Abstract
Gene expression is controlled by enhancers that activate transcription from the core promoters of their target genes. Although a key function of core promoters is to convert enhancer activities into gene transcription, whether and how strongly they activate transcription in response to enhancers has not been systematically assessed on a genome-wide level. Here we describe self-transcribing active core promoter sequencing (STAP-seq), a method to determine the responsiveness of genomic sequences to enhancers, and apply it to the Drosophila melanogaster genome. We cloned candidate fragments at the position of the core promoter (also called minimal promoter) in reporter plasmids with or without a strong enhancer, transfected the resulting library into cells, and quantified the transcripts that initiated from each candidate for each setup by deep sequencing. In the presence of a single strong enhancer, the enhancer responsiveness of different sequences differs by several orders of magnitude, and different levels of responsiveness are associated with genes of different functions. We also identify sequence features that predict enhancer responsiveness and discuss how different core promoters are employed for the regulation of gene expression.
Animal development is coordinated by differential gene expression that is commonly attributed to the dynamic and cell-type-specific activities of transcriptional enhancer sequences1,2. Enhancers are genomic regulatory elements that recruit transcription factors and cofactors to activate transcription from their target core promoters, short sequences at the 5′ ends of genes at which RNA polymerase II (Pol II) assembles and gene transcription initiates3,4. However, sensitive methods to assess endogenous transcription have revealed many positions outside gene starts that initiate transcription, blurring the distinction between promoters and other genomic regions5,6. We reasoned that one of the key functions of bona fide core promoters that is essential for differential gene expression is their ability to strongly respond to enhancers, that is, to efficiently convert the enhancers’ activating cues into productive gene transcription. However, despite the central importance of this enhancer responsiveness, it has remained unclear how many sequences in large animal genomes respond to a given enhancer, which sequences respond most strongly, and how wide the range of response strength is. Knowing the range of enhancer responsiveness and the sequence features of strongly versus weakly responding candidates is critical for determining how enhancer responsiveness is sequence-encoded and differentially employed for the regulation of gene expression. It is also important for our understanding of transcription and might explain the cause of endogenous transcription initiation within different genomic regions.
However, the quantitative assessment of enhancer responsiveness in a standardized manner is challenging and cannot be derived from measures of endogenous transcription. Endogenous initiation rates result from activation by different—possibly many7,8—enhancers of different strengths, and the combined contributions of core promoter and enhancer functionality cannot be deconvoluted. Therefore, the responsiveness of candidate sequences to an enhancer needs to be assessed in standardized reporter assays, under the influence of defined enhancers that are kept constant, a technique equivalent to the widely used enhancer activity assays that operate with constant core promoters2. Such assays have been performed for individual core promoters in human and Drosophila cells9–11, and systematic medium-scale tests of yeast core promoters12 and of mammalian core promoters in vitro13 exist. However, a method to systematically assess enhancer responsiveness for millions of candidate fragments across large genomes is lacking.
Here we present STAP-seq, a method to assess the sequence-intrinsic enhancer responsiveness of millions of candidate sequences at single-base-pair resolution and perform a genome-wide analysis of their enhancer responsiveness in Drosophila melanogaster cells. We find that in the presence of a single strong enhancer, thousands of sequences exhibit enhancer-responsive transcriptional activation with strengths that vary over three orders of magnitude. The strength of the response can be predicted from the candidates’ primary sequences, and the host genes of candidates that respond strongly or weakly to enhancers differ characteristically in their function. Positions within enhancers that initiate transcription endogenously exhibit low enhancer responsiveness compared to positions within bona fide core promoters. Overall, our systematic analysis identifies sequences that efficiently convert enhancer activity into transcription initiation events and shows how sequences with different enhancer responsiveness are employed for the regulation of gene expression.
Results
Determining sequence-intrinsic enhancer responsiveness
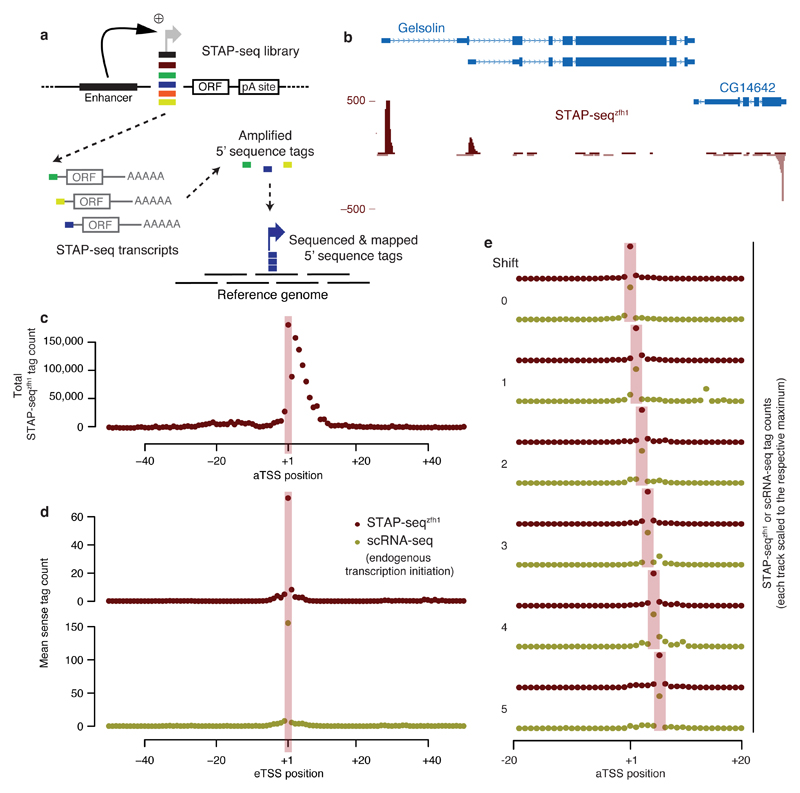
To identify genomic sequences that can initiate transcription in response to enhancers and to directly assess the strength of the response (that is, the candidate fragments’ inducibility or enhancer responsiveness), we developed STAP-seq (Fig. 1a). We separately determine the induced and basal activities for candidate DNA fragments at single-base-pair resolution in standardized reporter setups that do (STAP-seqenh) or do not (STAP-seqctrl) contain a single defined enhancer. For STAP-seqenh, we randomly sheared D. melanogaster genomic DNA into short fragments (median length, 192 bp; Supplementary Fig. 1a) and cloned them in bulk into a reporter plasmid at the position of the core promoter (also called the minimal promoter), between a defined strong developmental enhancer (from the transcription factor Zn finger homeodomain 1 (zfh1)10,14) and a protein-coding open reading frame (ORF; Fig. 1a). Core promoters are typically ~100 bp long, and the candidate fragments thus also included flanking sequences up- and/or downstream of potential core promoters15,16. This, however, should not influence our ability to assess enhancer responsiveness as negligible differences were observed between sequences within this length range10 (see also below for a demonstration that most candidates have low basal activities). If a fragment initiates transcription in response to the enhancer, it will produce reporter transcripts, and the number of reporter transcripts generated will directly reflect the fragment’s induced activity. Because all reporter transcripts will be identical except for short 5′ sequence tags that originate from the respective candidate, this allows the quantification of the candidate’s activity in the respective reporter setup (Fig. 1a). For STAP-seqctrl, we repeated the above with an enhancer-less reporter setup to assess the candidate fragments’ basal activities and subsequently their enhancer responsiveness (see below).
Figure 1. STAP-seq identifies position and orientation of transcription initiation within arbitrary candidate fragments.
(a) Experimental setup of STAP-seq. Short candidate DNA fragments are cloned into a reporter construct that provides an enhancer and a reporter gene (short open reading frame (ORF)). Active candidates initiate reporter transcripts that start with sequence tags depicting the exact TSS. These tags are then sequenced and mapped to the reference genome. (b) UCSC Genome browser screenshot depicting STAP-seq using the zfh1 enhancer. Tag coverage is shown in a strand-specific manner. (c) Cumulative STAP-seqzfh1 tag counts around FlyBase-annotated TSSs (aTSS). (d) Metagene profile of STAP-seqzfh1 tag counts and short-nuclear-capped-RNA-seq (scRNA-seq)18 signals at experimentally determined STAP-seq TSSs (eTSSs). (e) Agreement of STAP-seqzfh1 and scRNA-seq18 for eTSSs that are shifted with respect to aTSSs by 1–5 nt.
