Abstract
Summary
We present ‘Threshold-seq,’ a new approach for determining thresholds in deep-sequencing datasets of short RNA transcripts. Threshold-seq addresses the critical question of how many reads need to support a short RNA molecule in a given dataset before it can be considered different from ‘background.’ The proposed scheme is easy to implement and incorporate into existing pipelines.
Availability and Implementation
Source code of Threshold-seq is freely available as an R package at: http://cm.jefferson.edu/threshold-seq/
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
‘Short RNA-seq’ is widely employed to study categories of non-coding RNAs (ncRNAs), typically between 15 and 30 nucleotides (nts) in length (Bartel, 2004; Cloonan et al., 2011; Londin et al., 2015). The benefits of short RNA-seq include its comprehensive nature, relatively low cost, and the relative ease with which it can be implemented and executed. Unlike microarray approaches where one is constrained to quantifying the abundance of only the ncRNAs represented by the microarray’s probes, short RNA-seq quantifies any ncRNA that is present in a sample.
A number of factors establish the effective depth at which a given RNA sample is sequenced. These factors include RNA quality, library preparation, degree of multiplexing, DNA contamination, micro-organism contamination (e.g. by mycoplasma), etc. In the general case, the relative contributions of these factors cannot be quantified. Given these considerations, any two RNA-seq datasets will have different sequencing depths. The idea behind ‘normalization’ is to account for the uneven sequencing depths of datasets that are about to be compared, in order to ‘equalize’ the read counts of transcripts that are common to the datasets being compared. A popular normalization approach has been to express RNA transcript abundance in terms of ‘Reads Per Million Mapped’ reads (RPMM) values. RPMM can be applied to individual datasets. Several other normalization approaches have been suggested over the years; they are summarized and reviewed elsewhere (Dillies et al., 2013; Garmire and Subramaniam, 2012; Tam et al., 2015).
It is important to stress that normalization methods do not answer the question of how to establish the level of support below which one would likely be immersed in noise. The idea behind ‘thresholding’ is to separate molecules of putative biological relevance from those that likely result from degradation or aberrant transcription (‘background’ noise). A frequent and arbitrary threshold choice has been to use 1.0 RPMM. However, this choice will lead to complications if a dataset contains outlier features that receive a lot of support compared to the rest of the features that are present: such outliers can cause high variability in the number of features that are retained and analyzed (see Supplementary Fig. S2).
Here, we present Threshold-seq, a novel approach to thresholding short RNA-seq datasets. Threshold-seq adapts to sequencing depth variations and permits dynamic selection of dataset-specific thresholds that strike a balance between the competing requirements of sensitivity and specificity. Threshold-seq alleviates the above-mentioned influence of outliers by calculating a threshold through analysis of the number of distinct molecules that can be identified in a dataset. In other words, Threshold-seq does not rely only on the number of reads in the dataset.
2 Materials and methods
Threshold-seq proceeds as follows. After adapter removal and quality trimming, the sequenced reads are mapped uniquely to the genome of interest (Londin et al., 2015). No thresholding is applied at this step: all sequences that are supported by at least one read are kept: let K be the number of unique sequences that are kept. Note that K is not user-defined; it is determined from the dataset being analyzed. Each of the K sequences that are kept is then paired with a count that reflects its support in terms of mapped reads: thus, we generate a collection S of K pairs of the type (sequence, count). We iterate over the following three steps for a total of N times (default N = 1000). During the n-th iteration (1 ≤ n ≤ N):
we randomly resample K (sequence, count) pairs with replacement. i.e. we draw K (sequence, count) pairs from the original pool of K unique sequence-count values, creating a collection S′ that contains as many (sequence, count) pairs as the original collection S. However, owing to our use of replacement during resampling, the latter being a key element of our approach, S′ ≠ S.
for the resampled K (sequence, count) pairs, we approximate numerically the cumulative distribution CDF as follows: CDF n, where F(i) is the fraction of the resampled sequences that are paired with a count of exactly i reads;
for a user-defined interval [MIN, MAX] (default interval = [0.90, 0.99]), we report the abscissas xo, x1, x2, …, xmax at which CDFn(x0)=MIN, CDFn(x1)=MIN + ε, CDFn(x2)= MIN + 2ε, …, CDF n(xmax)=MAX (default ε = 0.5%).
We then identify CDFtarget as the smallest CDF value at which multiple values of x were reported across N iterations: CDFtarget is the smallest value within [MIN, MAX] at which biologically relevant sequences begin to differentiate themselves from background. Finally, we report as the threshold of choice, xthresh, the value mode(xn) over all values xn that satisfy CDFn(xn)=CDFtarget. In other words, xthresh is the most frequent abscissa at which the CDF reaches CDFtarget (illustrated in Supplementary Fig. S3) across N iterations.
To compare Threshold-seq with arbitrary thresholds, we used two public collections of datasets for which technical replicates are available: (i) five samples from the GEUVADIS RNA sequencing project (1000 Genomes Project; see also Lappalainen et al., 2013); and (ii) three samples that we published previously (Londin et al., 2015). The GEUVADIS datasets were sequenced at seven different sequencing centers and have different sequencing depths even though they are technical replicates (Supplementary Table S1). Our three samples were sequenced at two locations (our University and Applied Biosystems/Thermo Fisher Scientific—see also Supplement for more details). By working with these datasets, we can evaluate our method while removing biological variability.
