Preface
This review highlights how we can build upon the relatively new and rapidly developing field of bacterial communication or quorum sensing (QS). We now have a depth of knowledge about how bacteria use QS signals to communicate with each other and coordinate activities. There have been extraordinary advances in QS genetics, genomics, biochemistry, and diversity of signaling systems. We are beginning to understand the connections between QS and bacterial sociality. This foundation places us at the precipice of a new era where researchers can advance towards development of new medicines to treat devastating infectious diseases, and in parallel use bacteria to understand the biology of sociality.
Keywords: Sociomicrobiology, bacterial communication, Vibrio, Pseudomonas aeruginosa, orphan receptors, solo receptors, biogeography, quorum quenching, anti-virulence strategies
‘Competing is intense among humans, and within a group, selfish individuals always win. But in contests between groups, groups of altruistsa always beat groups of selfish individuals.’
E.O. Wilson
Main text
Humans have provided descriptions of the natural history of animals for millennia. Apart from basic anatomy and physiology, it was also noted that a number of animal species showed signs of “social intelligence”. Aristotle provided a description of animal social behavior in his 4th century BC book ‘History of Animals’, noting how ants march one after the other when putting away food, while bonitos swarm together when they catch sight of a dangerous creature. The application of evolutionary principles to social behaviors in the 19th century brought forth the field of sociobiology that was further explored and popularized in the 1970’s2. While the last 40 years have been a time of great advancement and debate in sociobiology, these ideas were largely unexplored in the field of microbiology until the 21st century. We now know that microbes are highly gregarious communicating organisms and bacterial communication can modulate a range of behaviors important for fitness (reproductive success).
Background and a Brief History of Quorum Sensing
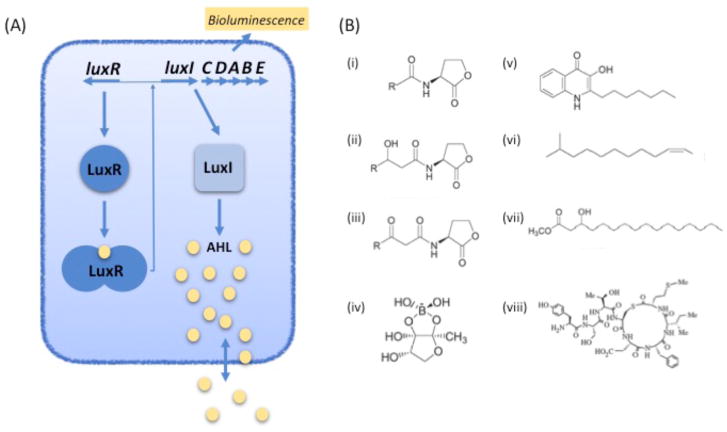
Bacterial QS involves self-produced extracellular chemical signals, which can accumulate in a local environment to levels that are required to activate transcription of specific genes3–5. The first hints about QS came in the late 1960s and early 1970s, when investigators showed that genetic competence in Streptococcus pneumoniae6 and luminescence in two species of marine bacteria7,8 required production of extracellular molecules. Cell-cell signaling via these molecules was proposed as a form of chemical communication, but these early publications were met with skepticism and generally ignored for the next 10–20 years. The 1980s brought two landmark discoveries: (i) the luminescence (lux) genes from the marine bacterium Vibrio fischeri were identified, and the genes required for what is now called quorum control of luminescence, luxI and luxR, were show to control lux gene transcription9,10; and (ii) the QS signal from V. fischeri was determined to be N-3-oxohexanoy-L-homoserine lactone (3OC6-HSL)11 (Fig. 1A). The luxI gene codes for the autoinducer synthase required for 3OC6-HSL production, and luxR codes for a 3OC6-HSL-responsive transcriptional activator of the lux genes (Fig. 1A).
Fig. 1. Canonical QS and the chemical diversity of signals.
(A) Quorum sensing in Vibrio fischeri. LuxI produces 3O-C6-HSL (AHL, yellow spheres), which specifically interacts with the LuxR transcriptional regulator when it reaches concentrations in the nM range. This leads to expression of the luxICDABE operon and bioluminescence. (B) Examples of quorum sensing signals from Gram-negative and Gram-positive bacteria. (i) AHL, N-acyl homoserine lactone; (ii) 3-Hydroxy-AHL, N-(3-hydroxyacyl)homoserine lactone; (iii) 3-oxo-AHL, N-(3-oxoacyl)-L-homoserine lactone. R can be a fatty acyl group of 4–18 carbons with or without one unsaturated carbon-carbon bond, the terminal carbon can be branched and some R groups are aromatic acids (p-coumaric acid or cinnamic acid); (iv) The V. harveyi AI-2, autoinducer-2, furanosyl borate ester form; (v) PQS, Pseudomonas quinolone signal, 2-heptyl-3-hydroxy-4(1H)-quinolone; (vi) DSF, diffusible factor, methyl dodecenoic acid; (vii) PAME, hydroxyl-palmitic acid methyl ester; (viii) Autoinducing peptide 1 (AIP-1) from Staphylococcus aureus.
General interest remained muted for another decade. In the 1990s DNA sequencing and comparative sequence analysis became everyday laboratory procedures, and gene pairs with homology to luxR and luxI began to attract the curiosity of some investigators. This led to an explosion of findings that other bacterial species controlled genes for conjugation, exoenzyme production and antibiotic synthesis with LuxI-LuxR-like systems3. A common theme emerged; the LuxI homologs catalyzed synthesis of an acylated homoserine lactone (AHL) and the LuxR homologs all showed specificity for their cognate AHL. This convergence of discoveries led to the QS concept (Fig. 1A); that the diffusible AHLs served as a proxy for cell density and allowed a bacterial species to produce costly extracellular public goods only when there was a sufficient biomass to benefit from the public goods4. Shortly thereafter the QS signal from S. pneumoniae (often referred to as a pheromone) was shown to be a small peptide12, and Staphylococcus aureus was shown to use small cyclic peptide pheromones to activate genes for production of extracellular toxins13. Quorum sensing was therefore shown to occur in Gram-positive and Gram-negative bacteria via diverse chemical signals (Fig. 1B). An early study showed that a luminescent marine bacterium called Vibrio harveyi could sense a self-produced signal and also a signal or signals produced by other bacterial species to induce light production7. This phenomenon is considered to be a type of QS, where cells of many species in a mixed microbial community sense the general bacterial population density via a molecule termed Autoinducer-2 (AI-2)14,15. The strengths and weaknesses of this concept are discussed later in this review. Further studies also described QS-like systems in eukaryotic microbes (the pathogenic fungi Candida and Histoplasma)16,17 and very recently in viruses18, thus providing clear examples of convergent evolution. Some QS signals are volatile, for example the DSF and PAME signals shown in Figure 1, and there is some evidence that volatile signaling can occur in a local atmosphere19. Functional studies followed the discovery of many of these systems and revealed that for many plant and animal pathogens, QS mutants showed greatly reduced virulence13,20,21. The early connection between QS and pathogen virulence brought forth excitement about the idea of targeting QS as a novel approach to treat bacterial infections (Box 1).
