Abstract
Motivation
Read assignment is an important first step in many metagenomic analysis workflows, providing the basis for identification and quantification of species. However ambiguity among the sequences of many strains makes it difficult to assign reads at the lowest level of taxonomy, and reads are typically assigned to taxonomic levels where they are unambiguous. We explore connections between metagenomic read assignment and the quantification of transcripts from RNA-Seq data in order to develop novel methods for rapid and accurate quantification of metagenomic strains.
Results
We find that the recent idea of pseudoalignment introduced in the RNA-Seq context is highly applicable in the metagenomics setting. When coupled with the Expectation-Maximization (EM) algorithm, reads can be assigned far more accurately and quickly than is currently possible with state of the art software, making it possible and practical for the first time to analyze abundances of individual genomes in metagenomics projects.
Availability and Implementation
Pipeline and analysis code can be downloaded from http://github.com/pachterlab/metakallisto
1 Introduction
The analysis of microbial communities via whole-genome shotgun sequencing has led to exceptional bioinformatics challenges (Chen and Pachter, 2005) that remain largely unsolved (Scholz et al., 2012). Most of these challenges can be characterized as ‘de novo’ bioinformatics problems: they involve assembly of sequences, binning of reads and annotation of genes directly from sequenced reads. The emphasis on de novo methods a decade ago was the result of a paucity of sequenced reference microbial and archaeal genomes at the time. However this has begun to change in recent years (Land et al., 2015). As sequencing costs have plummeted, the number of nearly complete genomes has increased dramatically, and while a large swath of the microbial world remains uncharacterized, there are now thousands of unique sequenced genomes suitable for the application of reference-based methods.
One of the fundamental metagenomics problems that is amenable to reference-based analysis is that of ‘sequence classification’ or ‘read assignment’. This is the problem of assigning sequenced reads to taxa. The MEGAN program (Huson et al., 2007) was one of the first reference-based read assignment programs and was published shortly after sequencing-by-synthesis methods started to become mainstream. It provided a phylogenetic context to mapped reads by assigning reads to the lowest taxonomic level at which they could be uniquely aligned, and became popular in part because of a powerful accompanying visualization toolkit. One of the drawbacks of MEGAN was that its approach to assigning ambiguously mapping reads limited its application to quantification of individual strains, an issue which was addressed in a number of subsequent papers, for example GRAMMy (Xia et al., 2011) and GASiC (Lindner and Renard, 2013), which were the first to statistically assign ambiguously mapped reads to individual strains. Unfortunately, these approaches all relied on read alignment, a computational problem that is particularly difficult in the metagenomic setting where reference genome databases can consist of more than a hundred million bases.
In a breakthrough publication in 2014 (Wood and Salzberg, 2014) it was shown that it is possible to greatly accelerate read assignment utilizing fast k-mer hashing to circumvent the need for read alignment. An implementation called Kraken was used to show that analyses that previously took hours were tractable in minutes, and the removal of the read alignment step greatly simplified workflows and storage requirements. However, the Kraken speed came at a cost. First, the growing size of reference genome databases means the indexing memory footprint rapidly increases beyond what end users can reasonably supply. Second, an examination of the Kraken algorithm and output reveals that the method takes a step back from GRAMMy and GASiC by discarding statistical assignment of reads at the strain level in favor of direct taxonomic assignment as in MEGAN. The net effect is that while Kraken is more accurate than MEGAN (Lindgreen et al., 2015), it is unsuitable for quantification. This is because unlike GASiC, Kraken is strictly designed to be a read assigner: its primary output is a file listing the taxonomic assignment for each read. A natural question to ask is whether the strengths of Kraken and GASiC can be combined, i.e. whether it is possible to leverage fast k-mer based hashing to map reads not at the taxonomic but at the strain level, while assigning the resulting ambiguously mapped reads using a statistical framework that allows for probabilistic assignment of reads.