We transfected D. melanogaster S2 cells with each reporter library, isolated polyadenylated RNA, and followed a modified CapSeq protocol17,18 to selectively capture the reporter mRNAs’ 5′ sequence tags, which enable the precise mapping of the transcription start sites (TSSs; i.e., the +1 nucleotides) throughout the genome. Briefly, all non-5′ capped RNA species were rendered ligation-incompetent by dephosphorylating their 5′ ends with calf intestinal phosphatase. Subsequently, the 5′ caps were removed with tobacco acid pyrophosphatase, and RNA oligonucleotides, each containing an 8-nucleotide (nt) random barcode as a unique molecular identifier, were ligated to the resulting 5′ phosphate RNA molecules. The 5′ sequence tags of the reporter transcripts were then selectively reverse transcribed, amplified, and paired-end sequenced. The paired-end reads were aligned to the D. melanogaster genome and the initiation events at each genomic position were quantified in a strand-specific manner by the number of unique sequence tags, as identified by the unique molecular identifiers. We performed two technical replicates for both STAP-seqenh and STAP-seqctrl, which in each case were highly similar (Pearson correlation coefficient (PCC) = 0.99 and 0.93, respectively; see below for replicates with independent libraries and transfections and an analysis of the variance of individual data points), and combined them for further analyses. We will now first discuss the induced activities obtained from STAP-seq with the zfh1 enhancer (STAP-seqzfh1) and compare them with measures of endogenous initiation (basal activities cannot be assessed in endogenous contexts) and with STAP-seq screens using different enhancers and another cell type, before we discuss enhancer responsiveness as the normalized ratio STAP-seqenh versus STAP-seqctrl.
STAP-seq identifies endogenous transcription start sites
STAP-seqzfh1 revealed a highly distinctive genomic profile of candidate transcription initiation events with specific signals that overlapped TSSs annotated by FlyBase (aTSSs; Fig. 1b). Indeed, even though candidates from across the entire genome were assayed, over half (55%) of all 28,509 genomic positions with ≥5 tags mapped to within 50 bp of an aTSS, and the degree of alignment with aTSSs improved with higher tag counts: 66% of all positions with ≥10 tags and 71% of all positions with ≥20 tags were within 50 bp, and 30% (≥5 tags), 39% (≥10 tags), or 45% (≥20 tags) were within 5 bp, respectively (Supplementary Fig. 2a). The latter corresponds to more than 140-fold enrichment over the genome (only 0.32% of the genome is within 5 bp of an aTSS; Supplementary Fig. 2a,b).
Furthermore, plotting the cumulative STAP-seqzfh1 tag count around all aTSSs revealed a strong enrichment, with the highest value precisely at the +1 position (Fig. 1c and Supplementary Fig. 2c). Data sets that measure endogenous transcription initiation18,19 and the analysis of the Initiator (Inr) motif, known to coincide with TSSs20, also supported STAP-seq-defined TSSs (experimentally defined TSSs or eTSSs; see Online Methods), even if they did not map to aTSSs (Fig. 1d,e and Supplementary Fig. 2d–h).
Overall, these results suggest that STAP-seq identifies positions that initiate transcription endogenously and that aTSSs at annotated gene starts are distinguished among the many genome-wide candidate fragments by their ability to efficiently convert enhancer activities into transcription initiation events. Because we tested short fragments in a defined reporter setup outside their endogenous sequence and chromatin contexts, these results confirm that the ability to convert enhancer activities into transcription initiation events and the precise position of transcription initiation are encoded in the DNA sequence.
Induced activities are consistent for three developmental enhancers
We next asked whether induced activities are influenced by the choice of enhancer. We therefore repeated STAP-seq with the zfh1 enhancer and with additional enhancers for a focused candidate library of reduced complexity, derived from 34 bacterial artificial chromosomes (BACs) that cover about 5% of the D. melanogaster genome and contain ~1,100 aTSSs. For these screens, we chose an intronic enhancer within sugarless (sgl) and an intergenic enhancer near hamlet (ham), both developmental enhancers weaker than the zfh1 enhancer10. In addition, we chose two housekeeping enhancers close to nucampholin (ncm) and to short spindle 3 (ssp3), respectively. As they were expected to specifically activate housekeeping- but not developmental-type core promoters10, they served as an outgroup in our subsequent analysis.
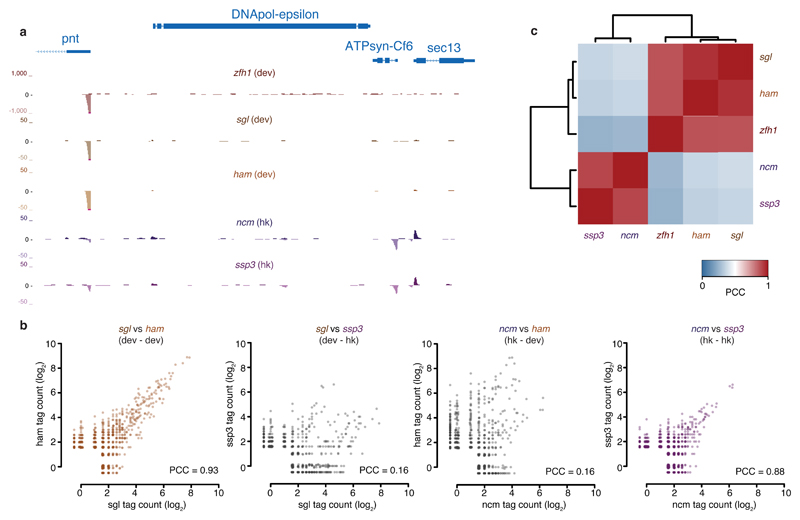
For the screens using developmental enhancers, strong signals were observed at aTSSs of developmentally regulated genes, while the housekeeping enhancers produced strong signals at aTSSs of housekeeping genes (Fig. 2a). This confirmed the expected core promoter specificity10 of the three developmental and the two housekeeping enhancers, and suggests that within each of these two broad transcriptional programs10—but not across programs—the tested fragments respond similarly to different enhancers.
Figure 2. Induced activities are consistent across developmental enhancers.
(a) UCSC genome browser screenshot showing STAP-seq signals of the focused screens using the indicated developmental (dev; zfh1, sgl, and ham) and housekeeping (hk; ncm and ssp3) enhancers. The depicted locus covers developmental and housekeeping genes. Pointed (pnt) codes for a transcription factor, whereas ATP synthase, coupling factor 6 (ATPsyn-Cf6), and Secretory 13 (sec13) code for components of ATP synthase and nuclear pore complex, respectively. (b) Scatterplots depicting the similarity of STAP-seq screens with two developmental (left) and two housekeeping (right) enhancers, respectively, and the dissimilarity between developmental- and housekeeping-enhancer screens (middle). (c) Bi-clustered heatmap depicting pairwise similarities. Pearson correlation coefficients (PCCs) of STAP-seq tag counts for three developmental and two housekeeping enhancers.
Indeed, the focused screens with all three developmental enhancers were highly similar (all PCCs ≥ 0.83; Fig. 2b and Supplementary Fig. 3a) and also agreed well with the genome-wide screen (PCC = 0.86 between focused and genome-wide STAP-seqzfh1; Supplementary Fig. 3b; for an analysis of the variance of individual data points, see Supplementary Fig. 3c). The most highly induced sequences in STAP-seqzfh1 were also the most highly induced ones when using the weaker developmental sgl and ham enhancers (Fig. 2b,c and Supplementary Fig. 3a), and their rankings agreed well (Spearman’s correlation coefficient (SCC) = 0.75 for sgl versus zfh1 and for ham versus zfh1). The similarity between the induced activities with each of the different developmental enhancers became particularly apparent in the comparison to the outgroup. Whereas the screens performed with the two housekeeping enhancers were highly similar (PCC = 0.88; Fig. 2b), they differed characteristically from the screens with the developmental enhancers (sgl versus ncm and ssp3 had PCC = 0.18 and 0.16, respectively; ham versus ncm and ssp3 had PCC = 0.16 and 0.14, respectively; Fig. 2b,c). Indeed, when we grouped all five screens by hierarchical clustering, the three developmental screens clustered tightly as did the two housekeeping screens, forming an outgroup as expected (Fig. 2c).