Box 1. The challenge to QS therapeutic development.
Early discovery that QS mutants of important plant and human pathogens are attenuated for virulence20,21 led quickly to the concept of using QS inhibitors to control some diseases110. In fact, a variety of small molecule inhibitors of QS signal receptors and LuxI-type QS signal synthases have been discovered, as well as enzymes, which degrade AHL signals90–92,99–104. Yet we face obstacles in moving from the bench to the clinic. Many of the obstacles are inherent to drug discovery such as lack of potency in animal models, delivery, toxicity, stability, and a narrow spectrum of activity. But what are other obstacles? Fundamental questions remain unanswered. At what point in an infection will QS inhibition be of value? Will QS inhibitors find general utility or will they function best as prophylactic agents? Can biological interference approaches be contemplated? Can we imagine introducing a benign bacterial species, which produces an AHL lactonase to interfere with a QS pathogen into a human? We note that such an anti-QS approach has been developed to control membrane fouling by biofilms in water purification plants (see image below)111. What pathogens should we target? What are the regulatory hurdles for a QS inhibition therapeutic to even enter a clinical efficacy trial? How quickly will resistant bacteria emerge? There is an idea that because QS inhibitors are anti-virulence agents rather than antibacterial agents, resistance is less likely to emerge. Laboratory experiments do not support this idea98 but in theory the route to the spread of resistance may be slow and depending on the resistance mechanism it may be self-limiting. The route to testing anti-QS approaches may be much less tortured in the case of crop diseases where any number of approaches can be easily tested in the laboratory or greenhouse. Ultimately, if QS inhibition is to gain therapeutic utility for human diseases, this will likely occur in the context of combination therapies with conventional antimicrobials.
Box 1 image.
Quorum-quenching membrane filtration (QQ-MBR) pilot plant system at sewage treatment facility in Seoul, South Korea. Professor Chung-Hak Lee of Seoul National University (center) has led a decade-long program to develop this technology
Sociomicrobiology: An evolutionary perspective on quorum sensing
Early research assumed that QS is a social trait (a trait that impacts both the individual performing a behavior and a recipient22), but this concept was not tested experimentally. In principle, QS could be non-social, for instance allowing individual bacteria to sense their physical environment (this has been termed diffusion sensing)23,24. However, there is now a large body of evidence showing that in certain environments, QS is social and that at the population level, QS regulates the production of extracellular public goods22,23,25–29. Importantly, whilst public goods directly benefit the producing cell, they also indirectly socially benefit surrounding cells. Because the production of a public good is costly, these behaviors are potentially exploitable by non-producing ‘cheats’, creating bacterial social dilemmas (Fig. 2). QS has now been shown to be exploitable by cheats in laboratory cultures26–29. One well-studied example is with the pathogenic bacterium Pseudomonas aeruginosa. This bacterium requires QS to induce production of extracellular proteases required for growth on proteins. QS mutants fail to grow by themselves on milk protein, but in co-culture with QS competent protease producing cells the mutants have a fitness advantage when at relatively low relative abundance25,26,29. In fact mixed infections of cooperating and cheating cells have been shown to be less virulent than single strain cooperator infections30. This has led to the idea of using bacterial cheats to help treat infections by reducing virulence or by introducing ‘Trojan Horse’ genes into virulent populations31.
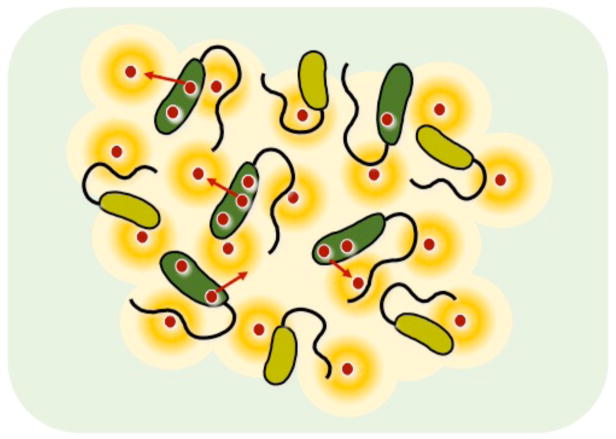
Fig. 2. Social cheating in QS populations.
Bacterial cells act as cooperators (green) when secreting QS-dependent public goods (e.g. protease, red spheres) into the surrounding environment and this imposes a fitness cost on individual cells. Cheater cells (yellow) do not secrete these enzymes and pay no fitness costs. Cheats can benefit from the action of public goods and therefore gain a fitness advantage in mixed populations with cooperators. Orange halos depict a nutrient source liberated by a public good (e.g. protease) from which all cells (cooperators and cheats) can benefit.
