Abstract
Organoarsenicals such as the methylarsenical methylarsenate (MAs(V)) and aromatic arsenicals including roxarsone (4-hydroxy-3-nitro-benzenearsenate or Rox(V)) have been extensively used as an herbicide and growth enhancers in animal husbandry, respectively. They undergo environmental degradation to more toxic inorganic arsenite (As(III)) that contaminates crops and drinking water. We previously identified a bacterial gene (arsI) responsible for aerobic demethylation of methylarsenite (MAs(III)). The gene product, ArsI, is an Fe(II)-dependent extradiol dioxygenase that cleaves the carbon–arsenic (C–As) bond in MAs(III) and in trivalent aromatic arsenicals. The objective of this study was to elucidate the ArsI mechanism. Using isothermal titration calorimetry, we determined the dissociation constants and ligand-to-protein stoichiometry of ArsI for Fe(II), MAs(III), and aromatic phenylarsenite. Using a combination of methods including chemical modification, site-directed mutagenesis, and fluorescent spectroscopy, we demonstrated that amino acid residues predicted to participate in Fe(II)-binding (His5–His62–Glu115) and substrate binding (Cys96–Cys97) are involved in catalysis. Finally, the products of Rox(III) degradation were identified as As(III) and 2-nitrohydroquinone, demonstrating that ArsI is a dioxygenase that incorporates one oxygen atom from dioxygen into the carbon and the other to the arsenic to catalyze cleavage of the C–As bond. These results augment our understanding of the mechanism of this novel C–As lyase.
Graphical Abstract
INTRODUCTION
Arsenic is a ubiquitous environmental toxin, deriving from natural geothermal and anthropogenic sources such as mining, smelting, and agriculture. The Environmental Protection Agency (EPA) and Agency for Toxic Substances and Disease Registry (ATSDR) consider arsenic to be the most prevalent environmental toxin and the most significant potential threat to human health and ranked it at the top of the U.S. Priority List of Hazardous Substances (https://www.atsdr.cdc.gov/SPL/index.html). Arsenic is classified as a Group 1 human carcinogen. In addition, exposure to arsenic causes health problems including cardiovascular disease,1,2 diabetes,3 hearing loss,4 peripheral neuropathy,5,6 and infant and childhood developmental problems.7
Arsenic toxicity depends on the species. Trivalent arsenicals are considerably more toxic than pentavalent arsenicals, and trivalent organoarsenicals are the most toxic.8–10 On the basis of their effects in cultured cells, the order of toxicity is methylarsenite (MAs(III)) > dimethylarsenite (DMAs(III)) > arsenite (As(III)) > arsenate (As(V)) > methylarsenate (MAs(V)), dimethylarsenate (DMAs(V)) > trimethylarsine oxide (TMAsO).9,10 In the 1940s Challenger proposed that the biological process of arsenic methylation involves a series of oxidative methylations, and the products are all pentavalent.11 Subsequently Hirano proposed a pathway in which the substrates and products of human As(III) S-adenosylmethyltransferase (AS3MT), the enzyme that methylates arsenic, are all trivalent.12 More recently we demonstrated that product of the first methylation step of both microbial and human As(III) S-adenosylmethyltransferases is MAs(III), and that MAs(V) forms by nonenzymatic oxidation in air, leading to a proposed catalytic pathway.13,14 We hypothesize that for billions of years bacteria have been methylating inorganic As(III) to MAs(III) and DMAs(III) and used these as primitive antibiotics before the Great Oxidation Event (GOE).15 Once oxygen formed in the atmosphere, these were oxidized to relatively nontoxic MAs(V) and DMAs(V). Methylarsenicals are also introduced anthropogenically, such as MAs(V) that is currently used as an herbicide on cotton fields or for postemergence control of crabgrass and other turf weeds in the United States (https://www.epa.gov/ingredients-used-pesticide-products/monosodium-methanearsonate-msma-organic-arsenical). Moreover, synthetic aromatic arsenicals such as roxarsone (4-hydroxy-3-nitrobenzenearsenate), Rox(V)), p-arsanilic acid (4-aminobenzenearsenate, p-ASA(V)), and nitarsone (4-nitrobenzenearsenate, Nit(V)) have been widely used in animal husbandry as antimicrobial growth enhancers.16 Even though the use of roxarsone was limited by the European Union in 1999,17 and the U.S. Food and Drug Administration withdrew approval for the use of Rox(V) and p-ASA(V) in 2011 and for Nit(V) in 2015 (https://www.fda.gov/animalveterinary/newsevents/cvmupdates/ucm440668.htm), they are still produced and widely used in China18–20 and other countries.16,21,22 Although a portion of these aromatic arsenicals is excreted unchanged from animals, metabolism of Rox(V) to 3-amino-4-hydroxybenzenearsenate and N-acetyl-4-hydroxy-m-arsanilic acid has been observed.23,24 When the resulting manure is used as fertilizer, these organoarsenicals and their metabolites are subsequently degraded into more mobile inorganic arsenicals that are taken up by plants and animals or enter the water table, contaminating our food and drinking water supplies.17,25
A major step in the degradation of organoarsenicals to inorganic arsenic is cleavage of the carbon–arsenic bond. We previously identified a two-step pathway for breakdown of MAs(V) by microbial communities, where some bacteria reduce MAs(V) to MAs(III) and others demethylate MAs(III) to inorganic As(III).26 We cloned the arsI gene responsible for MAs(III) demethylation from the environmental isolate Bacillus sp. MD1. ArsI is a nonheme iron-dependent type-I extradiol dioxygenase with C–As bond lyase activity. In addition, ArsI cleaves the C–As bond in a wide range of trivalent aromatic arsenicals, including Rox(III), Nit(III), and p-ASA(III).27 As of April 2017, 1305 putative ArsI orthologs were identified in 963 aerobic bacterial species by a BLink (Basic Local Alignment Search Tool Link to Protein Alignments and Structures) search, suggesting that arsI genes are widely distributed in the oxic environment. Degradation of environmental organoarsenicals has been also observed under anaerobic condition.21 For example, degradation of Rox(V) to 3-amino-4-hydroxybenzenearsenate and inorganic arsenate was demonstrated by pure cultures of the Gram-positive anaerobe Alkaliphilus oremlandii sp. nov. strain OhlLAs.28 On the basis of protein profiles, a novel pathway for anaerobic degradation of Rox(V) was proposed.29,30
To the best of our knowledge, ArsI is the only identified enzyme involved in organoarsenic degradation. We recently solved the crystal structure of a thermophilic ArsI ortholog from Thermomonospora curvata (TcArsI) in both the apo form and with metals.31 Although the overall structure is similar to that of other type-I extradiol ring-cleaving dioxygenases, ArsI possesses a unique flexible loop containing a cysteine pair that we propose binds substrate in bulk solvent and conveys it to the active site. In this study, we characterized the binding properties of metal and substrate and examined the requirement for residues involved in binding and activity. Finally, the results of electron spray ionization mass spectrometry (ESI-MS) analysis demonstrate that the products of Rox(III) degradation are As(III) and 2-nitrohydroquinone. These results support our propose that ArsI incorporates one atom from dioxygen onto the aromatic ring and the other onto the arsenic atom.27 Our results furnish molecular details of a pathway in aerobic degradation of organosarsenical herbicides and antimicrobial growth promoters and provide insight into how those processes contribute to the arsenic biogeochemical cycle.