These results show that different sequences responded to developmental versus housekeeping enhancers, recapitulating the previously reported enhancer–core-promoter specificity10. They demonstrate that STAP-seq is a tool that can probe enhancer responsiveness of millions of candidate fragments and identify strongly responding sequences for different types of enhancers. Notably, the responses were consistent across the three different developmental enhancers and across the two different housekeeping enhancers we tested, suggesting that enhancer responsiveness within a given transcriptional program is independent of the particular enhancers used and thus constitutes a functional sequence feature of general importance.
Activities are consistent across two different cell types
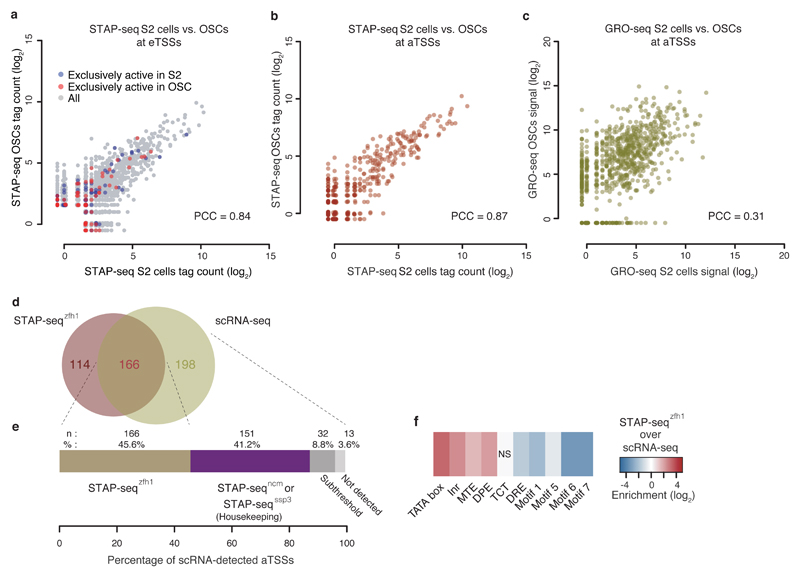
To investigate whether the different induced activities observed for different sequences are consistent across different cell types or vary with cell-type-specific gene expression, we repeated STAP-seq with the focused library in D. melanogaster ovarian somatic cells (OSCs)21. OSCs differ from S2 cells in gene expression and enhancer activities10,14, which required exchanging the S2-specific zfh1 enhancer with an OSC-specific developmental enhancer from traffic jam or tj14. Notably, STAP-seqzfh1 in S2 cells and STAP-seqtj in OSCs were highly similar (PCC = 0.85; Fig. 3a,b and Supplementary Fig. 4), reminiscent of the screens with three different developmental enhancers in S2 cells (Fig. 2b,c and Supplementary Fig. 3a), and much above the similarity of endogenous transcription between the two cell types as assessed by Global Run-On sequencing (GRO-seq)22,23 (PCC = 0.31 at aTSSs; Fig. 3c). Even sequences that are endogenously active exclusively in S2 cells or OSCs, respectively, behave similarly across the two cell types in STAP-seq (Kolmogorov–Smirnov test P value > 0.1; Fig. 3a and Supplementary Fig. 4).
Figure 3. Induced activities are consistent across cell types.
(a) Scatterplot depicting STAP-seq tag counts for STAP-seqzfh1 in S2 cells (x-axis) versus STAP-seqtj in OSCs (y-axis) and their similarity (PCC). TSSs that endogenously—as measured by GRO-seq22,23—are exclusively active in S2 cells or OSCs are labeled blue or red, respectively (see also Supplementary Fig. 4). (b,c) Scatterplots depicting comparisons between STAP-seq in b, and GRO-seq22,23 in c, in S2 cells and OSCs at aTSSs. (d) Venn diagram depicting the overlap of aTSSs detected by STAP-seqzfh1 and scRNA-seq18 in S2 cells for genomic regions covered in the focused STAP-seq screens. (e) Breakdown of aTSSs detected by scRNA-seq18: 45.6% are also detected by STAP-seqzfh1 and essentially all other aTSSs are detected by the focused STAP-seq screens with housekeeping enhancers. Only 13 aTSSs (3.6%) are not found by developmental and housekeeping STAP-seq screens combined. (f) Core promoter motif-enrichment analyses of aTSSs uniquely detected by either STAP-seqzfh1 or scRNA-seq18. NS, not significant.
Together, these results suggest that the enhancer responsiveness of a DNA sequence is an intrinsic property that is independent of cell-type-specific gene expression, confirming previous observations during transgene expression with widely used minimal promoters (e.g., the Drosophila synthetic core promoter in different tissues in transgenic flies8,24). These results also demonstrate the complementarity of STAP-seq and methods that assess endogenous transcription initiation, such as GRO-seq, that detects positions of transcriptionally-engaged Pol II, and short nuclear-capped RNA-seq (scRNA-seq) that measures short nascent transcripts in vivo (Fig. 3d–f and Supplementary Fig. 5); whereas endogenous transcription initiation reflects cell-type-specific gene expression, STAP-seq measures the sequence-intrinsic ability of DNA fragments to initiate transcription in response to an enhancer, that is, it measures the fragments’ enhancer responsiveness. Notably, enhancer responsiveness appears to be consistent across different enhancers and cell types.
A wide and continuous range of enhancer responsiveness
A notable aspect of the STAP-seq data is the very wide range of induced activities for the different candidate fragments. Whereas the vast majority of the tested genomic positions did not initiate transcription in STAP-seqzfh1 (0 tags), 1,864 eTSSs had ≥100 tags at their +1 positions, 136 had ≥1,000, and the strongest eTSS had 14,249—even though all fragments were tested using the same enhancer. The consistency of these differences between different enhancers and across different cell types suggests that the ability to efficiently convert enhancer-activity into transcription initiation events is a sequence-intrinsic property and an important contributor to transcription regulation.
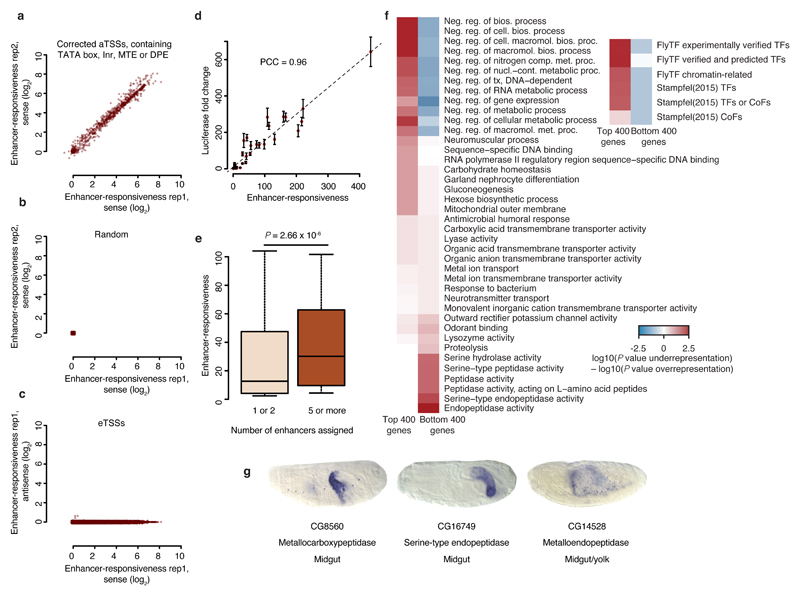
We therefore define the inducibility or enhancer responsiveness of a candidate sequence as the ratio of its induced versus basal activity, measured by STAP-seqenh and STAP-seqctrl, respectively. To assess enhancer responsiveness genome-wide, we repeated STAP-seq without an enhancer (STAP-seqctrl; Supplementary Fig. 1b), again obtaining two highly similar replicates (PCC = 0.93), and divided the induced by the basal activity for each eTSS, normalizing to spike-in controls present in both samples (Online Methods). This revealed a very wide range of developmental enhancer responsiveness with an up to 1,000-fold difference between the highest and lowest inducibility (Supplementary Fig. 6a–c; housekeeping enhancer responsiveness had a much reduced dynamic range, see Supplementary Fig. 6d). We also found a similarly wide range of responsiveness for the known aTSSs (Fig. 4a), particularly when we corrected their positions based on short nuclear-capped RNA-seq (scRNA-seq) data18,25 or restricted the analysis to corrected aTSSs containing exclusively TATA box, Inr, DPE, or MTE motifs (Supplementary Fig. 6b and Online Methods). By contrast, analyzing the same number of randomly selected positions (Fig. 4b) or antisense initiation at the +1 positions of eTSSs (Fig. 4c) revealed only very weak enhancer responsiveness.
