B. Whole-Brain Mapping of Neuronal Activity in the Learned Helplessness Model of Depression. Frontiers in neural circuits. 2016;10:3. doi: 10.3389/fncir.2016.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 2015;10:292–305. doi: 10.1016/j.celrep.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Koch C, Mihalas S. A Computational Analysis of the Function of Three Inhibitory Cell Types in Contextual Visual Processing. Front Comput Neurosci. 2017;11:28. doi: 10.3389/fncom.2017.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Fitz S, Johnson PL, Shekhar A. Repeated stimulation of CRF receptors in the BNST of rats selectively induces social but not panic-like anxiety. Neuropsychopharmacology. 2008;33:2586–2594. doi: 10.1038/sj.npp.1301674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends in neurosciences. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Penzo MA, Taniguchi H, Kopec CD, Huang ZJ, Li B. Experience-dependent modification of a central amygdala fear circuit. Nature neuroscience. 2013;16:332–339. doi: 10.1038/nn.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin-Kumar A, Rosenbaum R, Doiron B. Inhibitory stabilization and visual coding in cortical circuits with multiple interneuron subtypes. Journal of neurophysiology. 2016;115:1399–1409. doi: 10.1152/jn.00732.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris HM, Hashimoto T, Lewis DA. Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder. Cereb Cortex. 2008;18:1575–1587. doi: 10.1093/cercor/bhm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong HW, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, et al. An anatomic gene expression atlas of the adult mouse brain. Nature neuroscience. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Packer AM, Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J Neurosci. 2011;31:13260–13271. doi: 10.1523/JNEUROSCI.3131-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nature neuroscience. 2013;16:1068–1076. doi: 10.1038/nn.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H-J, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A. Cortical interneurons that specialize in disinhibitory control. Nature. 2013;503:521–524. doi: 10.1038/nature12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronneke A, Scheuer B, Wagener RJ, Mock M, Witte M, Staiger JF. Characterizing VIP Neurons in the Barrel Cortex of VIPcre/tdTomato Mice Reveals Layer-Specific Differences. Cereb Cortex. 2015;25:4854–4868. doi: 10.1093/cercor/bhv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nature methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C, Hof PR. Design-based stereology in neuroscience. Neuroscience. 2005;130:813–831. doi: 10.1016/j.neuroscience.2004.08.050. [DOI] [PubMed] [Google Scholar]
- Segovia S, Guillamon A. Sexual dimorphism in the vomeronasal pathway and sex differences in reproductive behaviors. Brain Res Brain Res Rev. 1993;18:51–74. doi: 10.1016/0165-0173(93)90007-m. [DOI] [PubMed] [Google Scholar]
- Shang C, Liu Z, Chen Z, Shi Y, Wang Q, Liu S, Li D, Cao P. BRAIN CIRCUITS. A parvalbumin-positive excitatory visual pathway to trigger fear responses in mice. Science. 2015;348:1472–1477. doi: 10.1126/science.aaa8694. [DOI] [PubMed] [Google Scholar]
- Simerly RB. Wired for reproduction: organization and development of sexually dimorphic circuits in the mammalian forebrain. Annual review of neuroscience. 2002;25:507–536. doi: 10.1146/annurev.neuro.25.112701.142745. [DOI] [PubMed] [Google Scholar]
- Spring S, Lerch JP, Henkelman RM. Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. NeuroImage. 2007;35:1424–1433. doi: 10.1016/j.neuroimage.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Swanson L. Brain Maps: Structure of the Rat Brain. 3. Amsterdam: Elsevier; 2004. A laboratory guide with printed and electronic templates for data, models and schematics. [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, et al. A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, De La Parra M, Sorokin JM, Taniguchi H, Huang ZJ, Callaway EM. Brain-Wide Maps of Synaptic Input to Cortical Interneurons. J Neurosci. 2016;36:4000–4009. doi: 10.1523/JNEUROSCI.3967-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace ML, Saunders A, Huang KW, Philson AC, Goldman M, Macosko EZ, McCarroll SA, Sabatini BL. Genetically Distinct Parallel Pathways in the Entopeduncular Nucleus for Limbic and Sensorimotor Output of the Basal Ganglia. Neuron. 2017;94:138–152. e135. doi: 10.1016/j.neuron.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-J. A microcircuit model of prefrontal functions: ying and yang of reverberatory neurodynamics in cognition. The frontal lobes. 2006:92–127. [Google Scholar]