To answer this question we turned to RNA-Seq (Cloonan et al., 2008; Lister et al., 2008; Mortazavi et al., 2008; Nagalakshmi et al., 2008), an experiment for which there has been extensive methods development that we hypothesized could be adapted and applied to metagenomics. Many of the challenges of metagenomic quantification translate to problems in RNA-Seq via a dictionary that replaces genome targets with transcript targets. For example, ambiguously mapped genomic reads that are difficult to resolve at the strain level in the metagenomics setting are analogous to reads that are difficult to assign to specific isoforms in RNA-Seq. Statistical questions at the heart of ‘comparative metagenomics’ (Huson et al., 2009; Rodriguez-Brito et al., 2006; Tringe et al., 2005) are analogous to the statistical problems in differential expression analysis. In fact, the only significant differences between metagenomics and RNA-Seq are that genome sequences are much larger than transcripts and reference databases are less complete. These differences have engineering implications, but statistically and computationally, metagenomics and transcriptomics are very much the same.
In this paper we show that technology transfer from RNA-Seq to metagenomics makes it possible to perform read assignment both rapidly and accurately. Specifically, we show that it is possible to accurately assign reads at the strain level using a fast k-mer based approach that goes beyond the hashing of Kraken and takes advantage of the principle of pseudoalignment (Bray et al., 2015). The idea of pseudoalignment originates with RNA-Seq, where it was developed to take advantage of the fact that the sufficient statistics for RNA-Seq quantification are assignments of reads to transcripts rather than their alignments. The same applies in the metagenomics setting, and we show that just as in RNA-Seq, application of the EM algorithm to ‘equivalence classes’ (Nicolae et al., 2011) allows for accurate statistical resolution of mapping ambiguities. We are able to maintain the speed of metagenomic-specific k-mer hashing programs, while also using a memory-efficient pre-screening step to greatly increase the effective number of metagenomes in our reference database without increasing computational requirements, from the few thousand used by CLARK and Kraken, to nearly 30 000. Using a published simulated dataset (Mende et al., 2012), a biological dataset from the human microbiome project and an implementation of pseudoalignment coupled to the EM algorithm in kallisto (Bray et al., 2015), we demonstrate significant accuracy and performance improvements in comparison to state of the art programs.
2 Approach
To test the hypothesis that RNA-Seq quantification methods can be applied in the metagenomics setting we began by examining the performance of eXpress, a program that implements a streaming EM algorithm for RNA-Seq read assignment from alignments, on simulated data (Roberts and Pachter, 2013). We chose eXpress because it utilizes traditional read alignments directly to a transcriptome but is more memory efficient than other approaches (e.g. RSEM (Li and Dewey, 2011)) and therefore more suitable in the metagenomics setting. Other RNA-Seq quantification tools such as Cufflinks (Trapnell et al., 2010) were not suitable for our needs because of their dependence on read alignments to genomes and not transcriptomes, a requirement that does not translate easily to the metagenomics setting.
To test eXpress we aligned a simulated dataset of Illumina-like reads from 100 microbial genomes to a reference database containing only those genomes, allowing us to compare results to a ground truth (the Illumina100 data) (Mende et al., 2012). We began by comparing eXpress to GASiC, which also utilizes read alignments for read assignment. The results are shown in Table 1. We found that eXpress outperforms GASiC at the exact genome, species, genus and phylum levels, which we believe is because the statistical model of eXpress takes into account data-dependent read error profiles in assigning reads.
Table 1.
Normalized count based classification accuracy at four taxonomic ranks
| Exact Genome |
Species |
Genus |
Phylum |
|||||
|---|---|---|---|---|---|---|---|---|
| AVGRE | RRMSE | AVGRE | RRMSE | AVGRE | RRMSE | AVGRE | RRMSE | |
| i100 | ||||||||
| kallisto | 0.97 | 5.42 | 0.14 | 0.36 | 0.13 | 0.38 | 0.09 | 0.10 |
| Bracken | – | – | 1.94 | 9.51 | 2.21 | 10.78 | 0.91 | 0.92 |
| CLARK | – | – | 12.28 | 22.73 | 10.32 | 18.22 | 7.52 | 7.88 |
| GASiC | 7.21 | 19.31 | 3.80 | 10.46 | 3.72 | 11.43 | 2.52 | 3.10 |
| eXpress | 2.57 | 11.92 | 0.40 | 0.61 | 0.34 | 0.57 | 0.13 | 0.18 |
| Ensembl | ||||||||
| kallisto | 17.15 | 39.32 | 1.26 | 3.01 | 0.98 | 2.17 | 0.72 | 0.76 |
| Bracken | – | – | 4.94 | 16.22 | 1.10 | 3.97 | 0.35 | 0.38 |
| CLARK | – | – | 59.15 | 72.40 | 52.68 | 67.04 | 45.44 | 56.76 |
CLARK and Bracken results are missing at the strain level because they do not output strain level counts. Calculated errors are Average Relative Error and Relative Root Mean Square Error.