Figure 4. Wide range of enhancer responsiveness and associated biological functions.
(a–c) Scatterplots showing the range of enhancer responsiveness at corrected aTSSs that contain exclusively TATA box, Inr, MTE, or DPE in a, random positions in b, and eTSSs in c, depicting replicate 1 versus 2 in a, and b, and sense versus antisense signals of replicate 1 in c. (d) Enhancer responsiveness according to STAP-seq versus luciferase induction by the zfh1 enhancer. Error bars, s.d.; n = 3. (e) Boxplot showing enhancer responsiveness for aTSSs of genes that are surrounded by 1 or 2 versus 5 or more enhancers (n = 1,325 and 139, respectively; Wilcoxon P value). Center line: median; limits: interquartile range; whiskers: 10th and 90th percentiles. (f) Heatmaps depicting enrichments for the most differentially enriched Gene Ontology (GO) categories and for defined sets of transcription factors among the 400 genes associated with the strongest or weakest eTSSs that contain exclusively TATA box, Inr, MTE, or DPE. (g) Berkeley Drosophila Genome Project (BDGP)39 in situ embryo images for genes representing the GO categories most strongly enriched near weak eTSSs.
To validate the different levels of enhancer responsiveness determined by STAP-seq, we tested 30 sequences (from ranks 2 to 4,675; 19 known aTSSs and 11 eTSSs that to our knowledge had not been described previously, including 2 candidates that overlap exons) and 16 negative controls (12 aTSSs and 4 candidates without TSS annotation) individually in luciferase assays. The enhancer responsiveness determined by STAP-seq and luciferase induction showed a high linear agreement (PCC = 0.96; Fig. 4d), much higher than the PCCs observed between the luciferase results and methods that measure endogenous transcription initiation (luciferase versus scRNA-seq18, CAGE26, and GRO-seq23 show PCCs of 0.35, 0.08, and 0.4, respectively). Together, these results validate the wide range of enhancer responsiveness and establish STAP-seq as a quantitative genome-wide assay to functionally quantify this measure, which does not necessarily correlate with endogenous transcription rates in any particular cell type.
Enhancer responsiveness correlates with gene function
The results above reveal that sequences in the genome vary widely in their ability to convert enhancer activities into transcription initiation events. Strong additive activation by multiple enhancers might require highly inducible TSSs, because weaker ones might otherwise limit transcription rates. Indeed, sequences proximal to aTSSs of genes with five or more developmental enhancers10,14 were significantly more inducible than those with only one or two developmental enhancers (Fig. 4e; P value = 2.66 × 10−6). Furthermore, sequences with different levels of enhancer responsiveness to the developmental zfh1 enhancer were also located near genes of different biological functions (Supplementary Fig. 7a). The overall most responsive sequences tended to be near genes involved in development, regulation of gene expression, and response to stimuli, whereas the overall weakest ones were next to housekeeping genes, as expected, given the incompatibility of the developmental zfh1 enhancer, used in STAP-seqzfh1, with the core promoters of housekeeping genes10. When we restricted the analysis to eTSSs that contain only TATA box, Inr, MTE, or DPE motifs (i.e., those that preferentially function with developmental enhancers10), we found that the most responsive eTSSs were enriched near genes coding for transcription factors, whereas weak ones were predominantly near genes for cell-type-specific enzymes (Fig. 4f). For example, CG8560, CG16749, and CG14528 are all annotated as peptidases and are expressed in midgut and/or yolk (Fig. 4g).
This suggests that highly responsive non-housekeeping core promoters might regulate genes that require rapid induction (e.g., transcription factors), whereas weakly responsive ones could be employed at genes with potentially lower transcription kinetics (e.g., enzymes; Fig. 4f,g). Together, these results suggest that core promoters with different levels of enhancer responsiveness are employed for the transcription of genes with different functions and different regulatory characteristics.
The DNA sequence predicts enhancer responsiveness
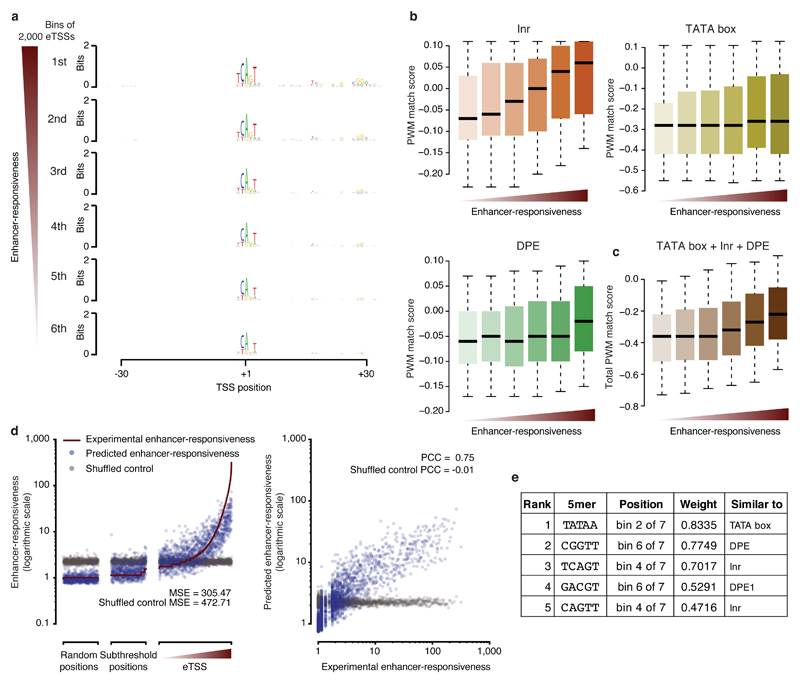
We next investigated whether sequences that respond very differently to developmental enhancers have recognizable features that can predict enhancer responsiveness. We binned eTSSs according to their responsiveness and visualized the nucleotide preferences, using the +1 positions of the eTSSs as anchor points. The resemblance with the established Inr motif20 correlated with responsiveness and was higher for strongly responsive sequences than for weaker ones (Fig. 5a). In addition, the most responsive sequences had a preference for guanine (G) around the +30 position and a consensus sequence that resembled the downstream promoter element (DPE) motif27. They also showed increasing information content around positions +15 to +20, where no known core promoter element resides, especially a prominent TC dinucleotide at position +17, just upstream of the motif-ten-element (MTE28; Fig. 5a).
Figure 5. Candidate sequences are predictive of responsiveness to developmental enhancers.
(a) Sequence logos summarizing position-specific nucleotide frequencies for eTSSs (bins of 2,000 sequences) ranked by decreasing enhancer responsiveness. (b) Position weight matrix (PWM) match scores for TATA box, Inr, and DPE motifs at eTSSs ranked by enhancer responsiveness (b), and aggregate quality scores of all three motifs from b (c). Center line: median; limits: interquartile range; whiskers: 5th and 95th percentiles. (d) Scatterplots of experimentally determined and predicted enhancer responsiveness for eTSSs, subthreshold positions, and random positions. Also included is predicted enhancer responsiveness after randomizing the assignment between the sequences and responsiveness (gray). MSE: mean square error. (e) Five most predictive 5mers, their positions (bin out of 7 bins along the sequence), and weights, as well as the most similar known core promoter motifs.
This observation prompted us to speculate whether the presence of core promoter motifs in the sequences (i.e., match quality/score or affinity) might determine the sequences’ enhancer responsiveness. Indeed, eTSSs with higher responsiveness showed greater similarity to the canonical Inr, TATA box, and DPE motifs20 (Fig. 5b), and the combined similarity scores for these motifs correlated with the responsiveness of eTSSs (Fig. 5c). This is also reflected by an enrichment of each of these motifs in eTSS sequences compared to random sequences, which increases toward more responsive eTSSs (Supplementary Fig. 7b), even though, for example, TATA-box- and DPE-containing core promoters are typically found at different genes with distinct expression properties29,30. Indeed, although both TATA box and DPE are increasingly enriched, they less frequently occur in the same eTSS, consistent with previous reports20 and with their non-overlapping spatiotemporal expression patterns and biological functions29,30 (Supplementary Fig. 7c). For more responsive eTSSs, the Inr, TATA box, and DPE motifs also aligned increasingly well to their consensus positions at +1, –27, and +30, respectively4, and the distance to these consensus positions increased for less responsive eTSSs (Supplementary Fig. 7d,e; the absence of the TATA box in the sequence logo in Fig. 5a, despite its enrichment, stems from a reduced positional constraint31, see Supplementary Fig. 7e).