MATERIALS AND METHODS
Reagents
Roxarsone and phenylarsenate (PhAs(V)) were purchased from Acros organics (Geel, Belgium) and p-ASA(V) was purchased from Alfa Aesar (Ward Hill, MA). Trivalent MAs(III), Rox(III), and p-ASA(III) were prepared by chemical reduction, as described previously27 and used for all the experiments except isothermal titration calorimetry (ITC). We confirmed by absorption spectroscopy that the chemical reduction method reduces As(V) to As(III) but does not reduce the nitro group of roxarsone. For ITC assays, the methylarsonous acid iodide derivative (MAs(III)I2) was synthesized as described.32 2-Nitrohydroquinone was purchased from Frinton Laboratories, Inc. (Hainesport, NJ). (NH4)2Fe(SO4)2·6H2O was used for preparation of Fe(II) solutions. Unless otherwise mentioned, all other chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Strains, Plasmid, and Preparation of Mutants
Escherichia coli strain TOP10 was used for molecular cloning (Supporting Information Table S1). E. coli strain BL21 (DE3) was used for protein expression and purification. A derivative of arsI from Bacillus sp. MD1 lacking the C-terminus from Glu125 to the end was used in this study and is simply termed arsI because those residues are not essential for enzyme activity.27 The arsI gene was amplified from Bacillus sp. MD1 genomic DNA27 with an annealing temperature of 65 °C using Pfu Turbo DNA polymerase (Agilent Technologies, Santa Clara, CA), with forward primer 5′-GGGGCATATGAAATATGCGCATGTGGGTCTT-3′ (NdeI site underlined) and reverse primer 5′-AAAAGGAATTCAA-CAGTTGTCTTTGTGTAG-3′ (EcoRI site underlined), double-digested by NdeI and EcoRI (New England BioLabs, Ipswich, MA) and cloned into vector plasmid pET28a (Novagen, Merck KGaA, Darmstadt, Germany), generating pET28a-arsI encoding ArsI with an N-terminal 6-histidine tag (Table S1). Mutations in this gene were generated by a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) with primers listed in Table S2. The codons for the Fe(II)-binding residues (His5, His62, and Glu115) were individually mutated to alanine codons, generating derivatives H5A, H62A, and E115A. The codons for the substrate-binding residues (Cys96 and Cys97) were sequentially mutated to serine codons, generating a C96/C97S double mutant derivative. All genes were confirmed by DNA sequencing.
Purification of ArsI
Cells of E. coli BL21(DE3) (Novagen) bearing wild type or mutant arsI genes were grown in lysogeny broth (LB) medium supplemented with 50 μg/mL kanamycin at 37 °C and induced at an A600 of 0.6 with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After incubation for an additional 4 h at 37 °C, the cells harvested by centrifugation at 2742g for 10 min were suspended in 5 mL per gram of wet cells in buffer A (50 mM morpholinopropane-1-sulfonic acid (MOPS), 0.5 M NaCl, 20% glycerol (v/v), and 1 mM tris(2-carboxyethyl)-phosphine (TCEP), pH 7.0) containing 20 mM imidazole. Cells were broken by a single passing through a French pressure cell at 20 000 psi, then treated immediately with diisopropyl-fluorophosphate (2.5 μL per gram wet cell) and by centrifugation at 125 000g for 1 h. The supernatant solution was loaded at a flow rate of 0.5 mL min–1 onto a 5-mL HisTrap HP column (GE Healthcare Bio Sciences, Pittsburgh, PA) pre-equilibrated with Buffer A containing 20 mM imidazole using an ÄKTA FPLC purification system (GE Healthcare Bio Sciences, Piscataway, NJ). The column was washed with more than 20 column volumes of buffer A containing 20 mM imidazole, and ArsI was eluted with Buffer A containing 0.2 M imidazole. The purified proteins were treated with 2 mM ethylenediaminetetraacetate (EDTA) to remove Ni(II) coeluted from the Ni-NTA column, then concentrated and buffer exchanged into Buffer A without imidazole. Purity was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 15% acrylamide gels,33 and protein concentrations were estimated by the method of Bradford.34 Purified proteins were rapidly frozen in aliquots and stored at −80 °C until used.
Isothermal Titration Calorimetric Assay of Ligand Binding to ArsI
Binding parameters for ArsI–ligand interactions were determined by isothermal titration calorimetry (ITC). To prevent Fe(II) oxidation, all solutions were prepared anaerobically in ITC buffer (50 mM MOPS, 0.5 M NaCl, pH 7.2) containing 5 mM TCEP. Solutions were deoxygenated using a Schlenk line,35 and samples were assayed using a VP-ITC microcalorimeter (MicroCal, LCC., Northampton, MA) housed in a Coy anaerobic chamber. Ligand solutions were titrated into the adiabatic cell containing 960 μL of 70 μM ArsI. For titration with Fe(II), a solution of 1 mM (NH4)2Fe(SO4)2·6H2O was added in an initial injection of 2 μL, with 24 subsequent 7-μL injections at 10-min intervals. Data were collected at 30 °C with a stirring speed of 350 rpm. Trivalent organoarsenicals are relatively stable compared to Fe(II), so those binding assays were carried out aerobically using a MicroCal iTC200 (GE Healthcare Bio Sciences). Titrations with organoarsenical substrates were performed with synthetic MAs(III)32 or commercial PhAs(III). These were used as substitutes for chemically reduced MAs(V) or Rox(V) because the reducing reagents used to prepare trivalent organoarsenicals generated heat during ITC titrations. ArsI (70 μM, final concentration) and ligand solutions (0.5 mM MAs(III), 0.5 mM PhAs(III), and 0.5 mM As(III)) were prepared in ITC buffer with 1 mM TCEP. Samples were degassed by bubbling with argon for 30 min at 4 °C. Each ligand solution was titrated into the protein solution (300 μL) in the adiabatic cell with an initial 0.4-μL injection followed by 24 subsequent injections of 1.2 μL at 4 min intervals. Data were collected at 30 °C with stirring at a speed of 1000 rpm. Spectra were analyzed using the Origin 7.0 software (TA Instruments, New Castle, DE). Binding constants were calculated with a one-site binding model. All experiments were performed in duplicate on independent samples.
Chemical Modifications of ArsI with Residue-Specific Reagents
Modification of purified wild-type ArsI with amino acid residue-specific reagents was examined as described.36 In brief, 1 μM enzyme was incubated in a buffer consisting of 50 mM MOPS, 150 mM KCl, 3 mM TCEP, 1 mM cysteine, and 0.1 mM Fe(II) containing the cysteine-specific reagents N-ethylmaleimide (NEM) (1 mM, final concentration) or iodoacetamide (IAA) (5 mM, final concentration), the histidine-specific reagent diethylpyrocarbonate (DEPC) (5 mM, final concentration), or the glutamic/aspartic acid-specific reagent 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide (EDC) (1 mM, final concentration) at 25 °C for 30 min, following which 1 μM MAs(III), final concentration, was added. After incubation at 30 °C for 30 min with shaking at 200 rpm, MAs(III) demethylation activity was analyzed as described below.
Assays of ArsI Activity
Demethylation of MAs(III) was analyzed in vitro with purified wild type ArsI and derivatives. MAs(III)-demethylating activity was assayed in a buffer consist of 0.1 M MOPS, 0.15 M NaCl (pH 7.2) containing 1 mM cysteine, 3 mM TCEP, and 0.1 mM Fe(II). The reaction was initiated by addition of 5 μM MAs(III), incubated at 30 °C with shaking at 200 rpm for 1 h and terminated with EDTA. Protein was removed by centrifugation through a 3-kDa cutoff Amicon Ultrafilter (Millipore, Billerica, MA), and the arsenic species in the filtrates were separated by high-pressure liquid chromatography (HPLC; Series 2000, PerkinElmer, Waltham, MA) using a C18 reverse phase column (250 × 4.6 mm; 5 μm; Thermo Fisher Scientific, Waltham, MA) with mobile-phase consisting of 3 mM malonic acid and 5% (v/v) methanol (pH 5.9, adjusted by tetrabutylammonium hydroxide) at a flow rate of 1 mL/min. Arsenic was determined by inductively coupled plasma mass spectroscopy (ICP-MS) (ELAN DRC-e, PerkinElmer). To determine arsenic recovery, the reactions were terminated by addition of H2O2 at 6% (v/v, final concentration), which oxidizes MAs(III) and As(III) to MAs(V) and As(V). Arsenic species were analyzed by HPLC-ICP-MS. The amount of each arsenic species was quantified from the area under the peak based on calibration with known amounts of authentic standards.37
Fluorescence Assays
A temperature-controlled Quanta-Master UV–vis QM-4 steady state spectrofluorometer (Photon Technology International, Birmingham, NJ) was used for fluorescence measurements. The fluorescence of 5-μM wild-type ArsI or derivatives was assayed in a buffer consisting of 50 mM MOPS (pH 7.2), 0.5 M NaCl, and 1 mM TCEP at excitation and emission wavelengths of 295 and 340 nm, respectively. Ligands (25 μM Fe(II) and/or one of the following arsenicals: As(III), MAs(III), MAs(V), PhAs(III), PhAs(V), p-ASA(III), p-ASA(V), Rox(III) or Rox(V) at the indicated concentrations) were added, and quenching of fluorescence was measured. For all assays, the temperature was 23 °C, and the spectrum of the buffer solution alone was subtracted to correct for background fluorescence and Raman scattering. Fluorescence intensities were corrected for volume changes.