A major problem with GASiC and eXpress is that the alignments they require are slow to generate. The alignments, made with Bowtie2 (Langmead and Salzberg, 2012), took days. As reported in (Wood and Salzberg, 2014) and the follow-up Bracken which has been specialized for quantification (Lu et al., 2016), significant speed-ups are possible using hashing methods. Braken and kallisto took similar amounts of time to index, but kallisto was significantly faster to quantify read abundances, with a run time of 5 min 55 s compared to 35 min for Bracken (Table 2). CLARK was faster in total time, but as seen in Table 1, kallisto performed noticably better than both Bracken and CLARK.
Table 2.
Indexing and quantification times for each program, when quantifying the Illumina 100 simulated dataset against the listed reference database
| Indexing | Quantification | |
|---|---|---|
| i100 | ||
| kallisto | 31 m 25 s | 5 m 55 s |
| Bracken | 22 m 27 s | 35 m 39 s |
| CLARK | – | 20 m 30 s |
| Ensembl | ||
| kallisto | 111 m | 60 m 40 s |
| Bracken | 235 m 35 s | 169 m 29 s |
| CLARK | – | 131 m 35 s |
Note that CLARK indexes and quantifies in a single step, so timing for each cannot be separated. All programs were run on a single core, on a cluster with 430 Gb of memory.
We next turned to a comparison of kallisto with Bracken and CLARK using the Illumina100 simulated data (i100) but using a full, more realistic reference database of 29 698 bacterial genomes from Ensembl (Kersey et al., 2016). In order to handle such a large database, which is significantly over the indexing threshold for all three programs, we first performed a pre-filtering step using recently-published metagenome distance estimator Mash (Ondov et al., 2016) (see methods for details). Mash filtered the 29 698 genomes down to 1027 genomes which were judged closest to the i100 reads being quantified; those 1027 genomes contained 83 out of the 100 ‘true’ strains present in the i100 dataset.
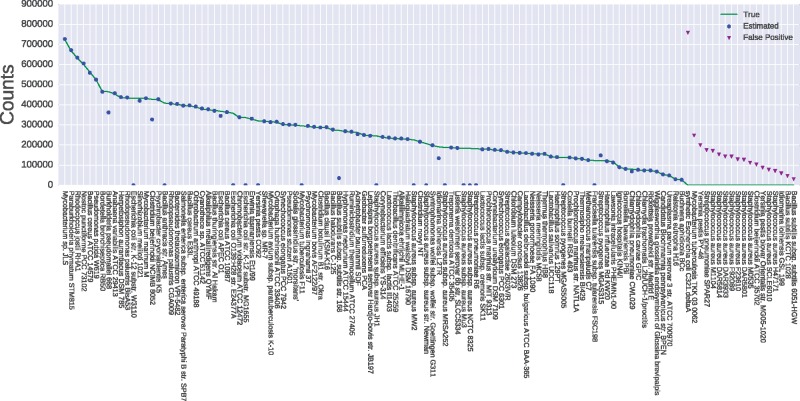
The results of estimating reads from all 100 genomes against the Ensembl-based index, listed in Table 1 (where the database is called ‘Ensembl’) and Figures 1 and 2, show that kallisto is significantly more accurate than CLARK at all taxonomic levels, and is only outmatched by Bracken at the genus level. The dramatic decrease in error from the exact genome to species level (from 17% to 1.26%) indicates that kallisto is correctly assigning the reads from the missing strains to closely related strains from the same species.
Fig. 1.