Given that QS can be readily exploited by cheats, why are numerous functional QS systems maintained in natural populations? This was a dilemma first discussed by Darwin: if cheats in a population have a fitness advantage over cooperators they should survive, take over the population and cooperation should be inherently unstable. Kin selection theory has been invoked to help explain this dilemma32,33. Put simply, by helping a relative reproduce, an individual indirectly passes its genes into the next generation. Kin selection has been proposed to be important in microbial social behaviors such as QS because of clonal reproduction and relatively local interactions26,34. Recently the importance of kin selection in maintaining cooperative behaviors in single and multicellular organisms has been theoretically challenged, and microbial systems provide powerful experimental platforms to help resolve this debate35–37. The concept of kin selection is impacted by spatial structure, which can maintain cooperation and public goods because it keeps cooperators and relatives close together. Surface associated microbial communities (termed biofilms, described in detail later) are a good example of spatially structured populations The biofilm lifestyle helps restrict the invasion of cheats and affects population dynamics28,38. In addition, recent studies of QS have revealed molecular mechanisms for stabilizing cooperator populations. We have learned that metabolic prudence can constrain cheaters. Here an expensive public good is co-regulated by QS and nutrient availability such that the public good is produced when two conditions are met; there is a quorum and an ample nutrient supply such that the cost of public goods production is not critical39. We have also learned that there are metabolic constraints on cheating where QS co-regulates private goods with public goods. Experimentally, transcriptomic studies revealed that although P. aeruginosa QS controls production of a battery of extracellular products, which may be considered public goods, it also controls some cell-associated products, one of which is a cellular enzyme required for growth on adenosine. As discussed above, when P. aeruginosa is grown on milk protein QS is required for expression of genes for extracellular proteases, and there is a fitness advantage for QS mutants. When P. aeruginosa is grown on milk protein plus adenosine cheats are unable to utilize the adenosine and their fitness advantage is nullified; a penalty has been placed on cheating40. Finally, we are beginning to understand policing, defined as an ability of bacterial cooperators or a host organism (host sanctioning), to hinder the fitness of cheats41–43. A recent report describes how QS regulation of pairs of genes coding for toxins and toxin immunity can serve as a policing mechanism. QS competent individuals deliver toxins to other individuals. QS competent cells are immune but QS mutants are not.
Although we now have a wealth of mechanistic data showing how QS systems function at the molecular level, the true biological function of many QS systems remains a research area of great opportunity. Evolution and ecological approaches can help address this knowledge gap. For example, are the chemically diverse QS molecules described in the literature always acting as signals? In an evolutionary context, a true signal evolved because it alters the behavior of a receiver and the receiver’s response must have co-evolved. This is distinct from a ‘cue’ where the production of a substance has not evolved because of its effect on a recipient. If the production of a substance forces a costly response from a receiver we can differentiate this from signaling and term it coercion or chemical manipulation44–46. The current literature sometimes conflates signaling, cueing and coercion, and whether bacteria are interacting via a signal, a cue or coercion can lead to different biological outcomes. This is becoming increasingly important as we begin to study microbiota interspecies interactions and as we seek to develop anti-QS therapeutics. It has been exciting for us to see these concepts introduced into the field of microbiology and to see the power of microbiology and microbial genetics brought to bear on questions about the mechanisms and consequences of communication and social interactions in ways that are immensely more difficult with higher organisms. Although there are certainly limitations in studies of social behavior and social evolution in bacteria there are important advantages. One can execute an experiment with hundreds of millions of individuals on the bench top. Experiments with ten generations of offspring can be done in a day. Correlating genes with social activity is routine. In this article, we focus a considerable amount of discussion on the pathogenic bacterium Pseudomonas aeruginosa. We do so in part because it is a particularly well-studied model in the QS field, and it is one on which the laboratories of all three co-authors have worked. There are mutant libraries of this bacterium. As a first analysis of gene-social function relationships one needs only to order a mutant and study it in the context of sociality.
How and why the distinction between signals, cues and coercion is important is exemplified by the AI-2 molecule described earlier (Fig. 1B). AI-2 is a furanosyl borate diester produced by V. harveyi14. The identification of the luxS gene, which is required for AI-2 production47, sparked a huge interest into AI-2 because this gene is found in both Gram-positive and Gram-negative bacteria15. This led to the hypothesis that AI-2 allows widespread communication between bacterial genera, a type of ‘bacterial Esperanto’48. However, evolution and signaling theory question whether AI-2 can be defined as a true interspecies signal. For this to be the case AI-2 must (1) diffuse from the producing cell; (2) interact with a receiver cell; (3) elicit a response from the receiver cell that has co-evolved with signal production by the producer; and (4) benefit both producer and receiver. Points 1 and 2 are met with respect to AI-2, but points 3 and 4 are often not met between two or more species. Despite AI-2 being produced by many genera there are few instances, including V. harveyi luminescence47 and the Lsr ABC transporter in Salmonella typhimurium49,50, linking it with direct activation of specific genes. While a number of studies have reported that AI-2 ‘signaling’ impacts specific bacterial phenotypes, many of these studies have relied on the use of luxS mutant strains. As LuxS is involved in recycling of S-adenosyl-L-methionine, of which AI-2 is a non-toxic metabolic byproduct, luxS mutant phenotypes may simply be due to metabolic perturbations51,52. In these cases, AI-2 cannot be considered a signal at either an intra or interspecies level. By understanding that in some cases AI-2 might serve as a signal where in other cases it is a cue, or even a waste product, studies of AI-2 should lead to evolutionary perspectives about how metabolic waste products can evolve to become signals, and about the evolution of chemical communication itself.
A next step: QS in natural habitats
The bulk of studies driving our current understanding of the molecular biology and evolution of QS utilize well-mixed laboratory cultures and growth environments not intended to mimic the natural environment. These in vitro systems provide reproducible conditions for biochemical and evolution studies, as well as the ability to grow large culture volumes often necessary for signal purification and identification. These systems have also provided a wealth of solid fundamental knowledge on which we can build to study the role QS plays in modulating the composition and function of natural microbial communities.
Working in natural microbial habitats is challenging and requires QS scientists to embrace the complexity of these environments while leveraging state of the art multi-disciplinary approaches. One elegant system that has been developed to begin to study QS within a natural bacterial community in an animal host is the mutualistic symbiosis between the squid Euprymna scolopes and its light organ symbiont V. fischeri. The squid, which inhabits coastal waters of the Hawaiian Islands is born with a sterile light organ. The light organ is colonized specifically by V. fischeri, which occurs in low abundance in the surrounding seawater. V. fischeri uses its QS system to activate genes for luminescence in the high-density light organ environment. The mutualism is simple in that there are but two partners, and it is amenable to laboratory manipulation. This model system has provided insight not only about the role of QS in an animal host-bacterial interaction, but also about how a microbiome, albeit a simple one, can influence host development (for recent reviews see53,54). Furthermore, juvenile squid are smaller than a pencil eraser and translucent, and bacteria tagged with GFP can be easily visualized in whole light organs. This has provided a means to study a difficult question discussed later in this review; can aggregates of bacteria employ QS signals to communicate with other aggregates, and if so is there a discrete distance over which this kind of communication can occur in a given condition (a calling distance)?