Enzymatic Degradation of Roxarsone
Purified ArsI was used in a 1:1 protein-to-substrate ratio. Rox(III) was freshly prepared as described previously.27 ArsI activity was assayed in a buffer consisting of 0.1 M MOPS, 0.15 M NaCl (pH 7.2) containing 1 mM cysteine, 3 mM TCEP, and 0.1 mM Fe(II). The reaction was initiated by addition of 200 μM Rox(III), and assayed at 30 °C with shaking at 200 rpm for 3 h. The reaction mixture then was filtered through a 3-kDa cutoff Amicon Ultrafilter to remove protein. Because the HPLC retention time for As(III) and Rox(III) is the same on a C18 column, the products were separated by HPLC using an Inertsil C4 column (150 mm × 2.1 mm; 5 μm; GL Sciences, Torrance, CA) and monitored by ICP-MS to confirm completion of the reaction.
Determination of the Organic Product of C–As Bond Cleavage
Liquid chromatography (LC) coupled with electron spray ionization mass spectrometry (ESI-MS) was used for the determination of the organic product of Rox(III) degradation. The enzyme-free filtrate prepared as described above (1 mL) was lyophilized and dissolved in 0.5 mL of water/isopropanol (50:50 v/v) solution. Samples (10 μL) were injected into a Thermo Surveyor Plus HPLC System (Thermo Fisher Scientific) equipped with a hydrophilic interaction liquid chromatography (HILIC) column (Merck, ZIC-pHILIC 150 × 4.6, 5 μm) protected with a guard column. A mobile phase gradient was used consisting of 10 mM ammonium formate dissolved in Optima LC-MS grade water with pH adjusted to 9 with ammonium hydroxide (aqueous) and Optima LC-MS grade acetonitrile (organic). The ions in the eluent from the HILIC column chromatography were analyzed by low-resolution (LR)-ESI-MS, with detection using an AB Sciex QTRAP 5500 Triple-Quadrupole mass spectrometer equipped with a Turbo V ESI ionization source (Danaher Corporation, Washington, DC). The instrument was operated under full scan (m/z 100–600), negative mode. Samples were also analyzed by high-resolution (HR)-ESI-MS using a Bruker Solarix 7.0T Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR) (Bruker, Billerica, MA) to analyze monoisotopic mass of indicated compounds. Samples of Rox(III), Rox(V), and commercial 2-nitrohydroquinone were analyzed by HR-ESI-MS as standards. Mass errors of the target compounds were calculated based on their theoretical molecular mass. The instrument was operated under negative mode, broadband scan (m/z 100–1400) at 1 Mword with 20 coadded scans. The dry gas temperature was 200 °C at a flow rate of 3.0 L/min, and the nebulizer gas pressure was 3.0 bar. The instrument was calibrated utilizing the ions from Agilent ESI-L tune mix observed in the LC-HR-MS analysis of the blank. This calibration was applied to the LC-HR-MS data.
Homology Model of the Bacillus ArsI with Bound Substrates
A homology model of the Bacillus ArsI was built on the crystal structure of TcArsI bound with 2-mercaptoethanol (PDB ID 5CB9) using a fully automated protein structure homology modeling server SWISS-MODEL (http://swissmodel.expasy.org/). The model quality was estimated based on the QMEAN scoring function. The sequence identity and RMSD of Bacillus ArsI and TcArsI are 43.6% and 0.45 Å, respectively. The final homology model incorporated residues from Met1 to Thr119. To place substrates in the model, in silico docking with the arsenical substrates Rox(III), p-ASA(III), and PhAs(III) was carried out using the PATCHDOCK server.38 The Fe(II)-bound model was derived from the structure of TcArsI (PDB ID 5CB9) by replacing the Ni(II) atom. PyMOL v1.3 was used to visualize the structural models.39
RESULTS
ITC Measurements of Ligand Binding to ArsI
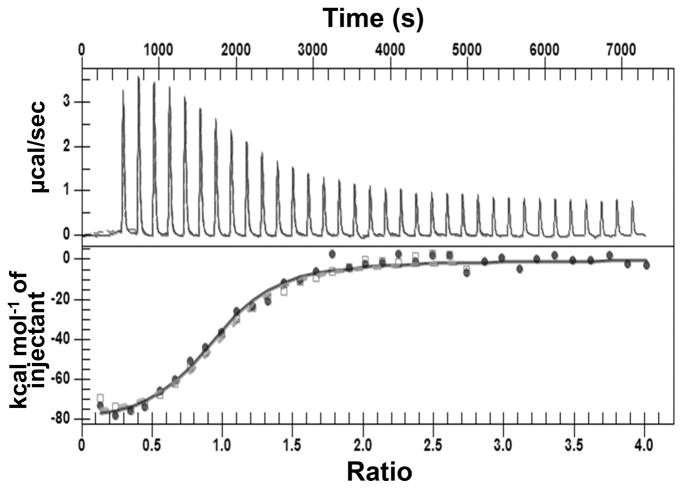
Binding of Fe(II) to ArsI was determined by ITC under anaerobic conditions to prevent air-induced Fe(II) oxidation (Figure 1 and Table 1). Fe(II) binds to ArsI with a dissociation constant (Kd) of 4.14 ± 0.08 μM at a ligand-to-protein stoichiometry (N) of 0.96 ± 0.03. This affinity is comparable with those for other nonheme Fe(II)-dependent dioxygenases.40
Figure 1.
Anaerobic binding of Fe(II) to ArsI. Anaerobic ITC data and the binding isotherm for ArsI with Fe(II) binding. Data were collected anaerobically at 30 °C with injections at 10 min intervals.
Table 1.
Thermodynamic Parameters of ArsI Determined by ITC
| ArsI-Ligand | Kd | N |
|---|---|---|
| Fe(II) | 4.14 ± 0.08 (μM) | 0.96 ± 0.03 |
| PhAs(III) | 0.62 ± 0.06 (nM) | 0.85 ± 0.05 |
| MAs(III) | 0.3 ± 0.09 (μM) | 0.65 ± 0.02 |
Binding of As(III), MAs(III), and PhAs(III), which are not as sensitive to oxidation as Fe(II), was analyzed aerobically by ITC (Table 1 and Figure S1). PhAs(III) was used as a substitute for Rox(III) because it is commercially available in the trivalent form, while Rox(III) is prepared by chemical reduction of Rox(V), and the reducing reagents generated too much heat to make titrations with Rox(III) interpretable. PhAs(III) bound with highest affinity (Kd = 0.62 ± 0.06 nM, N = 0.85 ± 0.05). MAs(III) bound with lower affinity (Kd = 0.30 ± 0.09 μM, N = 0.65 ± 0.02). There was essentially no binding of As(III) to ArsI, as would be expected since the product of the reaction must be easily released.