Results of kallisto on simulated reads pseudoaligned to the ensembl dataset at the exact genome level. The solid line indicates the actual counts simulated from each strain, while circle and triangle markers indicate the counts estimated by kallisto. Triangles are read counts assigned to strains that aren’t actually present in the dataset
Fig. 2.
Results of kallisto (top), Bracken (middle) and CLARK (bottom) on simulated reads pseudoaligned to the ensembl dataset at the species level
Even at the exact genome level (where neither Bracken nor CLARK offer estimates), kallisto performs well, given the restriction of missing 17% of the actual genomes present in the reads. To check the effect of the missing genomes on accuracy, we ran kallisto on the i100 reads only from the present 83 genomes and achieved an impressive AVGRE of 2.59% at the exact genome level. Even more promisingly, the species-level error of this 83-genome dataset is 0.77%, which is quite close to the 1.26% species-level error of the full 100-genome dataset. This further supports kallisto’s accuracy in assigning reads from missing genomes to closely related genomes.
All programs took significantly more time with the larger reference database. Mash took 362 minutes on a single core to index the full 30k Ensembl genomes, and another 130 minutes to compare the i100 reads against those genomes; these steps are easily parallelized to multiple cores. Once the pre-filtering was complete, kallisto was once again slower than CLARK but faster than Bracken.
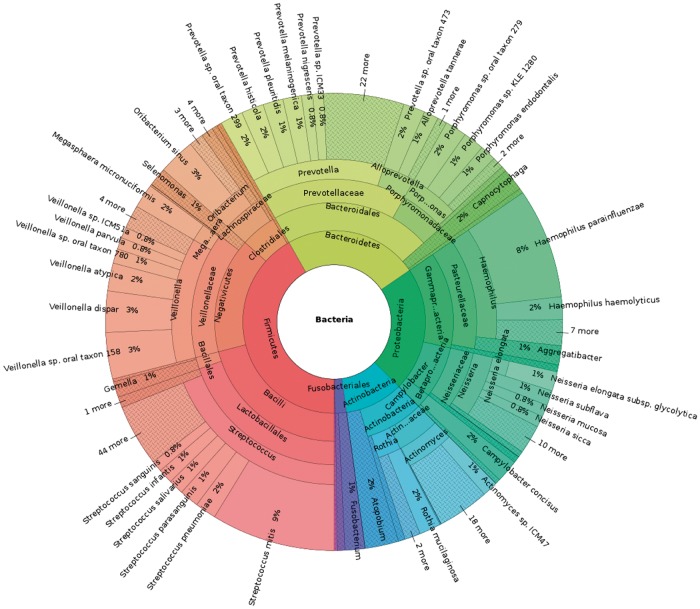
To test the performance of kallisto on biological data, we analyzed a set of saliva samples from the Human Microbiome Project. These three samples – SRS014468, SRS015055 and SRS019120 – consist of a total of 9.3 million 60–100 bp paired-end reads, collected from three separate individuals. We pooled them together to analyze the microbes present in the general saliva microbiome. Running the same Mash-based pipeline on 30k Ensembl genomes identified 744 likely genomes, and using kallisto to quantify the saliva reads against those genomes found primarily bacteria of the genera Streptococcus (17.5%), Prevotella (17.1%), Veillonella (11.2%) and Haemophilus (9.9%) as well as a number of less abundant genera (shown in Fig. 3). The most abundant species are those known to be abundant in the oral microbiome: Streptococcus mitis, Haemophilus parainfluenzae, Veillonella sp. oral taxon 158 and Prevotella histicola.
Fig. 3.
Results of kallisto on bacterial reads in human saliva samples at all taxonomic levels
3 Materials and methods
3.1 Illumina100 dataset
We tested kallisto and alternate programs on a set of simulated reads published in (Mende et al., 2012). The Illumina100 dataset consists of 53.33 million 75 bp reads, simulated by the iMESSi metagenomic simulator using an Illumina error model. The reads were simulated from a set of 100 unique bacterial genomes. The set is of genomes from 85 different species and 63 different genera, over a range of abundances from 0.86% to 2.2%.
Reads were trimmed with the program Trimmomatic (version 0.32) (Bolger et al., 2014) to a minimum length of 40 bp, using its adaptive trimming algorithm MAXINFO with a target length of 40 and default strictness. 40 reads were dropped due to quality issues.