An ultimate goal is to integrate our comprehensive understanding of QS derived from elegant laboratory culture studies, with ecological principles to illuminate the role of bacterial communication in natural habitats. Researchers are now beginning to study more habitats with more complex microbiota than the squid symbiosis. Here, we focus on three areas of emerging interest that have important and relatively unknown functions in natural ecosystems: (1) orphan LuxR homologs; (2) the link between QS and microbial biogeography, and (3) quorum quenching.
Orphan or solo LuxR homologs and interspecies interactions
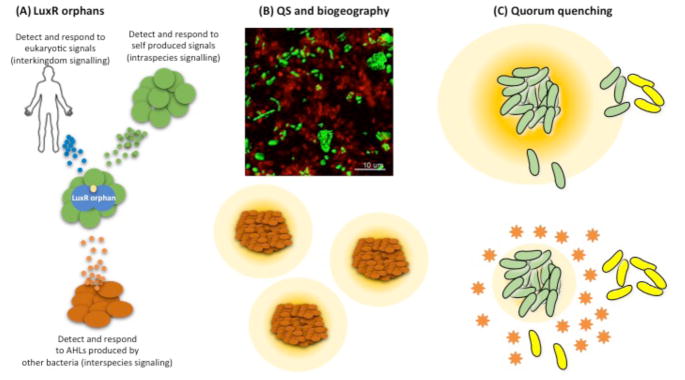
LuxR-type transcription factors consist of two domains, an N-terminal signal-binding domain and a C-terminal DNA-binding domain, and simple homology searches can be used to identify LuxR homologs in genomic sequences. Just such a search of the Salmonella typhimurium genome revealed the first example of what we now call orphan55 or solo56 LuxR homologs (Fig. 3A). The genome of S. typhimurium possesses a luxR homolog called sdiA but it does not possess a luxI-type gene, and S. typhimurium does not produce AHLs55,57,58. In fact, many Gram-negative bacteria possess luxR-type genes and do not possess luxI-type genes, and many more Gram-negative bacteria possess greater numbers of luxR-type genes than luxI-type genes. In the case of S. typhimurium, we have learned that SdiA responds to AHLs produced by other bacteria, leading to activation of specific genes58. There is also an sdiA gene in E. coli. The E. coli SdiA has been reported to respond to AHLs and mammalian host-produced small molecules59–61. These latter studies provided the first hints that some orphan LuxR homologs might be involved in sensing the host environment, directly, rather than serving as QS signal receptors (Fig. 3A).
Fig. 3. Factors that impact QS in natural environments.
(A) LuxR orphans (solos) can detect and respond to signals produced by eukaryotic hosts, self, or other microbial species. (B) Top: Aggregates of two P. aeruginosa strains (green, strain PA14 and red, strain PAO1) in synthetic cystic fibrosis sputum 101. Bottom: Diagram of P. aeruginosa aggregates (3 clusters of red rods) spatially separated and socially isolated (yellow halos represent QS signals and QS-controlled exoproducts). (C) Top: Diagram of a high-density aggregate of QS positive cells (green) secreting a diffusible QS signal into the surrounding environment (orange halo). The signal can activate QS in a nearby cells (green) but not in more distant cells (yellow). Bottom: quorum quenching (orange stars), either by the action of enzymes or due to environmental conditions, degrades the QS signal and limits the ability of an aggregate to induce QS in nearby cells.
Some orphan LuxR proteins, such as QscR in the opportunistic pathogen Pseudomonas aeruginosa, can respond to self-produced AHLs62–64, while others respond to non-AHL, self-produced signals. Two examples of the latter case involve members of the genus Photorhabdus. One species uses an orphan LuxR protein to detect self-produced α–pyrones65 and the other detects self-produced dialkylresorcinols and cyclohexanediones (Fig. 1B)66. Finally, there is a group of orphan LuxR homologs in some plant-associated bacteria that activate transcription of specific genes in response to small molecule(s) produced by the plant67–69. The identity of these small molecules remains elusive and they may in fact be produced by the plant microbiota. The orphan LuxR homologs and what we now know about them bring this research area to an interface with advances in microbiome research, and studies of this group of LuxR homologs represent a rapidly emerging area.
Advances in mammalian gut microbiome research are beginning to reveal how QS can influence microbiome species composition, how QS research might lead to ways to control infectious diseases like cholera, and how the host itself may have evolved mechanisms to affect bacterial QS to shape its microbiome. For example, the AI-2 molecule produced by many bacterial species, and discussed earlier, was recently shown to promote Firmicutes over Bacteroidetes gut colonization70 and production of AI-2 by a gut commensal bacterium can limit Vibrio cholerae infections71. Although the interactions of bacteria in the mammalian gut are much more complex than the two-partner squid symbiosis, it is becoming clear that the involvement of QS in these microbiomes can provide an opportunity to intervene in gut dysbiosis.
QS, biofilms and microbial biogeography
Biofilms are defined as high-density bacterial clusters frequently attached to surfaces and encased in an extracellular polymeric matrix72. Biofilm cells have several unique properties compared to their planktonic (free-living) counterparts, notably an enhanced tolerance to antimicrobials. Throughout the 1980s and 1990s, the prominence of biofilms in nature stimulated development of experimental laboratory systems. Several groups leveraged these systems for studying QS in biofilm communities. One common laboratory biofilm system is the flow-cell73, in which biofilms growing on glass coverslips covering small channels are imaged using a confocal scanning laser microscope. A nutrient medium is continuously pumped through the channels to feed the biofilms. Using flow-cell systems, several groups showed that QS can impact biofilm construction as well as the ability of the biofilm to tolerate antimicrobial treatment74. In some cases, QS is critical for building a biofilm, whereas in other cases it is important for biofilm disassembly. The link between QS and biofilm formation led to a flurry of studies to assess how microbial social behaviors impact this important mode of growth, but it was quickly discovered that this link was dependent on environmental conditions75. Not surprisingly, there is interplay between environmental cues and intercellular communication. This leads to the question: Is the link between QS and biofilms specific to flow-cell laboratory conditions or is it relevant in natural ecosystems, and if so, which ecosystems?