Identification of Residues in the Ligand Binding Sites
Our recent crystallographic study demonstrated that TcArsI, the ArsI ortholog from the thermophile T. curvata DSM 42183, shares overall structure similarity with the C-terminal catalytic domain of LapB from Pseudomonas sp. KL28, a representative of type I extradiol ring-cleaving dioxygenases.31 Analysis of crystal structures with metals suggests that TcArsI forms the active site by coordinating Fe(II) via the Gln8–His65–Glu117 triad, which corresponds to His5–His62–Glu115 in the Bacillus ArsI. This triad is conserved in ArsI orthologs and other type I extradiol ring-cleaving dioxygenases (Figure S2). In a number of gene products of ars operons, conserved cysteine residues often function as binding sites for substrates of trivalent arsenicals. In Bacillus ArsI there is a vicinal cysteine pair, Cys96–Cys97, which is conserved in other putative ArsI orthologs (Figure S2). We predicted that this vicinal pair forms the binding site for organoarsenical substrates.27
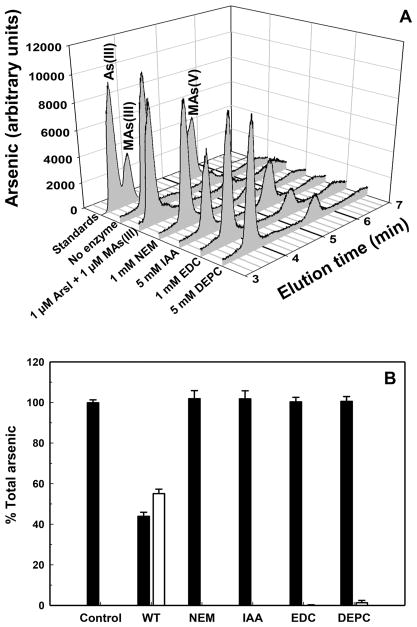
To examine the participation of specific types of residues in ArsI catalysis, the effects of reagents that chemically modify histidine residues (DEPC), aspartate or glutamate residues (EDC), or cysteine residues (NEM or IAA) on C–As lyase activity of purified wild-type ArsI were analyzed. Each reagent inhibited MAs(III)-demethylating activity (Figure 2). These results are consistent with the predicted His5–His62–Glu115 binding site for Fe(II) and the Cys96–Cys97 binding site for trivalent organoarsenicals. Note that the ArsI derivative used for this study has only two cysteine residues (Figure S2). Therefore, inhibition of the demethylation activity by cysteine modifiers is consistent with the involvement of the cysteine pair in catalysis.
Figure 2.
Effect of chemical modifiers on ArsI activity. (A) MAs(III) demethylation with purified ArsI was determined after modification with or without chemical modifiers as described in Materials and Methods. The products were determined by HPLC-ICP-MS. The reaction was initiated by addition of MAs(III). (B) ArsI cleavage of MAs(III) with or without chemical modifiers is expressed as a percentage of total arsenic. Demethylation was assayed in a mixture of 5 μM MAs(III), 1 μM purified ArsI, 3 mM TCEP, 1 mM cysteine, and 0.1 mM (NH4)2Fe-(SO4)2·6H2O in 50 mM MOPS buffer, pH 7.2, at 30 °C. Samples were withdrawn after 30 min, the reaction terminated by addition of 6% (v/v) H2O2, final concentration. Arsenic species were analyzed by HPLC-ICP-MS. The data are the means ± SE (n = 3).
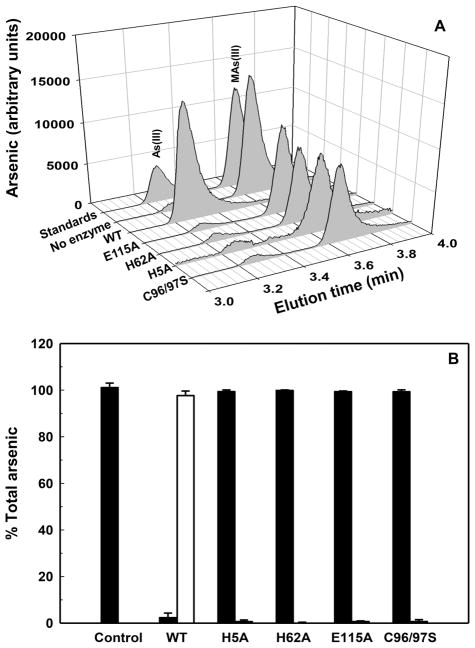
In contrast, ArsI contains multiple nonconserved histidine and acidic residues (Figure S2). The participation of specific residues in the Fe(II) and substrate binding sites was further investigated by mutagenesis. Each of the residues in the putative Fe(II)-binding site was changed to alanine, and the two cysteine residues in the predicted substrate binding site were both changed to serine residues, creating a double mutant lacking the vicinal pair. Each of the derivatives was produced in soluble form in amounts comparable to wild type ArsI. Each of the purified ArsI derivatives lost activity (Figure 3), consistent with a role of those residues in ArsI catalysis.
Figure 3.
Effect of amino acid substitutions on ArsI activity. (A) MAs(III) demethylation by purified wild type and mutants ArsI was determined. The reaction was initiated by addition of MAs(III), and activity determined by HPLC-ICP-MS, as described in Materials and Methods. (B) MAs(III) demethylation by wild type ArsI and mutants is expressed as a percentage of total arsenic. Demethylation was assayed in mixture of 5 μM MAs(III), 1 μM purified ArsI, 3 mM TCEP, 1 mM cysteine, and 0.1 mM (NH4)2Fe(SO4)2·6H2O in 50 mM MOPS buffer, pH 7.2, at 30 °C. Samples were withdrawn after 60 min, the reaction terminated by addition of 6% (v/v) H2O2, final concentration. Arsenic species were analyzed by HPLC-ICP-MS. The data are the means ± SE (n = 3).
Ligand Binding to ArsI Derivatives
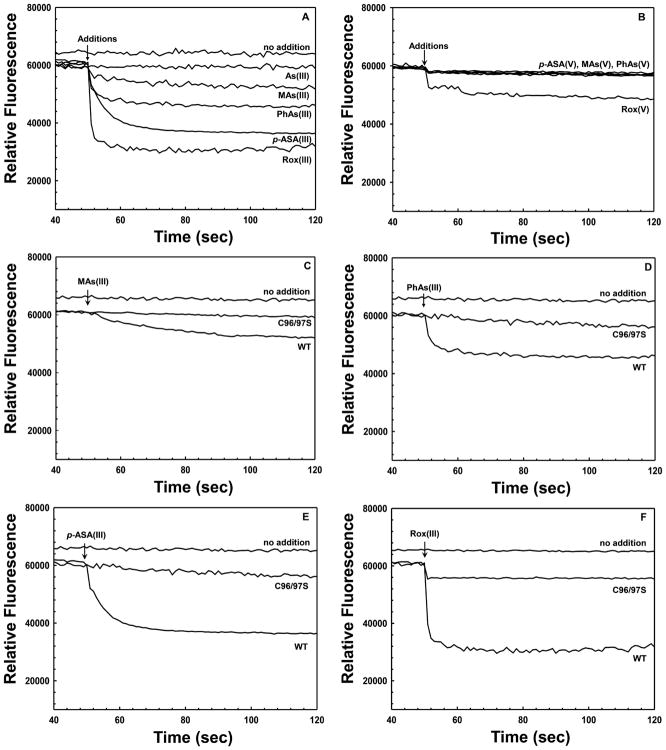
The ligand binding properties of the ArsI derivatives were examined using the quenching of intrinsic protein fluorescence. We have previously employed ligand-dependent changes in intrinsic tryptophan fluorescence to characterize binding of substrates to other proteins involved in arsenic detoxification.41,42 ArsI has two tryptophan residues, Trp105 and Trp114 (Figure S2), which are responsible for its intrinsic fluorescence. The intrinsic fluorescence was quenched by addition of Fe(II) (Figure 4) and trivalent organoarsenical substrates (Figure 5A). The product, inorganic As(III) (Figure 5A), or pentavalent organoarsenicals (Figure 5B), did not quench ArsI fluorescence. These results are consistent with the ITC results.