3 Results
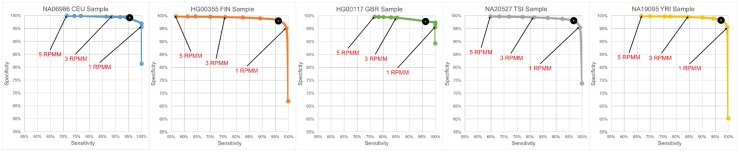
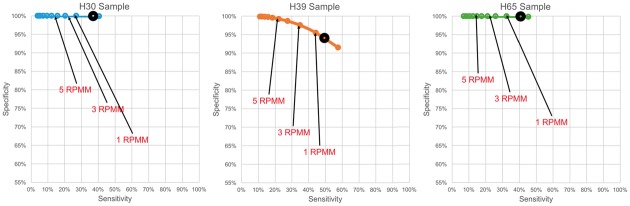
For each of the 44 analyzed datasets (5 biological samples × 7 technical replicates plus 3 biological samples × 3 technical replicates), we compare the results of Threshold-seq to several fixed RPMM threshold choices ranging from 0.5 to 5.0 (Figs 1 and 2). We also compare Threshold-seq to thresholds in terms of absolute read numbers (Supplementary Tables S1 and S2, Supplementary Figs S1 and S2). For each of the five GEUVADIS samples, we examined the corresponding seven technical replicates and identified: (i) collection A of all sequences that are supported by one or more reads and are present in all seven replicates of the sample; we refer to the set A as the set of positives; (ii) collection B of all sequences that are supported by one or more reads and are absent from at least one of the seven replicates; we refer to the set B as the set of negatives; and, (iii) collection C of all sequences that go above threshold in at least one of the seven replicates. Thus, for a given choice of threshold, the true positives will be equal to C ∩ A; the false positives will be equal to C ∩ B; and, the true negatives will be equal to B\C. In Figure 1, we plot sensitivity (X-axis) vs. specificity (Y-axis) for different choices of RPMM threshold, ranging from 0.5 to 5.0 (in steps of 0.5). For each of our three samples, we examined its three technical replicates and repeated the above analysis (Fig. 2). Note how Threshold-seq adaptively approaches the RPMM value that achieves a balance between sensitivity and specificity and how that value differs across the eight samples shown in Figures 1 and 2. Also, importantly, Threshold-seq is reliable over a large range of values for N (Supplementary Figs S3 and S4).
Fig. 1.
Comparison of Threshold-seq with arbitrary RPMM thresholds. The shown five samples were sequenced in seven technical replicates. In each case, we plot the obtained sensitivity (X-axis) vs. the obtained specificity (Y-axis) for different RPMM thresholds from 0.5 to 5 in increments of 0.5. Dark circles show the Threshold-seq equivalent metrics in each case
Fig. 2.
Comparison of Threshold-seq with arbitrary RPMM thresholds. The shown three samples were sequenced in three technical replicates. In each case, we plot the obtained sensitivity (X-axis) vs. the obtained specificity (Y-axis) for different RPMM thresholds from 0.5 to 5 in increments of 0.5. Dark circles show the Threshold-seq equivalent metrics in each case
4 Discussion
We presented Threshold-seq, a method for automatically establishing read thresholds when analyzing short RNA-seq datasets. Threshold-seq works on any short RNA-seq dataset regardless of choice of mapping program and of parameters. Threshold-seq can work with individual datasets; i.e. it does not require the availability of technical or of biological replicates. In Figure 1 and Supplementary Figure S1 we show that when using low absolute thresholds (e.g. 5 reads) or low RPM values (e.g. 1.0 RPM) more distinct sequences go above threshold (=high sensitivity) at the expense of low specificity; on the other hand, using a higher absolute threshold (e.g. 15 reads) or higher RPM (e.g. 5.0 RPM) improves specificity, at the expense of lower sensitivity. By resampling the distinct sequences of the dataset at hand, Threshold-seq achieves a good balance between sensitivity and specificity. Threshold-seq will capture those sequences of a sample that can be confidently assumed to represent biologically relevant features in the tissue/cell of origin, while remaining immune to any outliers that could be present in the data (see also Supplementary Fig. S2).
Funding
This work has been supported by a W. M. Keck Foundation grant (IR), NIH/NCI grant R21-CA195204 (IR), and by Institutional Funds.
Conflict of Interest: none declared.
Supplementary Material
References
- Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Cloonan N. et al. (2011) MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol., 12, R126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies M.A. et al. (2013) A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform., 14, 671–683. [DOI] [PubMed] [Google Scholar]
- Garmire L.X., Subramaniam S. (2012) Evaluation of normalization methods in mammalian microRNA-Seq data. RNA, 18, 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T. et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature, 501, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londin E. et al. (2015) Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. U. S. A., 112, E1106–E1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S. et al. (2015) Optimization of miRNA-seq data preprocessing. Brief. Bioinform., 16, 950–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.