- Wang X-J, Tegnér J, Constantinidis C, Goldman-Rakic P. Division of labor among distinct subtypes of inhibitory neurons in a cortical microcircuit of working memory. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1368–1373. doi: 10.1073/pnas.0305337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ. Neurophysiological and computational principles of cortical rhythms in cognition. Physiological reviews. 2010;90:1195–1268. doi: 10.1152/physrev.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans: the mind of a worm. Phil Trans R Soc Lond. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wolff SB, Grundemann J, Tovote P, Krabbe S, Jacobson GA, Muller C, Herry C, Ehrlich I, Friedrich RW, Letzkus JJ, et al. Amygdala interneuron subtypes control fear learning through disinhibition. Nature. 2014;509:453–458. doi: 10.1038/nature13258. [DOI] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. The Journal of comparative neurology. 2010;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GR, Murray JD, Wang XJ. A dendritic disinhibitory circuit mechanism for pathway-specific gating. Nat Commun. 2016;7:12815. doi: 10.1038/ncomms12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Hintiryan H, Gou L, Song MY, Bay M, Bienkowski MS, Foster NN, Yamashita S, Bowman I, Toga AW, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Automated CN-based cell counting with a 3D conversation and 3D volume measurements, Related to Figure 1.
(A–D) An example from SST-Cre:H2B-GFP brain (A), scale bar = 1mm. (B) High magnification image from yellow inset box in (A), scale bar = 50µm. (C) CN based cell detection. (D) Examples of watershed algorithm based cell separation, shown by green and red. CNs automated cell detection reached F score = 0.92 (precision = 0.97 and recall = 0.87) for H2B-GFP-based cell counting, and F score = 0.91 (precision = 0.86 and recall = 0.96) for tdTomato cytoplasmic-based cell counting (see the Methods).
(E) Manual 3D cell counts were obtained by a stereological principle: cells touching 3 sides highlighted in black lines of 3D space were included and the ones touching the other 3 sides highlighted in red lines were excluded in the counting (In the schema, the cells are elongated in the Z dimension, representing the point spread function effect of two-photon imaging. The 3D manual counts were used to derive the 2D to 3D conversion factor for generating cell densities per mm3 (see Methods).
(F) 3D volume measurement of anatomical structures. RSTP brain (F1) was registered to each brain (F2) to match its 3D shape (F3). 2D examples of before registration (F4) and after registration (F7) to SST-Cre:H2B-GFP brain (F6). RSTP label (F5) was transformed (F8) based on 3D image registration (F1–F3). Number of areas occupied each transformed label (F8) can then be calculated as volume.
(G) Whole brain flatmap, modified from the original version in http://larrywswanson.com/?page_id=1415
Figure S6. Cortical areas in L5 cell density spaces, Related to Figure 4
(A–D) Same analyses as in Figure 4, but with L5 density data. Segregation of cortical subnetworks is still present but is weaker in L5 data in comparison to L2/3 data.
Figure S7. Complete current response maps and spiking network simulation, Related to Figure 5
(A) E-E, PV-E, and SST-E current response maps (top to bottom) when external inputs target the E, PV, SST, or VIP population (left to right). The excitatory population rate response is proportional to the E-E current response.
(B) Excitatory population rate responses to external inputs targeting PV (left) or SST (right) population in the intact local circuit (top row), after removing all recurrent excitatory connections (middle row), or after weakening all connections by 70% (bottom row).