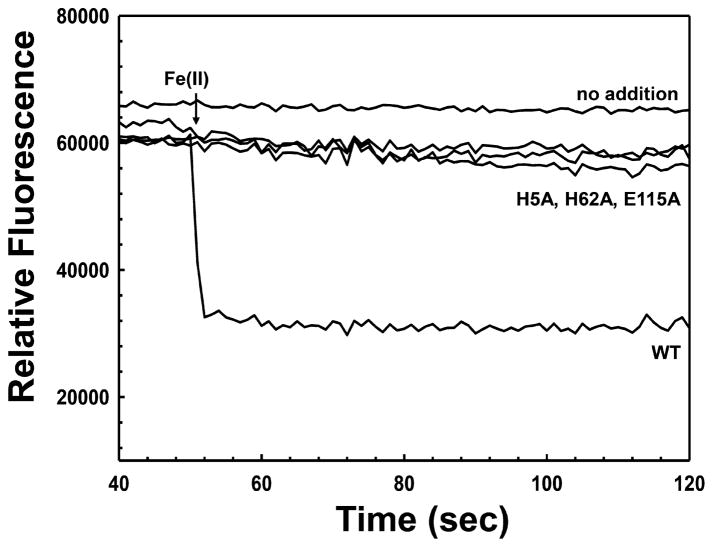
Figure 4.
Effect of Fe(II) on ArsI protein fluorescence. Fe(II)-dependent quenching intrinsic protein fluorescence of wild type ArsI and derivatives. Fluorescence was assayed at 23 °C with excitation and emission wavelengths of 295 and 340 nm, respectively, as described in Materials and Methods. (NH4)2Fe(SO4)2·6H2O prepared in degassed buffer (25 μM, final concentration) was added to 5 μM ArsI at indicated time (arrow).
Figure 5.
Effect of substrates on ArsI protein fluorescence. Fluorescence was assayed as described in the legend to Figure 4. Trivalent and pentavalent metalloids (25 μM, final concentration) were added to 5 μM ArsI at the indicated time (arrow). (A) Effect of trivalent arsenicals on ArsI protein fluorescence. (B) Effect of pentavalent arsenicals on ArsI protein fluorescence. (C) Effect of MAs(III) on protein fluorescence of wild type and C96/97S ArsI. (D) Effect of PhAs(III) on protein fluorescence of wild type and C96/97S ArsI. (E) Effect of p-ASA(III) on protein fluorescence of wild type and C96/97S ArsI. (F) Effect of Rox(III) on protein fluorescence of wild type and C96/97S ArsI.
Each derivative with single amino acid substitutions in Fe(II)-binding residues lost Fe(II)-induced fluorescence quenching (Figure 4), suggesting that the His5–His62–Glu115 triad is essential to maintain Fe(II) in the active site. In contrast, mutations in these Fe(II)-binding residues had no effect on quenching by organoarsenical substrates (Figure S3A–C). These results show that binding of substrate and metal is independent, which differs from other type I extradiol ring-cleaving dioxygenases that require Fe(II) to bind substrates.43
Trivalent aromatic arsenicals induced larger quenching than MAs(III) (Figure 5A), consistent with the higher affinity for PhAs(III) than MAs(III) found by ITC (Table 1). Addition of Rox(III) generated the greatest fluorescence quenching (Figure 5A). A C96/97S derivative lacking the cysteine residues of the putative substrate binding site no longer exhibited significant quenching with MAs(III) (Figure 5C) or any of the aromatic arsenical substrates (Figure 5D–F). Consistent with these results, the C96/97S derivative did not bind PhAs(III) when assayed by ITC. On the other hand, addition of Fe(II) to the C96/97S protein still quenched tryptophan fluorescence (Figure S3D). These results clearly demonstrate that the conserved cysteine pair is the substrate binding of ArsI, and that it is not required for binding of Fe(II).
2-Nitrohydroquinone is the Product of Rox(III) Degradation by ArsI
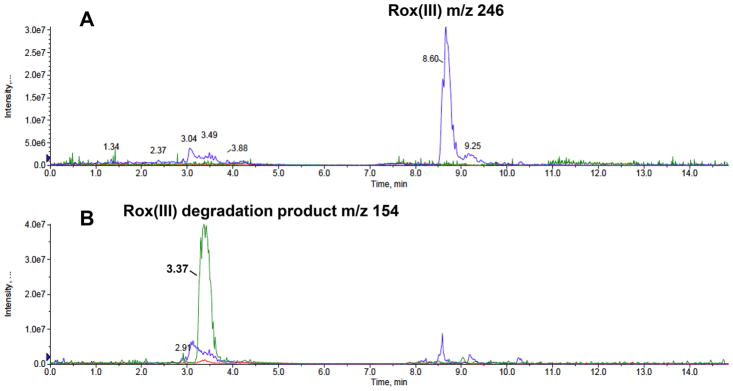
We previously showed that As(III) is the arsenical product of Rox(III) degradation by ArsI.27 However, the organic product has not been identified. We considered two possibilities. (1) Because most closely related orthologs are ring-cleaving dioxygenases,27,31 one possibility is that the aromatic ring is cleaved, leading to a straight-chain aldehyde product. (2) The other possibility is that the ring is left intact, leading to an oxidized ring product. To examine this question, the product of Rox(III) cleavage by ArsI was analyzed by ESI-MS. The product was generated by complete Rox(III) cleavage, following which the enzyme was removed. The compounds in the resulting reaction solution were separated by HILIC column chromatography, and analyzed by LR-ESI-MS with negative ion mode (Figure 6). The ion chromatograms of the m/z values for Rox(III) (m/z 246, blue line), 4-hydroxy-5-nitro-hexa-2,4-dienal (m/z 156, red line), 2-nitrohydroquinone (m/z 154, green line), and Rox(V) (m/z 262, gray line) were analyzed for their presence. As a control, in a reaction with Rox(III) in the absence of ArsI a single major peak of m/z 246 was observed (Figure 6A, blue line). In the sample after reaction of Rox(III) with ArsI two peaks, a small one of m/z 246 (blue line) corresponding to unreacted Rox(III) and a large peak of m/z 154 corresponding to the product 2-nitrohydroquinone (green line) were detected (Figure 6B).
Figure 6.
LR-ESI-MS analysis of the products of ArsI cleavage of Rox(III). The products were separated by HILIC column chromatography and analyzed by LR-ESI-MS (negative ion mode), as described under Materials and Methods. The ion chromatograms of the m/z values for Rox(III) (m/z 246, blue line), 4-hydroxy-5-nitro-hexa-2,4-dienal (m/z 156, red line), 2-nitrohydroquinone (m/z 154, green line), and Rox(V) (m/z 262, gray line) from the Rox(III) control (A) and the Rox(III) degradation sample (Rox(III) with ArsI) (B) are presented. The intensity of the signal observed at a chosen m/z value in the mass spectra was recorded as a function of retention time.