Moreover, the positional occurrence of specific 5mers relative to the eTSSs is predictive of the sequences’ enhancer responsiveness by a linear model32 using fivefold cross-validation, leading to a PCC of 0.75 between the predicted and experimentally determined enhancer responsiveness (Fig. 5d,e). The 5mers with the highest weights resemble known core promoter motifs and are specifically enriched at the canonical positions of these motifs (Fig. 5e). Together, these results suggest that enhancer responsiveness is determined by core promoter motif affinity (i.e., match quality) and positioning, providing a potential explanation for the positional preferences of these motifs.
Discussion
The ability to efficiently convert enhancer activities into transcription initiation events is of central importance for differential gene expression. Here, we develop a functional reporter assay, STAP-seq, to quantitatively assess enhancer responsiveness systematically for millions of candidate fragments across entire genomes. Thousands of short fragments of genomic DNA are able to specifically initiate transcription to very different levels when activated by a single strong enhancer. For annotated gene TSSs, the different levels of enhancer responsiveness correlate with the function and the number of enhancers of the respective host gene, suggesting that strongly responsive core promoters might be required to reach high transcription rates, whereas those weakly responsive might serve to limit transcription and thus enhancer additivity.
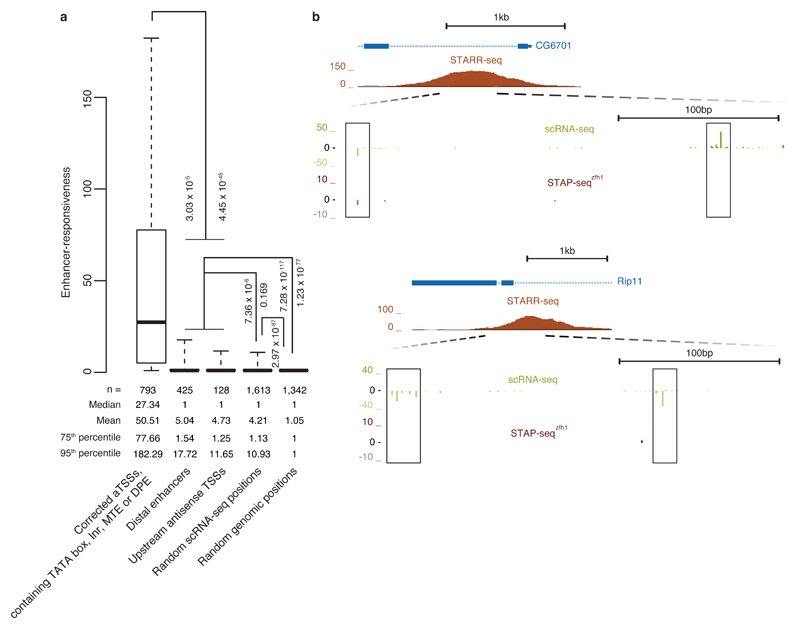
Our observations of both a continuum of responsiveness and its degree of variation across three orders of magnitude suggest that enhancer responsiveness is an important measure to characterize and classify transcriptional regulatory elements. The continuum of activity suggests that many sequences can initiate transcription at very low levels (at or below the thresholds used here) when brought into the vicinity of strong enhancers. This could explain recent observations that enhancers and positions upstream of active aTSSs can be sites of transcriptional initiation15,16,33–38. When we measured the sequence-intrinsic enhancer responsiveness of these positions with STAP-seq, we found it slightly higher than at control positions (random positions with endogenous initiation and arbitrarily chosen genomic positions), but substantially weaker than at positions within bona fide core promoters (Fig. 6). This finding suggests that in the vicinity of strong enhancers, accessible DNA might unavoidably initiate transcription, preferentially at sites of (degenerate) core promoter motifs, even if the respective DNA sequence is responsive to the enhancer only at the level of random sequences.
Figure 6. Positions of endogenous transcription initiation in developmental enhancers and upstream of aTSSs have weak sequence-intrinsic enhancer responsiveness.
(a) Boxplot depicting enhancer responsiveness of positions that initiate transcription in S2 cells (≥5 scRNA-seq18 tags; left-most four boxes) or are randomly selected from the D. melanogaster genome (rightmost box, ‘Random genomic positions’). ‘Corrected aTSSs, containing TATA box, Inr, MTE or DPE’, are position-corrected according to scRNA-seq18 as in Fig. 4a and Supplementary Fig. 6b. For ‘Distal enhancers’, we used STARR-seq enhancers14 that are more than 500 bp away from the nearest aTSS and for each enhancer considered the position with the highest scRNA-seq signal within ± 250 bp around the STARR-seq peak summit on either strand (disregarding enhancers for which this signal was below 5 tags). For ‘Upstream antisense TSSs’, we considered the position with the highest scRNA-seq signal upstream and antisense of aTSSs until the 3′end or—for divergent gene pairs—until 500 bp upstream of the 5′end (aTSS) of the next gene. ‘Random scRNA-seq positions’ are aTSS- and enhancer-distal and not closely spaced with respect to each other. Also shown are P values via one-sided Wilcoxon’s rank-sum test between the categories. Center line: median; limits: interquartile range; whiskers: 5th and 95th percentiles. (b) UCSC Genome browser screenshots exemplifying representative loci of endogenous transcription initiation within enhancers as measured by scRNA-seq18 that have only weak STAP-seq signals.
Our results further suggest that autonomously active promoters might consist of an enhancer-responsive core promoter and a TSS-proximal or TSS-overlapping enhancer. Whereas STAP-seqctrl had generally only a few tags, consistent with low basal activities of core promoters (Supplementary Fig. 8), the genomic positions of candidates with the highest activity in STAP-seqctrl frequently overlapped those of enhancers, predominantly housekeeping enhancers, suggesting that such autonomous promoter activities stem from proximal enhancers (Supplementary Fig. 9). This provides a simple explanation for the previously observed similarity between promoters and enhancers5,6 in that both have enhancer functionality yet differ in the presence of strongly responsive core promoters that support productive transcription.
Our results could explain the source of transcription initiation within enhancers and suggest how core promoters and proximal enhancers can form autonomously functioning promoters. Even though high enhancer activity and enhancer responsiveness can co-occur within a given DNA fragment, the two functions are generally uncoupled, re-emphasizing the difference and importance of the two key types of transcription regulatory elements and the functionalities they encode. STAP-seq will prove useful to assess enhancer responsiveness genome-wide and select the most responsive sequences, which should allow the highly efficient expression of transgenes, potentially beyond what is currently possible. STAP-seq will also be useful for studying the mechanisms of transcriptional initiation and its regulation, questions of fundamental importance especially today, when the key role of transcriptional regulation during development, evolution, and disease is becoming exceedingly clear.
Methods
STAP-seq screening vector
For STAP-seq in Drosophila cells we constructed a screening vector based on the pGL3-Promoter backbone (Promega; cat. no. E1751) by replacing the sequence between BglII and FseI with the following sequence, containing a ccdB suicide gene flanked by homology arms (used for cloning the candidates during library generation), an intron (mhc16), an ORF (truncated sgGFP, Qbiogene, Inc), followed by the pGL3′s SV40 late polyA-signal. The full sequence is available at www.addgene.org. The enhancers were cloned between the KpnI and BglII sites (for coordinates and sequences of the enhancers please see Supplementary Table 1). The control screens were performed with the STAP-seq vector not harboring any enhancer.
STAP-seq library generation
Genomic DNA (genome-wide libraries) or BAC DNA (focused libraries; Supplementary Table 2) was isolated as described previously10,14. The DNA was sheared by sonication (Covaris S220) and DNA fragments (100- to 250-bp length) were size-selected using a 1% agarose gel. Illumina NEBnext Multiplexing Adaptors (New England BioLabs (NEB); cat. no. E7335 or E7500) were ligated to 1 µg of size-selected DNA fragments using NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB; cat. no. E7645L) following the manufacturer’s instructions, except the final PCR amplification step. Ten PCR reactions (98 °C for 45 seconds (s); followed by 10 cycles of 98 °C for 15 s, 65 °C for 30 s, 72 °C for 10 s) with 1 µl adaptor ligated DNA as template were performed, using KAPA Hifi Hot Start Ready Mix (KAPA Biosystems; cat. no. KK2602) and primers (fw: TAGAGCATGCACCGGACACTCTTTCCCTACACGACGCTCTTCCGATCT and rev: GGCCGAATTCGTCGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT), which add a specific 15-nt extension to both adapters for directional cloning using recombination (Clontech In-Fusion HD; cat. no. 639650). Each five PCR reactions were pooled, purified, and size selected with Agencourt AMPureXP DNA beads (ratio beads/PCR 1.25; cat. no. A63881), followed by column purification (QIAquick PCR purification kit; cat. no. 28106.). Cloning of the fragments into the vector was performed as described previously14.