(C) Spiking neural network model consisting of 5,000 neurons. (D) Raster plot of all neurons at the spontaneous state. (E) From left to right: the PV-to-E, SST-to-E, and E-to-E current responses to external input currents driving PV neurons.
Table S1. Number of cells from the seven different subtypes across the entire brain, Related to Figure 1, 2, 3, S5, and Table S4
Table S1a tab includes cell counting (average ± standard deviation) data from the seven cell types.
Table S1b tab includes separate male and female cell counting (average ± standard deviation) data from the PV, SST, and VIP cell types with multiple comparison corrected p-value (called “q-value”) in each cell type.
Table S2. Brain volume measurement in the entire brain areas in male and female brain, Related to Figure 7 and S1.
Mean and standard deviation of brain volumes from male and female brain (N = 15 samples in each sex) is list across the entire brain areas.
Table S3. Density of cells from the seven different subtypes across the entire brain, Related to Figure 1, 2, 3, 4, S1, S5, and S6.
Table S4. Sexually dimorphic brain areas in SST and VIP neurons, Related to Figure 7
Cell counting (average ± standard deviation) of males and females (N = 5 mice per sex) and false discovery rate (FDR) adjusted statistical significance (q value) for multiple comparison correction in sexually dimorphic brain regions.
Movie S1. Quantitative display of the three major GABA subtypes and their overlay in evenly spaced voxel map, Related to Figure 1 and 2.
Movie S2. Quantitative display of the SST and VIP subtypes and their overlay in evenly spaced voxel map, Related to Figure 1 and 2.
Figure S2. Deep (5 and 6) and superficial (2/3) layer density of PV, SST, and VIP neurons, Related to Figure 2.
(A–B) Deep layer density distribution.
(A) Relative 3D density of PV+, SST+, and VIP+ neurons in the isocortex areas.
(B) Density mapping of PV+, SST+, and VIP+ neurons in the cortical flatmap (see also Table S3 for cell density in each anatomical area).
(C–D) Superficial layer density distribution.
(C) Relative 3D density of PV+, SST+, and VIP+ neurons in the isocortex areas.
(D) Density mapping of PV+, SST+, and VIP+ neurons in the cortical flatmap (see also Table S3 for cell density in each anatomical area).
Figure S3. STPT data in comparison to Allen Brain in situ hybridization data, Related to Figure 1.
Left two columns of panels: note the low density of PV+ cells in the infralimbic (ILA, row 1) and posterior agranular insular cortex (AIp, row 4) (top and bottom panels) compared to the motor (row 2) and somatosensory cortex (row 3) detected by both STPT imaging of the PV-Cre-H2BGFP+ cells (first column of panels) and Allen in situ hybridization (second column of panels).
Right two columns of panels: In contrast, a comparable SST+ cell density is detected by both methods in all four regions, resulting in high SST-to-PV ratio in the ILA and AIp compared to the motor and somatosensory areas.
Allen in situ data for the PV and the SST are from http://mouse.brain-map.org/experiment/show/79556738 and http://mouse.brain-map.org/experiment/show/1001, respectively.
Figure S4. Cortical density of SST and VIP subtypes, Related to Figure 1 and 2.
(A) Absolute 3D density of SST co-expressing either CR (SST:CR) or nNOS (SST:nNOS), and VIP co-expressing either CR (VIP:CR) or CCK (VIP:CCK). Also see Table S3 for the full dataset
(B) Density mapping of SST and VIP subtypes in the cortical flatmap
Figure S5. Stereotypic laminar distribution of interneuron subtypes, Related to Figure 3.
(A) Cell type distribution of 8 different bins equally spaced between pia surface and corpus callosum. (B–C) Cell density (mean ± standard deviation) of each cell types in auditory cortex (B) and the entire isocortex (C). The result was comparable to cortical layer based analysis in Figure 3.