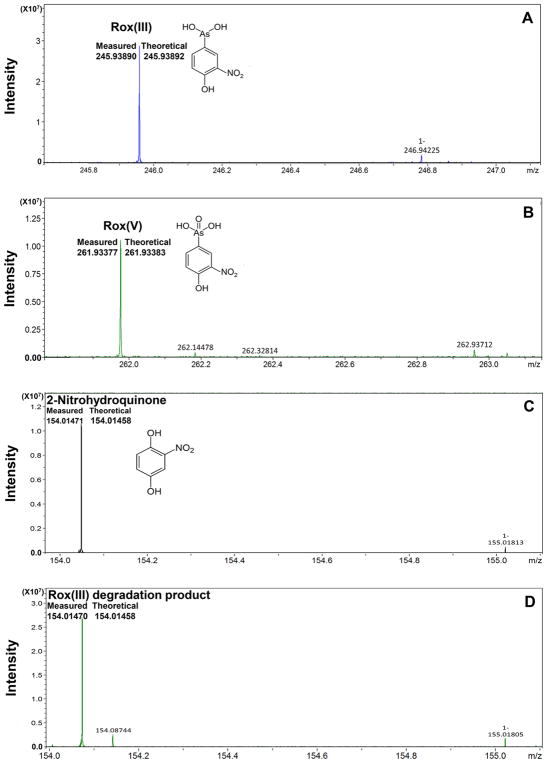
HR-ESI-MS was used to identify the monoisotopic mass of the product (Figure 7). The monoisotopic mass of the ion (in negative ion mode) at m/z 246 detected from the control sample corresponded well to the theoretical mass of Rox(III) with a mass error of ±0.08 ppm (Figure 7A). The monoisotopic mass of the ion at m/z 262 detected from the Rox(III) degradation sample corresponded to the theoretical mass of Rox(V) (Figure 7B) with a mass error of ±0.8 ppm, indicating that a portion of the Rox(III) oxidized to Rox(V) during the reaction, sample preparation, and/or analysis. Commercial 2-nitrohydroquinone used as a standard produced an m/z 154, and its monoisotopic mass matched well to the theoretical mass of 2-nitrohydroquinone with a mass error of ±0.84 ppm (Figure 7C). The sample containing the product gave a m/z 154, which agrees well to the theoretical mass of 2-nitrohydroquinone, with a mass error of ±0.76 ppm (Figure 7D). The small peaks at m/z 247 (Figure 7A), 263 (Figure 7B), and 155 (Figure 7C and 7D) correspond to the isotopes of Rox(III), Rox(V), and 2-nitrohydroquinone, respectively. In contrast, there was only a minor peak at m/z 156 (experimental m/z 156.0189), and the mass error calculated to the theoretical molecular mass weight of 4-hydroxy-5-nitro-hexa-2,4-dienal (156.0302) was ±72 ppm, demonstrating that the detected m/z 156 ion does not represent 4-hydroxy-5-nitro-hexa-2,4-dienal. Minor peaks in the Rox(III) degradation spectrum include one at m/z 228, which could correspond to the dehydrated Rox(III) ion. The 18 m/z difference between 246 and 228 is characteristic of the loss of a neutral mass of 18 by the molecular ion and is typical of a water molecule. In the same spectrum, the peak at 298 could be the chlorinated Rox(V) ion. The 35 m/z difference between 263 and 298 is typical of addition of a chlorine ion. The presence of dehydrated Rox(III) and chlorinated Rox(V) is likely ionization fragments of roxarsone formed in the HR-MS source. These results demonstrate ArsI cleaves the C–As bond of Rox(III) without ring cleavage, producing 2-nitrohydroquinone, not 4-hydroxy-5-nitro-hexa-2,4-dienal.
Figure 7.
Organic product of Rox(III) cleavage is 2-nitrohydroquinone. The products of ArsI cleavage of Rox(III) were separated by HILIC column chromatography and analyzed by HR-ESI-MS (negative ion mode), as described in Materials and Methods, to determine monoisotopic mass of the molecules of interest. (A) The experimental mass of the ion at m/z 246 detected in the Rox(III) control (245.93890) corresponded to its theoretical mass (C6H6O5NAs–, 245.93892) with a mass error of ±0.08 ppm. (B) The experimental mass of the ion at m/z 262 detected in the Rox(III) degradation sample (Rox(III) with ArsI) (261.93377) corresponded to the theoretical mass of Rox(V) (C6H6O6NAs–, 261.93383) with a mass error of ±0.76 ppm. (C) The experimental mass of commercial 2-nitrohydroquinone (154.01471) corresponded with its theoretical mass (C6H5O4N–, 154.01458) with a mass error of 0.84 ppm. (D) The experimental mass of the ion at m/z 154 detected in the Rox(III) degradation sample (Rox(III) with ArsI) (154.01470) agreed well with the theoretical mass of 2-nitrohydroquinone (154.01458) with a mass error of 0.78 ppm.
DISCUSSION
Roxarsone was first synthesized in 1923 (British patent GB 226255 19230718) and has been used commercially since 1944 (https://www.fda.gov/AnimalVeterinary/SafetyHealth/ProductSafetyInformation/ucm258313.htm#animal_drug_approvals) for prevention of coccidiosis in chickens and other farm animals.44 In 2010 nearly 90% of all broiler chickens in the U.S. were estimated to be treated with roxarsone.45 Roxarsone is excreted from chickens as a mixture of unaltered and partially metabolized species.23,24 The manure is used as fertilizer for commercial crops46 or disposed of in landfills.47 In 2011 the U.S. manufacturer, Alpharma, discontinued sales of roxarsone. Even though roxarsone is no longer used in the United States, it is still produced and used in other countries.16,21,22 When the chicken litter is used as a fertilizer or moved into landfills, roxarsone and its metabolites are broken down to inorganic arsenic, where the resulting inorganic arsenate and arsenite can be taken up by crop plants or, through agricultural runoff, enter the water supply of local communities.17,21 The entrance of breakdown products of roxarsone and other aromatic arsenicals represents a major anthropogenic source of arsenic exposure in the U.S. and elsewhere in the world.45
Environmental roxarsone breakdown in soil to inorganic arsenic occurs by photodegradation21 and by microbial metabolism.28 The fate of the aromatic substitute is not clear. In compost the nitro group of roxarsone has been shown to be reduced, producing 4-hydroxy-3-aminobenzenearsonic acid,48 presumably by microbial action. An environmental Clostridium isolate (A. oremlandii) is able to carry out reduction of the nitro group under anaerobic conditions,28 and Shewanella putrefaciens, a highly versatile environmental microbe, can carry out reduction under aerobic conditions.25 Further metabolism may include acetylation of the amine, producing N-acetyl-4-hydroxy-m-arsanilic acid.23
Subsequent roxarsone degradation requires breaking the C–As bond, which generates As(III) and a compound derived from the aromatic substituent. The only known enzyme that catalyzes C–As bond cleavage is the ArsI dioxygenase from the environmental isolate Bacillus sp. MD127 and its orthologs from the moderate thermophile T. curvata DSM 4318331 and the unicellular cyanobacterium Nostoc sp. PCC 7120.49 The objective of this study was to elucidate the properties of the enzyme and the chemical nature of the products.
From the results of binding studies employing ITC and ligand-dependent quenching of intrinsic protein fluorescence, we make several conclusions. First, from the crystal structure with bound Fe, it was not possible to distinguish between Fe(II) and Fe(III) in the metal binding site. Using anaerobic ITC under conditions that stabilize Fe(II) the affinity for Fe(II) was determined to be 4 μM. From mutagenesis, the binding site clearly includes the conserved His5–His62–Glu115 triad.
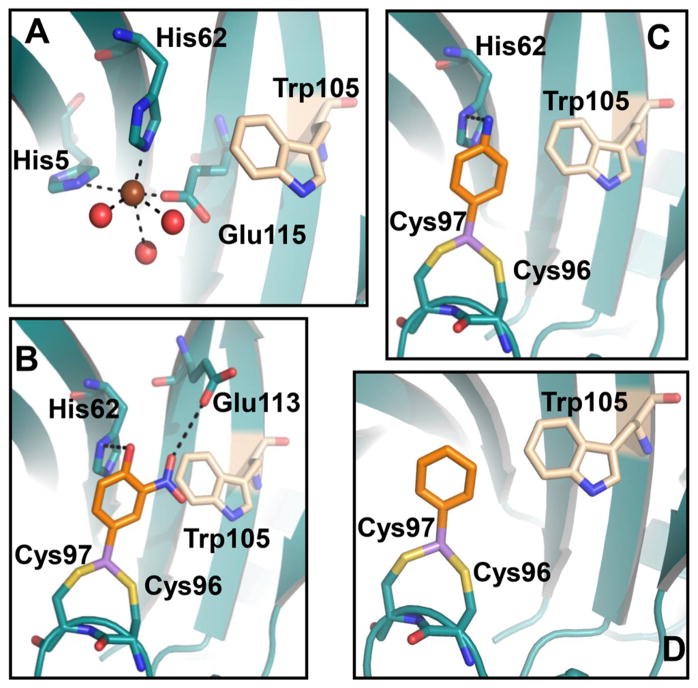
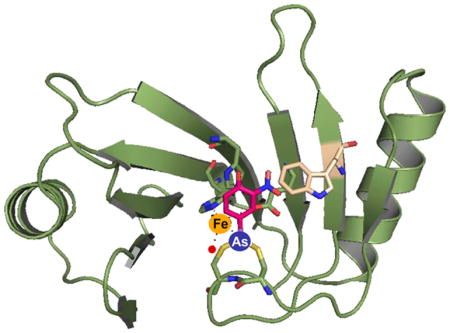
Using ITC the affinity for the aromatic arsenicals PhAs(III) and MAs(III) was determined to be 0.6 nM and 0.3 μM, respectively, and the product As(III) was not bound. From the quenching of intrinsic protein fluorescence, Rox(III) and other aromatic arsenicals were found to bind with higher affinity than MAs(III). It appears that man-made aromatic arsenicals are better substrates for ArsI than the natural substrate. Among these aromatic arsenicals, Rox(III) quenched ArsI fluorescence with the highest affinity. High-affinity substrate binding is due to presence of a substrate binding loop, a unique feature of ArsI orthologs. The loop contains a vicinal cysteine pair, and mutagenesis of the cysteines eliminates substrate binding, demonstrating that the cysteines are responsible for binding organo-arsenicals. Because there was essentially no binding of As(III), it is reasonable that the vicinal pair releases the product following C–As bond cleavage. To examine the question of selectivity for the aromatic arsenicals, we constructed a homology model of Bacillus ArsI built on the crystal structure of TcArsI (PDB ID 5CB9) (Figure 8). The model was populated with Fe(II) and three different trivalent aromatic arsenicals, Rox(III), p-ASA(III), or PhAs(III). Fe(II) is coordinated with the triad of His5, His62, and Glu115 (Figure 8A). Rox(III) has more interactions with the protein than the other two (Figure 8B). There is π–π stacking between the rings of Rox(III) and His62 that stabilizes bound Rox(III). Rox(III) also interacts with the hydrophobic side chain of Trp105, and forms hydrogen bonds between the hydroxyl group and imidazole nitrogen of His62, and the nitro groups with the carboxylate side chain of Glu113, respectively. p-ASA(III) retains the interaction with His62 but does not interact with either Trp105 or Glu113, and so is bound with lower affinity than Rox(III) (Figure 8C). PhAs(III) does not interact with any of the protein side chains, and so is bound with the lower affinity than the other two (Figure 8D).