STAP-seq spike-in controls
In order to control for transfection efficiency and to normalize all STAP-seq screens we used spike-in controls. We generated four STAP-seq spike-in control plasmids that are driven by the zfh1 enhancer and harbor a single sequence each, which were derived from the Drosophila pseudoobscura orthologs of even skipped (eve), CG32369, and two alternative TSSs from u-shaped (ush). Reads derived from those sequences map uniquely to the Drosophila pseudoobscura (dp3 assembly), but not to the D. melanogaster genome. We cloned the sequences into the STAP-seq vector using the same strategy as we used for library generation (see above; Supplementary Table 3). A mix of the four spike-in control plasmids was added to the genome-wide STAP-seq libraries before transfection at a final dilution of 1:1,000,000. For the focused libraries (BAC) we only used the eve spike-in control plasmid at a dilution of 1:100,000.
Cell culture and transfection
S2 cells were obtained from Life Technologies and cultured as described previously14. Transfection of the STAP-seq libraries was performed with 1 × 108 (focused libraries) or 1.2 × 109 cells (genome-wide libraries) at 70–80% confluence using the MaxCyte STX Scalable Transfection System. Cells were transfected at a density of 1 × 109 cells per milliliter in MaxCyte HyClone buffer using OC-100 or OC-400 processing assemblies and 50 µg library per milliliter of cells. S2 were pulsed with the pre-set program Optimization 1. Cells were transferred to a cell culture flask and mixed with 10% DNaseI (2,000 U/ml) and incubated for 30 min at 27 °C, before resuspension in full medium. Cells were incubated after electroporation for 24 h before RNA isolation. OSCs21 originally isolated by the M. Siomi laboratory (Keio University School of Medicine) were obtained from the laboratory of J. Brennecke (Institute of Molecular Biotechnology (IMBA)) and cultured as described previously14. Transfection of the focused library was performed using all cells from a 70–80% confluent square dish (24.5 cm × 24.5 cm) in an OC-400 processing assembly in 400 µl MaxCyte HyClone buffer mixed 1:1 with the OSC culture medium without supplements and 20µg of library (pre-set program Optimization 5). Cells were transferred to a cell culture flask and mixed with 10% DNaseI (2,000 U/ml) and incubated for 30 min at 27 °C, before resuspension in full medium. The cells were plated on a square dish (24.5 cm × 24.5 cm) for 24 h after electroporation. For the focused screens, we performed three STAP-seqzfh1(PCC ≥ 0.97 with each other), and two each for STAP-seqsgl (PCC = 0.95), STAP-seqham(0.98), STAP-seqssp3(0.87), STAP-seqncm(0.78), and STAP-seqtj(0.98). All cell lines used are checked for mycoplasma contamination on a regular basis.
STAP-seq RNA processing
24 h after electroporation total RNA was isolated followed by polyA+ RNA purification and DNaseI treatment, as described previously14. 10–20 µg (focused) or 200 µg (genome-wide) of DNaseI-treated RNA was incubated with calf intestinal alkaline phosphatase (CIP; NEB cat. no. M0290L). Per 1 µg RNA, 0.5 µl CIP was used. The reactions were cleaned up using Qiagen RNeasy MinElute reaction clean-up kit (cat. no. 74204) according to the manufacturer’s protocol, adding beta-Mercaptoethanol to the RLT buffer. Subsequently all RNA was processed during all further reactions. The CIP-treated RNA was then incubated with 0.05 µl Tobacco Alkaline Phosphatase (TAP; Epicentre, discontinued, now available as Cap-Clip Acid Pyrophosphatase (cat. no. C-CC15011H) from CELLSCRIPT) per 1 µg RNA to remove the 5′ cap of all 5′-capped RNA species. The reactions were cleaned up using Agencourt RNAClean XP (BeckmanCoulter, cat. no. A63987) at a ratio of 1.8 of beads to RNA. To the 5′ ends of the TAP-treated RNA 10 µM RNA oligonucleotide (GUUCAGAGUUCUACAGUCCGACGAUCNNNNNNNN) was ligated per 1 µg RNA at 16 °C for 16 h using 0.2 µl T4 RNA Ligase 1 (ssRNA Ligase, NEB, cat. no. M0204L). The eight random nucleotides at the 3′ end of the 5′ RNA linker are used as a Unique-Molecular-Identifier (UMI) to count reporter mRNAs (see below), but also minimizes sequence preferences during the T4 RNA Ligase 1 reaction40. The reactions were cleaned up using Agencourt RNAClean XP (BeckmanCoulter, cat. no. A63987) at a ratio of 1.0 of beads to RNA. First strand cDNA synthesis was performed with 1µl of Invitrogen’s SuperscriptIII (50 °C for 60 min, 70 °C for 15 min; cat. no. 18080085) using a reporter-RNA-specific primer (CAAACTCATCAATGTATCTTATCATG) for 2.5–5 µg of polyA+ RNA in 20 µl total volume. Five reactions were pooled and 1 µl of 10 mg/ml RNaseA was added (37 °C for 1 h) followed by bead purification (Agencourt AMPureXP DNA beads (ratio beads/RT reaction 1.8). We amplified the total amount of reporter cDNA obtained from reverse transcription (above) for Illumina sequencing. For the focused libraries we performed two PCR reactions using the KAPA real-time library amplification kit (KAPA Biosystems, cat. no. KK2702) according to the manufacturer’s protocol. The genome-wide screens were amplified using KAPA Hifi Hot Start Ready Mix (KAPA Biosystems; cat. no. KK2602) in 32 PCRs. As forward primer we used AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA and as reverse primer NEBNext Multiplex Oligos for Illumina (NEB; cat. no. E7335 or E7500). PCR products were purified with Agencourt AMPureXP DNA beads (ratio beads/PCR 1.25).
Illumina sequencing
All samples were sequenced by the VBCF’s NGS unit on an Illumina HiSeq2500 platform, following manufacturer’s protocol. All deep sequencing data are available at www.starklab.org and are deposited in GEO.
Luciferase reporter assays
We replaced the SV40 promoter of the pGL3-promoter plasmid (Promega) by the candidate sequences (see Supplementary Table 4 for coordinates and primers) between the BglII and SbfI restriction sites. As in STAP-seq the zfh1 enhancer (inserted in the KpnI restriction site) was used to drive transcription from the candidates. To determine the basal activities of the candidates they were also cloned into the luciferase vector not harboring any enhancer. Individual constructs were tested by co-transfecting 100,000 cells with 95 ng of the respective pGL3 firefly construct and 5 ng of a Renilla control plasmid (driven by the ubiquitin-63E promoter) that is based on the pRL plasmid (Promega) using FuGENE HD Transfection Reagent (Promega; cat. no. E2312). Using the Promega Dual Luciferase Assay kit (cat. no. E1960), we measured luciferase activity at a Bio-Tek Synergy H1 fluorescence plate reader.
Candidate selection for Luciferase validation
The candidate TSSs were selected to not have basal activity (STAP-seqctrl ≤ 3 tags) but are active in STAP-seqzfh1 (≥5 tags). According to this rule we selected 30 positive regions across the entire range of strength (from rank 2 to rank 4,675; 19 aTSS and 11 eTSS, including 2 candidates that overlap exons). We in addition selected 12 aTSS for which we did not observe any STAP-seq signal and 4 genomic regions without any TSS annotation as negative controls.
STAP-seq NGS data processing
Paired-end STAP-seq reads were trimmed to 44 bp, with the first 8 bp as the unique molecular barcode identifier (UMI). The reads were mapped using the remaining 36 bp to dm3 and dp3 (for spike-in controls) genome assemblies using Bowtie41 version 0.12.9 as in ref. 10. The dp3 spike-in controls were selected from their dm3 orthologs that allows unambiguously unique mapping to dp3 with the same mapping parameter. For paired-end reads that are mapped to the same positions, we collapsed those that have identical UMIs as well as those for which the UMIs differed by 1 bp to ensure the counting of unique reporter mRNAs (we removed all mapped reads that had N’s in their UMIs). Tag counts at each position represent the sum of the 5′-most position of collapsed fragments. For focused STAP-seq screens in S2, we subsampled all reads at 700,000 reads before mapping and removed fragments outside of the focused regions (see Supplementary Table 2 for full list of BACs) from analysis (Supplementary Table 5). For analysis of STAP-seqzfh1 versus STAP-seqtj, we subsampled 300,000 of the mapped BAC fragments.