We have learned that 3D biofilm architecture in flow-cells often does not mimic biofilm structure in natural ecosystems76,77. Indeed, some naturally occurring microbial biofilms are composed of high-density, micron-scale aggregates containing hundreds to thousands of cells. These aggregates exhibit remarkable micron-scale spatial organization, and this biogeography is critical for fitness of the microbial community78,79. The fact that natural aggregates share traits with laboratory biofilms, including enhanced antimicrobial tolerance, led to the hypothesis that QS is involved in aggregate formation. This hypothesis is supported by studies of several bacteria including Pantoea ananatis, Rhodobacter sphaeroides, Burkholderia thailandensis, and E. coli. In P. ananatis and R. sphaeroides, QS inhibits formation of large aggregates via unknown mechanism(s), while aggregate formation is promoted by QS in Burkholderia thailandensis and E. coli80,81. In the case of E. coli, aggregate formation is promoted via active movement of individual cells towards aggregates excreting AI-281.
Although QS appears to play some role in aggregate formation, is it also important for the precise spatial organization of aggregates, and how does aggregate formation influence QS? To answer these questions, it is not only necessary to understand the relationship between aggregate size and QS-controlled behaviors but also the effective ‘calling distance’ of signals produced by an aggregate. Elucidating the number of cells required to reach a quorum has been actively pursued over the past 10 years. As predicted by the QS hypothesis, confining single bacterial cells in femto- to picoliter aqueous volumes has provided evidence that single cells can QS, although it is not clear whether there are fitness benefits to doing so82,83 (for reference the volume of an E. coli cell is on the order of about a half to one or two femtoliters). However, these closed systems do not allow exchange of solutes outside the confinement volume, which ultimately results in lack of robust bacterial growth. As microfluidics and laser printing technologies have continued to develop, it has become possible to address these sorts of questions by confining small numbers of bacteria in diffusive picoliter-scale hydrogel traps. One such study in which V. harveyi was trapped showed that aggregates with diameters of about 25 microns demonstrated robust QS while those ≤10 microns showed little QS-dependent gene expression84. Similar results were observed for P. aeruginosa, which had been confined in micro-3D printed bacterial ‘lobster traps’. As few as 500 P. aeruginosa cells were shown to produce the QS-controlled exoproduct pyocyanin when confined within 8 picoliter traps, indicating that aggregates of this size are capable of initiating social behaviors in an open system85.
A next step in understanding bacterial communication revolves around whether aggregates can communicate with each other: What is the ‘calling distance’ of QS signals, how far can two aggregates be from each other and still interact? Several theoretical studies have focused on this question, and the general consensus is that communication at distances greater than 10–100 microns is unlikely86–88. One empirical study provided a cursory set of experiments aimed at assessing communication between spatially organized aggregates85. This study employed state-of-the-art laser-printing technology to trap P. aeruginosa in aggregates separated by 8 microns. The ability of one aggregate to communicate with another via AHL QS was assessed for aggregates of different sizes. Signaling across this 8-micron distance required signal-producing aggregates to contain at least 2,000 cells. Although P. aeruginosa aggregates greater than 2,000 cells have been observed in natural ecosystems including chronic infections76,89, most aggregates in these communities are generally smaller and are often spaced further than 8 microns apart (Fig. 3B). These data suggest that in some environments, P. aeruginosa QS may primarily function as an intra-aggregate communication system. The squid-V. fischeri symbiosis provides a unique opportunity to address questions of inter-aggregate signaling in a natural host-associate habitat (see Box 2). We believe that there is much yet to learn about how micron-scale biogeography influences how bacteria interact. This will be critical as we begin to manipulate microbial communities such as the human gut microbiome by influencing QS either chemically or using probiotic approaches.
Box 2. Biogeography of V. fischeri in a squid light organ.
As described in the text the squid-V. fischerilight organ mutualism has proven to be a useful two-partner model for fundamental studies of quorum sensing in a real-world host-microbe setting. It is also becoming an important model for biogeography studies concerning the ability of spatially segregated aggregates to communicate with one another. The light organ consists of two lobes, and each lobe possesses three crypts. Descending into a lobe the crypts become progressively smaller. If the experimenter presents a new born squid with a mixture of V. fischerimutants or strains in the surrounding seawater different crypts can be colonized with different mutants or strains. We know this because V. fischeri can be tagged with genes coding for different fluorescent proteins and whole light organs can be imaged by fluorescence microscopy (see image below). AHL QS signals can diffuse through the crypt barriers. Can an AHL signal produced by a wildtype V. fischeriin one crypt activate the luminescence genes in an adjacent crypt? Can wildtype in a small crypt activate bacteria in a larger crypt or perhaps maybe signaling is undirectional depending on crypt size? Can a signal-producing luminescence mutant activate luminescence by a signal-negative mutant in an adjacent crypt, and will the luminescent strain compensate for the luminescence mutant by producing more light itself? The answers to these questions will be fascinating, and coupled with laboratory experiments on physically separated aggregates of bacteria will provide critical information as we begin to understand the social interactions in more complex ecosystems like the human gut.
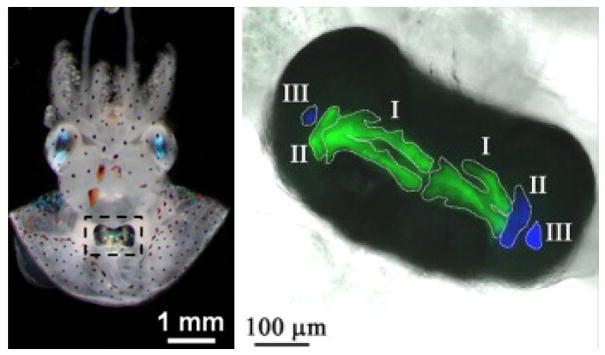
Box 2 Images.