3.2 Taxonomic identification
We analyzed each program’s output at four taxonomic ranks: phylum, genus, species and ‘exact genome’ level. The latter tests the abundance estimation of the actual Illumina100 genomes, which are a combination of strains and substrains and thus aren’t taxonomically well defined. The other three ranks are as assigned by NCBI’s Taxonomy Database, as of August, 2016.
3.3 Count estimation accuracy calculation
Using a simulated dataset with known abundances allowed us to benchmark programs by comparing program outputs with true values for each genome. While kallisto is able to output length-corrected individual genome abundances, most of the programs we compared with only output counts, so for consistency we analyzed the accuracy of assigned or estimated counts for each program. We normalized the estimated counts by the percent of assigned reads in order to be able to compare relative count estimates between programs.
We primarily used the error measures AVGRE (Average Relative Error), which computes the mean of the difference between truth and estimate, and RRMSE (Relative Root Mean Square Error), which computes the root mean square average of the difference between truth and estimate, to judge the accuracy of our estimates. Formally, with n true genomes/species/genera/phyla, true counts τi (1 ≤ i ≤ n) and estimated counts ti at the rank, and A aligned reads out of T total reads we computed
The scripts used to compile the results are available at https://github.com/pachterlab/metakallisto.
3.4 Reference genome database
Two reference genome databases were used, for all programs: one, referred to as ‘i100’ in the text, consists solely of the 100 genomes from which the Illumina 100 dataset was simulated from. These genomes were indexed (by kallisto, GASiC and eXpress) or loaded as a custom database (for CLARK and Kraken) without any pre-processing.
In addition, we tested the more realistic case of aligning against a large bacterial database – Ensembl’s bacterial genomes as of version 30, referred to as ‘Ensembl’ in the text. All 29 698 bacterial genomes were downloaded, combined with the i100 genomes, and used as-is with Mash (see below). For abundance estimation with Bracken, CLARK and kallisto, constituent contigs, chromosomes and plasmids were concatenated together with a series of 10 ambiguous bases represented as N, and NCBI’s taxonomic ID was manually added to the headers for Kraken’s use.
3.5 Mash genome pre-filtering
To lower the number of genomes to abundance estimate against to a reasonable level, we ran the Illumina100 dataset against all 30 000 ensembl genomes using Mash, a genome distance calculator. We used only the top 10 genomes from each species that were judged closest to the reads in subsequent abundance estimation, to get a reasonable number of genomes for indexing.
The scripts used to filter the genomes based on Mash results are available at http://github.com/pachterlab/metakallisto.
4 Conclusion
The idea of translating RNA-Seq methodology to and from metagenomics was, to our knowledge, first proposed in (Paulson et al., 2013) where statistical methods for identifying differential abundances in microbial marker genes were developed. In that paper, there were comparisons between the proposed metagenomics method and RNA-Seq differential analysis methods implemented in DESeq (Anders and Huber, 2010) and edgeR (Robinson et al., 2010). Notably, the central idea of the paper, the specific consideration of zero inflated distributions to account for undersampling, is also used in single cell expression analysis (McDavid et al., 2013).
Our results show that RNA-Seq methods for quantification are also applicable in the metagenomics setting, and our results with kallisto demonstrate that it is possible to accurately and rapidly quantify the abundance of individual strains. With a few exceptions, e.g. (Bradley et al., 2015), most metagenomic analyses have focused on higher taxonomy, a point highlighted in the recent benchmarking paper (Lindgreen et al., 2015) which compares predictions at the phylum level because ‘[comparisons at that level are] less prone to differences’. The phylum level is four levels removed from genus, let alone species or strain. Our results suggest that the door is now open to metagenome analyses at the highest possible resolution.
While our benchmarks are primarily based on simulated data, our experiments are much more realistic than previous analyses. For example, the Kraken and CLARK papers report results on simulations with ten genomes, whereas we have simulated from 100 genomes and mapped against nearly 30 000. One of the difficulties we faced in our analyses was the technical issue of taxonomic naming and annotation in collating results. This seemingly trivial matter is complicated by the lack of attention paid to low taxonomic level analysis in previous studies. Hopefully our results will spur others to standardize and organize analysis scripts so that low taxonomic level analysis can become routine.