Genomic distribution
We assigned a unique annotation for each nucleotide in the genome via the following priority order: ± 5, ± 10, ± 50 bp around aTSS, CDS, 5′-untranslated region (UTR), 3′-UTR, intron, intergenic region. We then assigned each eTSSs to one of these categories by the annotation of the +1 position of the eTSSs.
eTSSs calling
We selected candidate positions as those with at least 5 tag counts in the STAP-seqzfh1 experiment. At 5 tag counts >98% of eTSSs could be recovered in the replicates. For normalization, we used the tag counts at +1 position of the dp3 eve (focused STAP-seqzfh1) or together with the tag counts from the two most prominent initiation positions from the ush-1 spike-in TSSs (genome-wide STAP-seqzfh1; Supplementary Table 3). We used these tag counts as the probability P in determining the binomial distribution, and determined the enhancer responsiveness of each candidate position as the corrected ratio of STAP-seqzfh1/STAP-seqctrl tag using a pseudo count of 1. We considered only positions that were more than 1.5-fold enriched in STAP-seqzfh1 over STAP-seqctrl with P value ≤ 0.05, before merging those that were within ± 10 bp around each other. We determined within each of these regions the position with the highest enhancer responsiveness, with which we ranked the eTSSs, as the +1 positions.
Metagene plots
We obtained raw reads from scRNA-seq obtained from ref. 18 (GSM463298), and mapped the 38 bp reads as 36-nt reads using Bowtie41 with the following parameters: -p 4 -q -v 3 -m 1–best–strata –quiet. PEAT data were obtained from ref. 19 (https://ohlerlab.mdc-berlin.de/research/Download_The_Data_97/). Raw tag counts were directly derived from the strand-specific log2-transformed bigwig files. To test the accuracy of identification of the highest position as the +1 position in STAP-seq (Supplementary Fig. 2c), we bootstrapped the STAP-seqzfh1 tag counts, normalized to the spike-ins, at each position 100 times, calculated the mean, and plotted the 5th, 50th, and 95th percentiles.
Sequence logo
We downloaded genomic sequence using fastacmd version 2.2.10, and used WebLogo version 2.8 (ref. 42).
Scatterplots
For tag count scatterplots, we considered only positions that have at least 3 tag counts. For enhancer responsiveness scatterplots, we calculated the corrected ratio of STAP-seqzfh1/STAP-seqctrl tag using a pseudo count of 1, and computed the log2 values. We added a pseudo count of 0.7071 to any zero values, and a dithering factor of 0.1 for nonzero coordinates that are plotted more than once. For comparison between STAP-seq and GRO-seq, we took the sum of STAP-seq tag counts within ± 5 bp of aTSSs, or the sum of GRO-seq fragment 5′ positions in a window spanning 101 bp downstream. To depict variation of STAP-seq, we plotted the sample s.d. from three focused STAP-seqzfh1 replicates at eTSS positions, called on the combined three replicates, on a scatterplot on two of the replicates with no dithering. For scatterplot of enhancer responsiveness in focused screens, we summed up the tag counts of positions ± 50 bp around aTSSs for the induced and basal activities, and calculated the enhancer responsiveness as above.
Luciferase assay analysis
We first normalized firefly over Renilla luciferase values for each of the three independent transfections per construct individually and then calculated the mean and s.d. for these normalized values. Finally, we used these means and s.d. to calculate the fold change of the zfh1 enhancer-driven luciferase signals over the enhancer-less control.
STAP-seq correlation heatmap
For all pairs of enhancers, we computed pairwise Pearson correlation coefficients (PCCs) between the respective STAP-seq tag counts at positions that are covered by at least 3 tag counts in either enhancer. We performed hierarchical clustering (complete linkage) in R, directly using the correlation values as similarities as previously10.
Cell-type specificity analysis
We obtained GRO-seq raw reads performed in S2 (ref. 23) (GSM577244) and OSCs22 (GSM1027403) and mapped them as 36-nt reads using Bowtie41 with the following parameters: -p 4 -q -v 3 -m 1–best–strata–quiet. We considered all positions that were ≥ 3 tag counts in either STAP-seqzfh1 S2 or STAP-seqtj OSC and determined the sum of GRO-seq fragment 5′ positions in a window spanning 101 bp downstream. We classified a position to be exclusively active in a cell type if it had GRO-seq start fragments at least 15 in a cell type but not in the other. As this gives more OSC active TSSs, we considered only OSC TSSs with the highest GRO-seq signal to be the same number as in S2 cells. For the cumulative distribution plot, we randomized “all” positions and took the same number as S2 active positions. We performed two-sided Kolmogorov-Smirnov between S2 and OSC active positions.
Global comparison of STAP-seqzfh1 and other endogenous methods
We used our previously published RNA-seq14. We obtained raw reads from two CAGE replicates in S2 (SRX142946 and SRX144189, modENCODE submission 5331)26 and mapped the 27-nt reads using Bowtie41 with the following parameters: -p 4 -q -v 2 -m 1–best–strata –quiet. As CAGE reads often carries nontemplate G nucleotide at the 5′end43, we shifted the start position of the fragment 1 bp downstream when the first position mapped to a G in the genome, and combined the two replicates. We computed Pearson correlation coefficients (PCCs) only for aTSSs that in either experiment had the following cutoffs: around ±5 bp aTSSs with sum of signal ≥3 for STAP-seqzfh1, ≥5 for scRNA and CAGE, or ≥15 for 101 bp downstream of aTSSs for GRO-seq. For comparisons with RNA-seq, we took the aTSS with the highest signal from each gene, and considered only genes with ≥3 RPKM. We used our core promoter motif counts and calculated their enrichment as previously10. To specifically compare STAP-seqzfh1 and scRNA-seq18 (Fig. 3d–f and Supplementary Fig. 5), an aTSSs was considered to be detected if within ±5 bp of either side it has ≥3 STAP-seqzfh1 tag counts, or ≥5 by scRNA-seq tag counts in total. We calculated the binomial distribution of DHS-seq14 within ±250-bp window and called it to be closed for a P value > 0.05. We obtained bigwig RAMPAGE data44 (GSE36212), and determined the mean RAMPAGE signal at shifted controls 200 bp upstream of aTSSs to be 10, and therefore used this value as cutoff. For aTSS detection in the focused screens, an aTSSs was considered to be detected in a housekeeping screen if it had ≥3 tag counts in either ncm or ssp3 screens within ±50 bp to account for broad initiation of housekeeping genes, or if it had 1 or 2 tag counts to be considered to be detected at subthreshold level.
Density plot of enhancer responsiveness
Kernel density was calculated using density function in R from the log2 values of the enhancer responsiveness. We first calculated density parameters using aTSSs, for which we obtained bandwidths of 0.0903 and 0.09597, respectively, for replicates 1 and 2. We plotted the density using polygon function, and added pseudo-positions at the ends of the estimated kernel density: x coordinates corresponded to the estimated end positions, and y coordinates were from the minimum y coordinates from the estimate.
Number of enhancer per gene analysis
We assigned a gene to an eTSS if the eTSS lies within ± 20 bp from the 5′ end of a transcript from that gene. We removed eTSSs and genes that are assigned more than once. We used annotation from D. melanogaster FlyBase release 5.50. As a gene could have multiple aTSS, we only considered aTSS with the highest enhancer responsiveness. To count the number of enhancers per gene, we used our previous assignment10. Briefly, developmental enhancers were assigned to a gene provided that they fall anywhere within 5 kb upstream from the TSS to 2kb downstream from the gene end of the longest isoform.
Correction of aTSS +1 positions
For “corrected aTSSs”, we realigned the +1 positions to be the highest scRNA position that is at least 5 within a window of up to 20bp on either side of the aTSS. For aTSSs that contain TATA box, Inr, MTE or DPE, we considered only “corrected aTSSs” that contain either of these motifs but not TCT, DRE, Ohler motifs 1, 5, 6, and 7 as previously10.