Left, Juvenile squid with infected light organ (boxed). The mantle has beed dissected to expose the light organ. Right, Fluorescence microscopy of light organ crypts colonized with a mix of YFP-labeled V. fischeri FQ-A001 and CFP labeled strain ES114. The three crypts in each lobe are outlined and labeled I, II and II. Images provided by T. Miyashiro Penn State University. Details of this type of experiment have been published previously112.
Quorum quenching in natural ecosystems and as a therapeutic modality
As it is clear that many natural microbial communities are polymicrobial and spatially structured, it is important to consider how ecological interactions between species shape the evolution of signaling. Important considerations include abiotic and biotic factors that interfere with QS through degradation of signals (Fig. 3C), a process termed quorum quenching (QQ)90. Signal degradation can result from the chemical characteristics of an environment such as pH, or due to the action of enzymes produced by microbes or animals. In the latter case, two main types of AHL-degrading enzymes, lactonases and acylases, have been described. Lactonases hydrolyse the HSL ring of an AHL to produce corresponding acyl homoserines90, whereas acylases cleave the AHL amide bond generating the corresponding fatty acid and homoserine lactone91. Determining the impact of QQ on natural microbial communities, given the complexities of spatial structure and signal calling distance, remains a key challenge for the future. For example, are QQ enzymes produced for competition, cooperation or for the private benefit of producing cells? It is notable that humans also possess lactonases (the paraoxonase (PON) family of enzymes), which are better known for their ability to hydrolyze organophosphate toxins and low density lipoproteins92–95. PON lactonase activity is considered to be the ancestral activity92,94, and Drosophila PONs appear to serve a host-defense function96. It is possible that such enzymes provide the host a means of manipulating the microbiome through modulation of microbial social interactions.
Regardless of the true functions of QQ enzymes, QQ presents an attractive and progressive route for treating the rising number of antimicrobial resistant infections. A recent report commissioned by the Wellcome Trust estimated that by 2050, antimicrobial resistance (AMR) could cause 10 million additional deaths annually, and a cumulative loss to the worlds GDP of $100 trillion97. QQ enzymes and other QS-blocking approaches do not kill pathogens but instead block virulence factor production, and such anti-virulence agents have been proposed to impart less selective pressures that lead to the development of resistant mutants98 (see Box 1). The basis for QS inhibition as a therapeutic approach dates back to the 1990s, when a brominated furanone produced by the Australian macro-alga Delisea pulchra was shown to antagonize AHL-controlled phenotypes in a number of bacterial species99. Importantly, halogenated furanones have been shown to be effective in vivo, resulting in clearance of P. aeruginosa from the lungs of infected mice100. Subsequent studies identified a number of small molecule inhibitors of AHL QS, many of which have potent activity against several bacterial pathogens laboratory culture99–104.
What diseases might be treated by QS inhibition? Obviously, we would like to be able to treat infections that are not resolved by current therapies. The chronic P. aeruginosa lung infections, which plague people with the genetic disease cystic fibrosis (CF), have been an inviting target, and targeting chronic P. aeruginosa infections presents a clear example of why fundamental research and therapeutic development are co-dependent and must proceed hand-in-hand. These chronic infections are not resolved by antibiotics, and the chronic microbial communities residing within the CF lung are comprised of bacterial aggregates89. Because QS activates a battery of P. aeruginosa virulence factors, and P. aeruginosa QS mutants have reduced virulence in animal models, LasR, the master QS signal receptor became a focus of small-molecule inhibitor screens103,105,106. Subsequent ecological studies showed that many patients harbor P. aeruginosa LasR mutants107,108. These ecological studies were of course discouraging, but further investigation showed most LasR mutant CF isolates have co-opted a second QS system, the RhlR system, to replace LasR in ways, which remain unclear109. Perhaps a RhlR inhibitor or an inhibitor that targets both RhlR and LasR might be an appropriate CF therapeutic, and one has recently been reported94. The lesson is that we need continued basic research about QS in natural human habitats to know how, when or what to target with a QS inhibitor.
In conclusion, there has been an explosion of activity on bacterial QS and the field continues to expand rapidly. One area ripe for advancement involves complex adaptive microbial communities. QS likely controls behaviors critical for development and success of these communities in diverse environments like the human gut microbiome and chronic infections of humans. Elucidating the roles and functions of QS in natural ecosystems requires a continued willingness to embrace the complexity of these microbial communities and incorporate systems level ecological principles. We will continue to see incorporation of the fundamental biology of bacterial social dynamics into thinking about how to interfere with certain infectious diseases, and there will be a continued use of bacteria to understand the biology of communication and sociality. We have come to understand that there are many different small molecule-dependent interactions between microbes and between microbes and their hosts. There are certainly more to be discovered and there is an opportunity to sort out fundamental differences between these diverse systems. Understanding these issues will be critical as we move towards translating basic studies of QS to meet future needs including functional studies of the human microbiome.
Acknowledgments
We acknowledge support for of our research programs from US Public Health Service (USPHS) Grants GM59026 and P30DK089507 (to EPG), and R01GM116547 and NIH R01DE023193 (to MW).
Footnotes
The ethological definition of altruism is a behavior by an individual that increases fitness of another individual while decreasing fitness of the actor1.