A significant difficulty, for all analyses at the species or strain level, is distinguishing between those strains that share their core genome. For instance, our results highlight the difficulty in distinguishing Escherichia coli strains from Shigella strains: both kallisto and Bracken underestimate E.coli abundance and overestimate the abundance of several false-positive Shigella strains. This is unsurprising, as these two species are so genetically similar that the latter is sometimes considered a strain of the former (Zuo et al., 2013). As the number of highly-similar strains increases, the need for new algorithms that can distinguish strains on the basis of a few differentiating reads becomes more pressing.
In addition, the sheer size of growing reference databases will lead to continued challenges in quantification and downstream analysis. While the two-step Mash-kallisto workflow we have described here can scale for the time being, novel algorithmic ideas are needed to that can leverage large databases for individual genome analysis, yet efficiently discard irrelevant information.
Acknowledgements
We thank readers of preprints of this manuscript for helpful suggestions that have improved our method and its description in the paper.
Funding
H.P. was supported by an NSF graduate research fellowship. P.M. was partially supported by a Fulbright fellowship. L.S and L.P. were partially supported by NIH R01 HG006129 and NIH R01 DK094699.
Conflict of Interest: none declared.
References
- Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol., 11, R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M. et al. (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics, btu170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P. et al. (2015) Rapid antibiotic resistance predictions from genome sequence data for S. aureus and M. tuberculosis. Nat. Commun., 6, 10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. et al. (2015). Near-optimal RNA-Seq quantification. Nature Biotechnol, 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Chen K., Pachter L. (2005) Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol., 1, 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N. et al. (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods, 5, 613–619. [DOI] [PubMed] [Google Scholar]
- Huson D.H. et al. (2007) MEGAN analysis of metagenomic data. Genome Res., 17, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H. et al. (2009) Methods for comparative metagenomics. BMC Bioinformatics, 10, S12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P.J. et al. (2016) Ensembl genomes 2016: more genomes, more complexity. Nucleic Acids Res., 44, D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land M. et al. (2015) Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics, 15, 141–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen S. et al. (2015) An evaluation of the accuracy and speed of metagenome analysis tools. bioRxiv, 017830. [DOI] [PMC free article] [PubMed]
- Lindner M.S., Renard B.Y. (2013) GASiC: Metagenomic abundance estimation and diagnostic testing on species level. Nucleic Acids Res., 41, e10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R. et al. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell, 133, 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. (2016) Bracken: Estimating species abundance in metagenomics data. bioRxiv.
- McDavid A. et al. (2013) Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics, 29, 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende D.R. et al. (2012) Assessment of metagenomic assembly using simulated next generation sequencing data. PLoS ONE, 7, e31386.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A. et al. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U. et al. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science, 320, 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae M. et al. (2011) Estimation of alternative splicing isoform frequencies from RNA-Seq data. Algorithms Mol. Biol., 6, 9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov B.D. et al. (2016) Mash: fast genome and metagenome distance estimation using minhash. Genome Biol., 17, 14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J.N. et al. (2013) Differential abundance analysis for microbial marker-gene surveys. Nat. Methods, 10, 1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pachter L. (2013) Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods, 10, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D. et al. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Brito B. et al. (2006) An application of statistics to comparative metagenomics. BMC Bioinformatics, 7, 162.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz M.B. et al. (2012) Next generation sequencing and bioinformatic bottlenecks: the current state of metagenomic data analysis. Curr. Opin. Biotechnol., 23, 9–15. [DOI] [PubMed] [Google Scholar]
- Trapnell C. et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol., 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe S.G. et al. (2005) Comparative metagenomics of microbial communities. Science, 308, 554–557. [DOI] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol., 15, R46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L.C. et al. (2011) Accurate Genome Relative Abundance Estimation Based on Shotgun Metagenomic Reads. Plos One, 6, e27992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo G. et al. (2013) Shigella strains are not clones of Escherichia coli but sister species in the genus Escherichia. Genomics Proteomics Bioinf., 11, 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]