Gene Ontology (GO) and TF enrichment analysis
We used aTSS-to-eTSS assignments as above. We ranked the genes based on their enhancer responsiveness and divided them into two categories: the top and bottom 1,000 genes. We assessed whether genes assigned to an eTSS were enriched for any GO terms45 by calculating hypergeometric P values and enrichment for all terms. For all terms that were enriched more than twofold among the top and bottom gene sets and were present at least 4 times in all assigned eTSSs, we sorted for their enrichment and counts. For each category, we calculated log10 (P value under-representation)—log10 (P value over-representation), and sorted the terms in a descending order of difference between values from the classes. The color intensity of the heatmaps represents log10 (P value under-representation)—log10 (P value over-representation). To investigate the relationship between eTSSs that contain TATA box, Inr, MTE, or DPE, and their biological function, we considered only eTSSs that have any of these motifs but not TCT, DRE, Ohler motifs 1, 5, 6, and 7, terms that were enriched more than 1.5-fold among the top and bottom gene sets and were present at least twice in all assigned eTSSs. For TF enrichment analysis, we used our curated TF and cofactor lists46 as well as sets of factors annotated by the Drosophila Transcription Factor Database (http://www.flytf.org/)47 (“experimentally verified site-specific TFs”, “equivalent to release v1—trusted TFs”, and “proteins involved in chromatin-related processes”).
Analysis of enhancer responsiveness and length of fragments
For +1 of eTSS positions, as well as STAP-seqzfh1 positions that are ≥1–5 tag counts, we intersected with the sequenced input STAP-seqzfh1 fragments. We considered only fragments that intersect on the same strand and cover ±30 bp around the positions, and determined the longest ones. We also divided the fragments into 4 groups around the median: 80 to 140 bp, 141 to 190 bp, 191 to 240 bp, and 241 to 300 bp, and determined the eTSSs that obey the same intersection rule as above.
Core promoter element enrichment and position heatmaps
We scanned for motif occurrences using MAST from the MEME suite48 (version 4.9.0) and used parameters that ensured specificity and sensitivity (for enrichment heatmap) or sensitivity only (for position heatmaps) for each motif as previously10. For enrichment heatmap, we calculated enrichment and hypergeometric distribution, and considered an enrichment to be significant for P value ≤ 0.05.
Core promoter element quality and position boxplots
We scanned for motif occurrences as above to obtain motif match scores, and added the individual match scores to derive the aggregate match scores. We determined the median position for the core promoter element from these positions, and computed the deviation of each sequence for the position boxplot.
k-mer-based prediction of enhancer responsiveness
We considered all 13,218 eTSSs, and also included two categories of control positions with no STAP-seqctrl signal: 6,405 each for subthreshold positions with STAP-seqzfh1 less than 5, and random positions with no STAP-seqzfh1 signal (from random positions for motif enrichment analysis). This selection covers a large span of responsiveness and made the sequence-based discrimination tractable. We counted the occurrences of 5mers in seven equally spaced sectors around the +1 position: –50 to –37, –36 to –23, –22 to –9, –8 to +6, +7 to + 20, +21 to +34, +35 to +48. To simplify the learning, we de-noised the data by binning every ten consecutive sequences: the k-mer counts were summed in each bin and a median of responsiveness was considered as the responsiveness of the bin, resulting in 10× smaller data set and larger k-mer counts. We log10-transformed the responsiveness to decrease their dynamic range and exponential growth, and used an L1-regularized linear model (ref. 32, implemented in the scikit-learn Python package49). We kept the regularization coefficient α fixed at 10−3, and estimated the mean squared error and correlation coefficient of the predicted responsiveness using a fivefold cross-validation.
Enhancer responsiveness analysis at positions of endogenous distal transcription
For distal enhancers, we defined the position within ± 250 bp of distal enhancer summits as previously defined10 with the highest scRNA signal ≥5. For antisense upstream TSSs, we looked only upstream of positions of active aTSSs (total scRNA signal within ±5 bp either side of the +1 position to be ≥5) of the longest 5′ isoform of each gene, and considered the highest positions that were not in the gene body of another gene, or 500 bp upstream of another aTSSs, and had scRNA signal ≥5. For random scRNA positions, we first selected positions with scRNA signal ≥5 that did not overlap 500 bp of all aTSSs, developmental enhancers, as well as antisense upstream TSSs on both strands, merged those that were within 10 bp of each other, and considered only the highest positions. For random genomic positions, we considered 2,000 positions and controlled for their chromosome distributions. For all of the positions defined as above, we removed those that have gaps in the input coverage in the same strand, and calculated the sum of STAP-seqzfh1 and STAP-seqctrl within ±5 bp and calculated enhancer responsiveness as above (see eTSSs calling). We calculated the P value via one-sided Wilcoxon’s rank-sum test.
Generation of random TSSs (motif enrichment)
We aimed the random positions to be the same number of all aTSSs, considered positions that do not overlap 1 kb surrounding aTSS and eTSSs. We further merged positions that are within ± 50 bp of each other, recentered the position, and removed positions with undefined nucleotides (Ns) within ± 50 bp.
Generation of random TSSs (scatter and density plots)
We generated random positions that do not overlap ±50 bp of eTSS and aTSSs, aimed to be the same number of all aTSSs.
Coordinate intersections
We performed genomic coordinate intersections using the BEDTools suite50 version 2.17.0.
Statistics
We performed all statistical calculations and created graphical displays with R51.
Supplementary Material
Acknowledgments
We thank L. Cochella and members of the Stark group for comments on the manuscript and Life Science Editors (http://lifescienceeditors.com) for editorial support. We are grateful to P. Heine and E. Jans (MaxCyte) for help setting up efficient plasmid transfection. Deep sequencing was performed at the Vienna Biocenter Core Facilities GmbH (VBCF) Next-Generation Sequencing Unit (http://vbcf.ac.at). The Stark group is supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 647320) and by the Austrian Science Fund (FWF, F4303-B09). Basic research at the IMP is supported by Boehringer Ingelheim GmbH and the Austrian Research Promotion Agency (FFG).
Footnotes
Data Availability
Accession codes. GEO: GSE78886. Vectors are available from Addgene (http://www.addgene.org/Alexander_Stark)
Author Contributions
C.D.A., M.A.Z., and A.S. conceived the project. C.D.A., M.P., and M.R. performed the experiments with the help of K.S., and M.A.Z. the computational analyses. T.K. performed the k-mer based predictions. C.D.A., M.A.Z., and A.S. wrote the manuscript. A.S. supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 3.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 4.Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol. 2012;1:40–51. doi: 10.1002/wdev.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim T-K, Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 8.Kvon EZ, et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 9.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods. 2006;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 10.Zabidi MA, et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ede C, Chen X, Lin M-Y, Chen YY. Quantitative analyses of core promoters enable precise engineering of regulated gene expression in mammalian cells. ACS Synth Biol. 2016;5:395–404. doi: 10.1021/acssynbio.5b00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubliner S, et al. Core promoter sequence in yeast is a major determinant of expression level. Genome Res. 2015;25:1008–1017. doi: 10.1101/gr.188193.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 15.Duttke SHC, et al. Perspectives on unidirectional versus divergent transcription. Mol Cell. 2015;60:348–349. doi: 10.1016/j.molcel.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson R, et al. Human gene promoters are intrinsically bidirectional. Mol Cell. 2015;60:346–347. doi: 10.1016/j.molcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu W, et al. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nechaev S, et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni T, et al. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods. 2010;7:521–527. doi: 10.1038/nmeth.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohler U, Liao G-C, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito K, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 22.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Core LJ, et al. Defining the status of RNA polymerase at promoters. Cell Reports. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 28.Lim CY, et al. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engström PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponjavic J, et al. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc B. 1996;58:267–288. [Google Scholar]
- 33.Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam MTY, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scruggs BS, et al. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol Cell. 2015;58:1101–1112. doi: 10.1016/j.molcel.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomancak P, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCHH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kodzius R, et al. CAGE: cap analysis of gene expression. Nat Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 44.Batut P, Dobin A, Plessy C, Carninci P, Gingeras TR. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013;23:169–180. doi: 10.1101/gr.139618.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stampfel G, et al. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature. 2015;528:147–151. doi: 10.1038/nature15545. [DOI] [PubMed] [Google Scholar]
- 47.Adryan B, Teichmann SA. FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006;22:1532–1533. doi: 10.1093/bioinformatics/btl143. [DOI] [PubMed] [Google Scholar]
- 48.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 49.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 50.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.