References
- 1.Bell G. Selection: The mechanism of evolution. Oxford University Press; 2008. pp. 367–368. [Google Scholar]
- 2.Wilson E. Sociobiology: The new synthesis. Harvard University Press; 1975. [Google Scholar]
- 3.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. This review paper coined the term ‘quorum sensing’ to describe AHL-mediated bacterial communication. This term has come into common usage to cover other mechanisms involved in population density sensing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 6.Tomasz A. Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature. 1965;208:155–159. doi: 10.1038/208155a0. This paper was the first to provide evidence that an extracellular ‘hormone’ promoted bacterial group behavior. [DOI] [PubMed] [Google Scholar]
- 7.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. This paper showed that the sudden increase of bioluminescence by mid-logarithmic Vibrio fischeri and Vibrio harveyi cultures required transcription and a self-produced factor, and this phenomenon was referred to as autoinduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg E, Hastings J, Ulitzer S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 9.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 10.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberhard A, et al. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. This paper determined the first acyl-homoserine lactone chemical structure. [DOI] [PubMed] [Google Scholar]
- 12.Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci U S A. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. This paper determined the structure of AI-2. [DOI] [PubMed] [Google Scholar]
- 15.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 16.Hornby JM, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol. 2001;67:2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kugler S, Schurtz Sebghati T, Groppe Eissenberg L, Goldman WE. Phenotypic variation and intracellular parasitism by Histoplasma capsulatum. Proc Natl Acad Sci U S A. 2000;97:8794–8798. doi: 10.1073/pnas.97.16.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erez Z, et al. Communication between viruses guides lysis-lysogeny decisions. Nature. 2017;541:488–493. doi: 10.1038/nature21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groenhagen U, et al. Production of bioactive volatiles by different Burkholderia ambifaria strains. J Chem Ecol. 2013;39:892–906. doi: 10.1007/s10886-013-0315-y. [DOI] [PubMed] [Google Scholar]
- 20.Pearson JP, Feldman M, Iglewski BH, Prince A. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun. 2000;68:4331–4334. doi: 10.1128/iai.68.7.4331-4334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirhonen M, Flego D, Heikinheimo R, Palva ET. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 23.West SA, Winzer K, Gardner A, Diggle SP. Quorum sensing and the confusion about diffusion. Trends Microbiol. 2012;20:586–594. doi: 10.1016/j.tim.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 25.Darch SE, West SA, Winzer K, Diggle SP. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci U S A. 2012;109:8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. These papers showed that while QS can provide a benefit at the group level, cheaters can avoid the cost of QS and can therefore spread in a population. [DOI] [PubMed] [Google Scholar]
- 27.Pollitt EJ, West SA, Crusz SA, Burton-Chellew MN, Diggle SP. Cooperation, quorum sensing, and evolution of virulence in Staphylococcus aureus. Infect Immun. 2014;82:1045–1051. doi: 10.1128/IAI.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popat R, et al. Quorum-sensing and cheating in bacterial biofilms. Proc Biol Sci. 2012;279:4765–4771. doi: 10.1098/rspb.2012.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. These papers showed that while QS can provide a benefit at the group level, cheaters can avoid the cost of QS and can therefore spread in a population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rumbaugh KP, et al. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19:341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 31.Brown SP, West SA, Diggle SP, Griffin AS. Social evolution in micro-organisms and a Trojan horse approach to medical intervention strategies. Philos Trans R Soc Lond B Biol Sci. 2009;364:3157–3168. doi: 10.1098/rstb.2009.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton WD. The genetical evolution of social behaviour. II. J Theor Biol. 1964;7:17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 34.Rumbaugh KP, et al. Kin selection, quorum sensing and virulence in pathogenic bacteria. Proc Biol Sci. 2012;279:3584–3588. doi: 10.1098/rspb.2012.0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbot P, et al. Inclusive fitness theory and eusociality. Nature. 2011;471:E1–4. doi: 10.1038/nature09831. author reply E9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen B, Nowak MA, Wilson EO. Limitations of inclusive fitness. Proc Natl Acad Sci U S A. 2013;110:20135–20139. doi: 10.1073/pnas.1317588110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowak MA, Tarnita CE, Wilson EO. The evolution of eusociality. Nature. 2010;466:1057–1062. doi: 10.1038/nature09205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 39.Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol. 2011;79:166–179. doi: 10.1111/j.1365-2958.2010.07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dandekar AA, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338:264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- 42.Majerczyk C, Schneider E, Greenberg EP. Quorum sensing control of Type VI secretion factors restricts the proliferation of quorum-sensing mutants. Elife. 2016;5 doi: 10.7554/eLife.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci U S A. 2015;112:2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diggle SP, Gardner A, West SA, Griffin AS. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Philos Trans R Soc Lond B Biol Sci. 2007;362:1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 46.Maynard Smith J, Harper D. Animal signals. Oxford Univ. Press; 2003. [Google Scholar]
- 47.Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winans SC. Bacterial esperanto. Nat Struct Biol. 2002;9:83–84. doi: 10.1038/nsb0202-83. [DOI] [PubMed] [Google Scholar]
- 49.Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003;50:1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- 50.Taga ME, Semmelhack JL, Bassler BL. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol Microbiol. 2001;42:777–793. doi: 10.1046/j.1365-2958.2001.02669.x. [DOI] [PubMed] [Google Scholar]
- 51.Rezzonico F, Duffy B. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 2008;8:154. doi: 10.1186/1471-2180-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can’t talk now - gone to lunch! Curr Opin Microbiol. 2002;5:216–222. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- 53.McFall-Ngai M. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 2014;12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McFall-Ngai MJ. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmer BM, van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramoni S, Venturi V. LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology. 2009;155:1377–1385. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- 57.Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol. 2001;183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith JN, Ahmer BM. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J Bacteriol. 2003;185:1357–1366. doi: 10.1128/JB.185.4.1357-1366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hughes DT, et al. Chemical sensing in mammalian host-bacterial commensal associations. Proc Natl Acad Sci U S A. 2010;107:9831–9836. doi: 10.1073/pnas.1002551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen Y, et al. Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. MBio. 2015;6 doi: 10.1128/mBio.02429-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Houdt R, Aertsen A, Moons P, Vanoirbeek K, Michiels CW. N-acyl-L-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol Lett. 2006;256:83–89. doi: 10.1111/j.1574-6968.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- 62.Chugani SA, et al. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 64.Chugani S, Greenberg EP. An evolving perspective on the Pseudomonas aeruginosa orphan quorum sensing regulator QscR. Front Cell Infect Microbiol. 2014;4:152. doi: 10.3389/fcimb.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brachmann AO, et al. Pyrones as bacterial signaling molecules. Nat Chem Biol. 2013;9:573–578. doi: 10.1038/nchembio.1295. [DOI] [PubMed] [Google Scholar]
