Abstract
Summary
The ease with which phylogenomic data can be generated has drastically escalated the computational burden for even routine phylogenetic investigations. To address this, we present phyx: a collection of programs written in C ++ to explore, manipulate, analyze and simulate phylogenetic objects (alignments, trees and MCMC logs). Modelled after Unix/GNU/Linux command line tools, individual programs perform a single task and operate on standard I/O streams that can be piped to quickly and easily form complex analytical pipelines. Because of the stream-centric paradigm, memory requirements are minimized (often only a single tree or sequence in memory at any instance), and hence phyx is capable of efficiently processing very large datasets.
Availability and Implementation
phyx runs on POSIX-compliant operating systems. Source code, installation instructions, documentation and example files are freely available under the GNU General Public License at https://github.com/FePhyFoFum/phyx
Supplementary information
Supplementary data are available at Bioinformatics online.
Introduction
Phylogenetic and phylogenomic analyses now involve massive datasets which makes traditional approaches for the analysis and manipulation of data onerous undertakings. A number of phylogenetic toolkits exist including but not limited to ETE (Huerta-Cepas et al., 2016), newick utilities (Junier and Zdobnov, 2010), Mesquite (Maddison and Maddison, 2016), ape (Popescu et al., 2012), phyutility (Smith and Dunn, 2008) and DendroPy (Sukumaran and Holder, 2010). Of course, no individual software package is exhaustive in its functionality (i.e. methods and supported file formats), so these packages largely complement one another, both in terms of novel and overlapping functionalities (i.e. confirming computed values), with differences (e.g. in memory requirements and speed) sometimes making certain packages more conducive to particular workflows. However, despite the rich diversity of existing tools, there is a niche to be filled for programs that are conducive to high throughput processes and the convenience of a POSIX-style interface.
In an effort to provide a more flexible and efficient software package for processing phylogenetic data and for conducting phylogenomic research we present phyx, a set of programs to carry out a wide range of phylogenetic tasks. Written in C ++ and modeled after Unix/GNU/Linux command line tools, individual programs perform a single task, have individual manual (i.e. man) pages and operate on standard I/O streams. A result of this stream-centric approach is that, for most programs, only a single sequence or tree is in memory at any moment. Thus, large datasets can be processed with minimal memory requirements. phyx’s ever-growing complement of programs currently consists of 35+ programs (see Table 1 for a subset) focused on exploring, manipulating, analyzing and simulating phylogenetic objects (alignments, trees and MCMC logs). As with standard Unix command line tools, these programs can be piped (together with non-phyx tools), allowing the easy construction of efficient analytical pipelines. phyx also logs all program calls to a plain text file, which is an executable record that can be submitted as part of a manuscript for reviewing and replicability purposes. phyx thus provides a convenient, lightweight and inclusive toolkit consisting of programs spanning the wide breadth of programs utilized by researchers performing phylogenomic analyses.
Table 1.
Selected phyx programs and their functions. See github for additional details and full program list
| Program | Function |
|---|---|
| pxlssq/pxlstr | List attributes of alignments/trees |
| pxrms/pxrmt | Remove taxa from alignments/trees |
| pxrls/pxrlt | Relabel taxa in alignments/trees |
| pxboot | Alignment bootstrap/jackknife resampling |
| pxclsq | Remove missing/ambiguous sites from an alignment |
| pxs2fa/phy/nex | Convert alignment to fasta/phylip/Nexus format |
| pxlog | Concatenate and resample MCMC parameter/tree logs |
| pxfqfilt | Filter fastq files by quality |
| pxrr | Reroot/unroot trees |
| pxtlate | Translate nucleotide sequences |
| pxsw/pxnw | Pairwise sequence alignment |
| pxstrec | Ancestral state reconstruction, stochastic mapping |
| pxbdfit/pxbdsim | Birth-death tree inference/simulator |
| pxseqgen | Simulate nucleotide/protein sequences on user tree |
2 Materials and methods
2.1 File processing, manipulation and conversion
File manipulation and conversion is a tedious and error-prone, but often required, component of phylogenetic analysis, made more burdensome by the volume of data available in current phylogenomics studies. phyx supports the popular formats for sequence alignments (fasta, fastq, phylip and Nexus) and trees (newick and Nexus), and provides lightweight, high-throughput utilities to convert data among formats without the user needing to provide the format of the original data as phyx will attempt to auto-detect the original format. Alignments can be further manipulated by removing individual taxa, resampling (bootstrap or jackknifing), sequence recoding, translation to protein, reverse complementation, filtering by quality scores or the amount of missing data, and concatenation across mixed alignment formats.