Figure 8.
Model of interaction of Bacillus ArsI with (A) Fe(II), (B) Rox(III), (C) p-ASA(III), and (D) PhAs(III). Fe(II) (brown sphere) directly coordinates with protein through residues His5, His62, and Glu115 and water molecules (red sphere) (A). The aromatic ring of Rox(III) stacks with the five-membered ring of His62 and the hydrophobic side chain of Trp105. The hydroxyl and nitro groups make contact with His62 and Glu113, respectively (B). p-ASA(III) interacts in a similar manner but has no contact with Glu113 (C). PhAs(III) has no contact with either Glu113 or His62 (D). Dotted lines represent hydrogen bonding. Homology modeling was performed as described in Materials and Methods.
On the basis of the crystal structures of ArsI with the loop in three different conformations, we proposed a loop-gating mechanism that controls the catalytic reaction.31 The substrate-binding loop acts as a gate that opens as the loop moves to solvent, where substrate is bound, in turn, the loop moves to the active site, where catalysis occurs, closing the gate. The flexible substrate-binding loop allows substrate to bind independently of the metal. In other type I extradiol ring-cleaving dioxygenase there is no flexible binding site, rather substrate binds directly to bound metal and to residues near the metal binding site to initiate the catalysis.50 For example, in protocatechuate 3,4-dioxygenase, substrates bind to tyrosyl ligands near the bound metal.51,52 Thus, in most extradiol dioxygenases metal must bind before substrate, while ArsI binds them independently, a unique feature of ArsI.
In conclusion, we propose that ArsI is a significant mechanism of decomposition of environmental organoarsenicals, whether biologically produced MAs(III) or the anthropogenically introduced MAs(III) herbicide, and importantly, it is a significant step in the oxic breakdown of synthetic aromatic arsenical farm animal feed supplements such as roxarsone. As such, we predict that this C–As lyase has a substantial impact on the arsenic biogeocycle.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R01 GM055425 to B.P.R., R01 DK068139 to T.S., and pilot project funding from the Herbert Wertheim College of Medicine, Florida International University 800008403 to M.Y. We thank Professor Konstantinos Kavallieratos, Department of Chemistry and Biochemistry, Florida International University, for suggestions and advice. Mass spectroscopy was performed by the Florida International University Advanced Mass Spectroscopy Facility.
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.7b03180.
Strains and plasmids; oligonucleotide primers for mutagenesis; isothermal titration calorimetric data for ArsI; multiple alignment of ArsI orthologs; ligand-dependent quenching of protein fluorescence of ArsI mutants (PDF)
References
- 1.Mateen FJ, Grau-Perez M, Pollak JS, Moon KA, Howard BV, Umans JG, Best LG, Francesconi KA, Goessler W, Crainiceanu C, Guallar E, Devereux RB, Roman MJ, Navas-Acien A. Chronic arsenic exposure and risk of carotid artery disease: The strong heart study. Environ Res. 2017;157:127–134. doi: 10.1016/j.envres.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman JD, Navas-Acien A, Kuo CC, Guallar E, Howard BV, Fabsitz RR, Devereux RB, Umans JG, Francesconi KA, Goessler W, Best LT, Tellez-Plaza M. Peripheral arterial disease and its association with arsenic exposure and metabolism in the strong heart study. Am J Epidemiol. 2016;184(11):806–817. doi: 10.1093/aje/kww002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drobna Z, Del Razo LM, Garcia-Vargas GG, Sanchez-Pena LC, Barrera-Hernandez A, Styblo M, Loomis D. Environmental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico. J Exposure Sci Environ Epidemiol. 2013;23(2):151–155. doi: 10.1038/jes.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bencko V, Symon K. Test of environmental exposure to arsenic and hearing changes in exposed children. Environ Health Perspect. 1977;19:95–101. doi: 10.1289/ehp.771995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee SC, Rahman MM, Chowdhury UK, Sengupta MK, Lodh D, Chanda CR, Saha KC, Chakraborti D. Neuropathy in arsenic toxicity from groundwater arsenic contamination in West Bengal, India. J Environ Sci Health, Part A: Toxic/Hazard Subst Environ Eng. 2003;38(1):165–183. doi: 10.1081/ese-120016887. [DOI] [PubMed] [Google Scholar]
- 6.Tyler CR, Allan AM. The effects of arsenic exposure on neurological and cognitive dysfunction in human and rodent studies: a review. Curr Environ Health Rep. 2014;1(2):132–147. doi: 10.1007/s40572-014-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodrigues EG, Bellinger DC, Valeri L, Hasan MOSI, Quamruzzaman Q, Golam M, Kile ML, Christiani DC, Wright RO, Mazumdar M. Neurodevelopmental outcomes among 2- to 3-year-old children in Bangladesh with elevated blood lead and exposure to arsenic and manganese in drinking water. Environ Health. 2016;15(1):44. doi: 10.1186/s12940-016-0127-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirano S, Kobayashi Y, Cui X, Kanno S, Hayakawa T, Shraim A. The accumulation and toxicity of methylated arsenicals in endothelial cells: important roles of thiol compounds. Toxicol Appl Pharmacol. 2004;198(3):458–467. doi: 10.1016/j.taap.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Petrick JS, Ayala-Fierro F, Cullen WR, Carter DE, Vasken Aposhian H. Monomethylarsonous acid (MMA(III)) is more toxic than arsenite in Chang human hepatocytes. Toxicol Appl Pharmacol. 2000;163(2):203–207. doi: 10.1006/taap.1999.8872. [DOI] [PubMed] [Google Scholar]
- 10.Styblo M, Del Razo LM, Vega L, Germolec DR, LeCluyse EL, Hamilton GA, Reed W, Wang C, Cullen WR, Thomas DJ. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Arch Toxicol. 2000;74(6):289–299. doi: 10.1007/s002040000134. [DOI] [PubMed] [Google Scholar]
- 11.Challenger F. Biological methylation. Sci Prog. 1947;35(139):396–416. [PubMed] [Google Scholar]
- 12.Hayakawa T, Kobayashi Y, Cui X, Hirano S. A new metabolic pathway of arsenite: arsenic-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol. 2005;79(4):183–191. doi: 10.1007/s00204-004-0620-x. [DOI] [PubMed] [Google Scholar]