- 66.Brameyer S, Kresovic D, Bode HB, Heermann R. Dialkylresorcinols as bacterial signaling molecules. Proc Natl Acad Sci U S A. 2015;112:572–577. doi: 10.1073/pnas.1417685112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramoni S, et al. Bacterial subfamily of LuxR regulators that respond to plant compounds. Appl Environ Microbiol. 2011;77:4579–4588. doi: 10.1128/AEM.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez JF, Venturi V. A novel widespread interkingdom signaling circuit. Trends Plant Sci. 2013;18:167–174. doi: 10.1016/j.tplants.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 69.Schaefer AL, et al. A LuxR homolog in a cottonwood tree endophyte that activates gene expression in response to a plant signal or specific peptides. MBio. 2016;7 doi: 10.1128/mBio.01101-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 71.Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Costerton JW, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 73.Crusz SA, et al. Bursting the bubble on bacterial biofilms: a flow cell methodology. Biofouling. 2012;28:835–842. doi: 10.1080/08927014.2012.716044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 75.Shrout JD, et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 76.Bjarnsholt T, et al. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 77.Roberts AE, Kragh KN, Bjarnsholt T, Diggle SP. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol. 2015;427:3646–3661. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 78.Stacy A, et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chandler JR, et al. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J Bacteriol. 2009;191:5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laganenka L, Colin R, Sourjik V. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat Commun. 2016;7:12984. doi: 10.1038/ncomms12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boedicker JQ, Vincent ME, Ismagilov RF. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew Chem Int Ed Engl. 2009;48:5908–5911. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carnes EC, et al. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat Chem Biol. 2010;6:41–45. doi: 10.1038/nchembio.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao M, et al. A crucial role for spatial distribution in bacterial quorum sensing. Sci Rep. 2016;6:34695. doi: 10.1038/srep34695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Connell JL, Kim J, Shear JB, Bard AJ, Whiteley M. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc Natl Acad Sci U S A. 2014;111:18255–18260. doi: 10.1073/pnas.1421211111. This paper provided the first assessment of the calling distance of AHL signals produced by P. aeruginosa aggregates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boyer M, Wisniewski-Dye F. Cell-cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol Ecol. 2009;70:1–19. doi: 10.1111/j.1574-6941.2009.00745.x. [DOI] [PubMed] [Google Scholar]
- 87.Decho AW, Frey RL, Ferry JL. Chemical challenges to bacterial AHL signaling in the environment. Chem Rev. 2011;111:86–99. doi: 10.1021/cr100311q. [DOI] [PubMed] [Google Scholar]
- 88.Prosser JI. Ecosystem processes and interactions in a morass of diversity. FEMS Microbiol Ecol. 2012;81:507–519. doi: 10.1111/j.1574-6941.2012.01435.x. [DOI] [PubMed] [Google Scholar]
- 89.Kragh KN, et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun. 2014;82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dong YH, et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411:813–817. doi: 10.1038/35081101. This paper provided evidence that engineering a plant host to enzymatically degrade QS signals can reduce disease caused by a bacterial pathogen. [DOI] [PubMed] [Google Scholar]
- 91.Lin YH, et al. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol. 2003;47:849–860. doi: 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- 92.Elias M, Tawfik DS. Divergence and convergence in enzyme evolution: parallel evolution of paraoxonases from quorum-quenching lactonases. J Biol Chem. 2012;287:11–20. doi: 10.1074/jbc.R111.257329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li XC, Wang C, Mulchandani A, Ge X. Engineering soluble human paraoxonase 2 for quorum quenching. ACS Chem Biol. 2016;11:3122–3131. doi: 10.1021/acschembio.6b00527. [DOI] [PubMed] [Google Scholar]
- 94.Ozer EA, et al. Human and murine paraoxonase 1 are host modulators of Pseudomonas aeruginosa quorum-sensing. FEMS Microbiol Lett. 2005;253:29–37. doi: 10.1016/j.femsle.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 95.Yang F, et al. Quorum quenching enzyme activity is widely conserved in the sera of mammalian species. FEBS Lett. 2005;579:3713–3717. doi: 10.1016/j.febslet.2005.05.060. [DOI] [PubMed] [Google Scholar]
- 96.Stoltz DA, et al. Drosophila are protected from Pseudomonas aeruginosa lethality by transgenic expression of paraoxonase-1. J Clin Invest. 2008;118:3123–3131. doi: 10.1172/JCI35147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O’Neill J. The review on antimicrobial resistance: Tackling drug-resistant infections globally. 2016 [Google Scholar]
- 98.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 99.Givskov M, et al. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. This paper discovered a class of compounds produced by the Australian macroalga Delisea pulchra that inhibit QS-controlled processes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hentzer M, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Christensen QH, Grove TL, Booker SJ, Greenberg EP. A high-throughput screen for quorum-sensing inhibitors that target acyl-homoserine lactone synthases. Proc Natl Acad Sci U S A. 2013;110:13815–13820. doi: 10.1073/pnas.1313098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Muh U, et al. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob Agents Chemother. 2006;50:3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O’Loughlin CT, et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Starkey M, et al. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 2014;10:e1004321. doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gerdt JP, McInnis CE, Schell TL, Rossi FM, Blackwell HE. Mutational analysis of the quorum-sensing receptor LasR reveals interactions that govern activation and inhibition by nonlactone ligands. Chem Biol. 2014;21:1361–1369. doi: 10.1016/j.chembiol.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Borlee BR, Geske GD, Blackwell HE, Handelsman J. Identification of synthetic inducers and inhibitors of the quorum-sensing regulator LasR in Pseudomonas aeruginosa by high-throughput screening. Appl Environ Microbiol. 2010;76:8255–8258. doi: 10.1128/AEM.00499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bjarnsholt T, et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One. 2010;5:e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hoffman LR, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros. 2009;8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feltner JB, et al. LasR Variant Cystic Fibrosis Isolates Reveal an Adaptable Quorum-Sensing Hierarchy in Pseudomonas aeruginosa. MBio. 2016;7 doi: 10.1128/mBio.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Greenberg EP. Bacterial communication and group behavior. J Clin Invest. 2003;112:1288–1290. doi: 10.1172/JCI20099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bouayed N, Dietrich N, Lafforgue C, Lee CH, Guigui C. Process-oriented review of bacterial quorum quenching for membrane biofouling mitigation in membrane bioreactors (MBRs) Membranes (Basel) 2016;6 doi: 10.3390/membranes6040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Verma SC, Miyashiro T. Niche-Specific Impact of a Symbiotic Function on the Persistence of Microbial Symbionts within a Natural Host. Appl Environ Microbiol. 2016;82:5990–5996. doi: 10.1128/AEM.01770-16. [DOI] [PMC free article] [PubMed] [Google Scholar]