Processing large data matrices is only one step required for phylogenomic analyses. In order to perform downstream analyses (e.g. orthology detection (Yang and Smith, 2014), mapping gene trees to species tree (Smith et al., 2015), or gene tree/species tree reconciliation (Mirarab et al., 2014)) it is now also essential to be able to manipulate individual gene trees constructed from these data. phyx enables fast, efficient manipulations such as pruning individual taxa, extracting subclades and rerooting/unrooting trees. Finally, Bayesian MCMC analyses involving phylogenies have become common in the biological sciences, and often involve large log files generated from replicated analyses. phyx enables both the concatenation and resampling (burnin and/or thinning) of MCMC tree or parameter logs for downstream summary.
2.2 Analysis and simulation
In addition to file manipulation, phyx provides a growing number of tools for data analysis and simulation. Analytical capabilities presently include pairwise sequence alignment using either the Needleman-Wunsch or Smith-Waterman algorithms, tree inference using the neighbour-joining criterion, ancestral state reconstruction and stochastic mapping of discrete characters, fitting of Brownian or OU models to continuous characters, fitting birth-death models to trees, and computing alignment column bipartitions either in isolation or on a user tree.
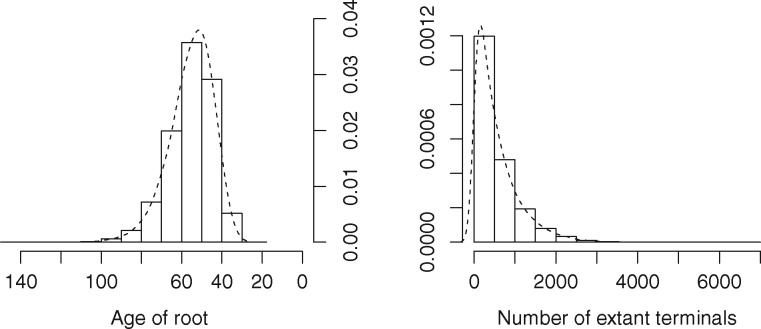
Data simulation is an essential tool with which to explore model sensitivity and adequacy through parametric bootstrapping or posterior predictive analyses (Bollback, 2002). phyx currently enables simulation of both birth-death trees (see example in Fig. 1) and nucleotide or protein alignments given a tree and substitution model parameters.
Fig. 1.
Parametric bootstrapping of a diversification process. The primate phylogeny of Springer et al. (2012) was fit to a birth-death model (pxbdfit). To explore the breadth of plausible diversification outcomes the maximum likelihood parameters (b: 0.339487, d: 0.268944) were used to simulate (pxbdsim) 25 000 phylogenies conditioned on either the extant diversity (367, left) or root age (66.7066 Ma, right) of the empirical tree
2.3 Comparison to existing programs
While we view phyx as a complement to existing tools, we demonstrate the relative performance (speed and memory requirements) of some phyx programs for common phylogenomics tasks in the Supplementary Data, available at Bioinformatics online.
3 Conclusion
phyx was designed to complement existing phylogenetic toolkits by enabling the exploration, manipulation, analysis and simulation of phylogenetic objects directly from the command line. Moreover, by conforming to a stream-centric approach, memory requirements are reduced significantly so that large volumes of data can be processed on even personal laptop computers.
Supplementary Material
Acknowledgements
We thank Ya Yang, Jeff Johnson and three anonymous reviewers for helpful suggestions that significantly improved the manuscript, and Ning Wang, Jeff Johnson and Ross Mounce for testing.
Funding
SAS and JWB were supported by the NSF AVATOL Grant 1207915, and JFW was supported by NSF DEB Grant 1354048.
Conflict of Interest: none declared.
References
- Bollback J.P. (2002) Bayesian model adequacy and choice in phylogenetics. Mol. Biol. Evol., 19, 1171–1180. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J. et al. (2016) ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol., 33, 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier T., Zdobnov E.M. (2010) The newick utilities: high-throughput phylogenetic tree processing in the unix shell. Bioinformatics, 26, 1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W.P., Maddison D.R. (2017) Mesquite: a modular system for evolutionary analysis. Version 3.2, http://mesquiteproject.org.
- Mirarab S. et al. (2014) ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics, 30, i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu A.A. et al. (2012) ape 3.0: New tools for distance-based phylogenetics and evolutionary analysis in r. Bioinformatics, 28, 1536–1537. [DOI] [PubMed] [Google Scholar]
- Smith S.A., Dunn C.W. (2008) Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics, 24, 715–716. [DOI] [PubMed] [Google Scholar]
- Smith S.A. et al. (2015) Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evol. Biol., 15, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M.S. et al. (2012) Macroevolutionary dynamics and historical biogeography of primate diversification inferred from a species supermatrix. PLoS ONE, 7, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J., Holder M.T. (2010) DendroPy: a python library for phylogenetic computing. Bioinformatics, 26, 1569–1571. [DOI] [PubMed] [Google Scholar]
- Yang Y., Smith S.A. (2014) Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: Improving accuracy and matrix occupancy for phylogenomics. Mol. Biol. Evol., 31, 3081–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.