- 13.Dheeman DS, Packianathan C, Pillai JK, Rosen BP. Pathway of human AS3MT arsenic methylation. Chem Res Toxicol. 2014;27(11):1979–1989. doi: 10.1021/tx500313k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marapakala K, Packianathan C, Ajees AA, Dheeman DS, Sankaran B, Kandavelu P, Rosen BP. A disulfide-bond cascade mechanism for As(III) S-adenosylmethionine methyltransferase. Acta Crystallogr, Sect D: Biol Crystallogr. 2015;71(3):505–515. doi: 10.1107/S1399004714027552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Pawitwar SS, Rosen BP. The organoarsenical biocycle and the primordial antibiotic methylarsenite. Metallomics. 2016;8(10):1047–1055. doi: 10.1039/c6mt00168h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nachman KE, Baron PA, Raber G, Francesconi KA, Navas-Acien A, Love DC. Roxarsone, inorganic arsenic, and other arsenic species in chicken: a US-based market basket sample. Environ Health Perspect. 2013;121(7):818–824. doi: 10.1289/ehp.1206245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher DJ, Yonkos LT, Staver KW. Environmental concerns of roxarsone in broiler poultry feed and litter in Maryland, USA. Environ Sci Technol. 2015;49(4):1999–2012. doi: 10.1021/es504520w. [DOI] [PubMed] [Google Scholar]
- 18.Yao L, Huang L, He Z, Zhou C, Li G. Occurrence of arsenic impurities in organoarsenics and animal feeds. J Agric Food Chem. 2013;61(2):320–324. doi: 10.1021/jf3045022. [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Yao L, He Z, Zhou C, Li G, Yang B, Deng X. Roxarsone and its metabolites in chicken manure significantly enhance the uptake of As species by vegetables. Chemosphere. 2014;100:57–62. doi: 10.1016/j.chemosphere.2013.12.074. [DOI] [PubMed] [Google Scholar]
- 20.Kazi TG, Shah AQ, Afridi HI, Shah NA, Arain MB. Hazardous impact of organic arsenical compounds in chicken feed on different tissues of broiler chicken and manure. Ecotoxicol Environ Saf. 2013;87:120–123. doi: 10.1016/j.ecoenv.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Mangalgiri KP, Adak A, Blaney L. Organoarsenicals in poultry litter: detection, fate, and toxicity. Environ Int. 2015;75:68–80. doi: 10.1016/j.envint.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Nachman KE, Love DC, Baron PA, Nigra AE, Murko M, Raber G, Francesconi KA, Navas-Acien A. Nitarsone, inorganic arsenic, and other arsenic species in turkey meat: exposure and risk assessment based on a 2014 US market basket sample. Environ Health Perspect. 2017;125(3):363–369. doi: 10.1289/EHP225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Peng H, Lu X, Liu Q, Huang R, Hu B, Kachanoski G, Zuidhof MJ, Le XC. Arsenic metabolites, including N-acetyl-4-hydroxy-m-arsanilic acid, in chicken litter from a roxarsone-feeding study involving 1600 chickens. Environ Sci Technol. 2016;50(13):6737–6743. doi: 10.1021/acs.est.5b05619. [DOI] [PubMed] [Google Scholar]
- 24.Moody JP, Williams RT. The metabolism of 4-hydroxy-3-nitrophenylarsonic acid in hens. Food Cosmet Toxicol. 1964;2:707–715. doi: 10.1016/s0015-6264(64)80422-3. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Rosen BP. Organoarsenical biotransformations by Shewanella putrefaciens. Environ Sci Technol. 2016;50(15):7956–7963. doi: 10.1021/acs.est.6b00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshinaga M, Cai Y, Rosen BP. Demethylation of methylarsonic acid by a microbial community. Environ Microbiol. 2011;13(5):1205–1215. doi: 10.1111/j.1462-2920.2010.02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshinaga M, Rosen BP. A C-As lyase for degradation of environmental organoarsenical herbicides and animal husbandry growth promoters. Proc Natl Acad Sci U S A. 2014;111(21):7701–7706. doi: 10.1073/pnas.1403057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher E, Dawson AM, Polshyna G, Lisak J, Crable B, Perera E, Ranganathan M, Thangavelu M, Basu P, Stolz JF. Transformation of inorganic and organic arsenic by Alkaliphilus oremlandii sp. nov. strain OhILAs. Ann N Y Acad Sci. 2008;1125:230–241. doi: 10.1196/annals.1419.006. [DOI] [PubMed] [Google Scholar]
- 29.Thomas JA, Chovanec P, Stolz JF, Basu P. Mapping the protein profile involved in the biotransformation of organoarsenicals using an arsenic metabolizing bacterium. Metallomics. 2014;6(10):1958–1969. doi: 10.1039/c4mt00185k. [DOI] [PubMed] [Google Scholar]
- 30.Chovanec P, Stolz JF, Basu P. A proteome investigation of roxarsone degradation by Alkaliphilus oremlandii strain OhILAs. Metallomics. 2010;2(2):133–139. doi: 10.1039/b915479e. [DOI] [PubMed] [Google Scholar]
- 31.Nadar VS, Yoshinaga M, Pawitwar SS, Kandavelu P, Sankaran B, Rosen BP. Structure of the ArsI C-As lyase: insights into the mechanism of degradation of organoarsenical herbicides and growth promoters. J Mol Biol. 2016;428(11):2462–2473. doi: 10.1016/j.jmb.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stice S, Liu G, Matulis S, Boise LH, Cai Y. Determination of multiple human arsenic metabolites employing high performance liquid chromatography inductively coupled plasma mass spectrometry. J Chromatogr B: Anal Technol Biomed Life Sci. 2016;1009:55–65. doi: 10.1016/j.jchromb.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigues AV, Kandegedara A, Rotondo JA, Dancis A, Stemmler TL. Iron loading site on the Fe-S cluster assembly scaffold protein is distinct from the active site. BioMetals. 2015;28(3):567–576. doi: 10.1007/s10534-015-9846-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nogales J, Canales Á, Jiménez-Barbero J, Serra B, Pingarrón JM, García JL, Díaz E. Unravelling the gallic acid degradation pathway in bacteria: the gal cluster from Pseudomonas putida. Mol Microbiol. 2011;79(2):359–374. doi: 10.1111/j.1365-2958.2010.07448.x. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Packianathan C, Rossman TG, Rosen BP. Nonsynonymous polymorphisms in the human AS3MT arsenic methylation gene: implications for arsenic toxicity. Chem Res Toxicol. 2017;30(7):1481–1491. doi: 10.1021/acs.chemrestox.7b00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL User’s Manual. DeLano Scientific; San Carlos, CA: 2001. [Google Scholar]
- 40.Leitgeb S, Straganz GD, Nidetzky B. Biochemical characterization and mutational analysis of the mononuclear non-haem Fe2+ site in Dke1, a cupin-type dioxygenase from Acinetobacter johnsonii. Biochem J. 2009;418(2):403–411. doi: 10.1042/BJ20081161. [DOI] [PubMed] [Google Scholar]
- 41.Marapakala K, Qin J, Rosen BP. Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry. 2012;51(5):944–951. doi: 10.1021/bi201500c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou T, Rosen BP. Tryptophan fluorescence reports nucleotide-induced conformational changes in a domain of the ArsA ATPase. J Biol Chem. 1997;272(32):19731–19737. doi: 10.1074/jbc.272.32.19731. [DOI] [PubMed] [Google Scholar]
- 43.Nozaki M. Oxygenases and dioxygenases. Biochemistry. 1979;78:145–186. doi: 10.1007/BFb0048193. [DOI] [PubMed] [Google Scholar]
- 44.Mehlhorn H. Encyclopedia of Parasitology. 3. Vol. 1. Springer; New York: 2008. p. 860. [Google Scholar]
- 45.Nigra AE, Nachman KE, Love DC, Grau-Perez M, Navas-Acien A. Poultry consumption and arsenic exposure in the US population. Environ Health Perspect. 2017;125(3):370–377. doi: 10.1289/EHP351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutherford DW, Bednar AJ, Garbarino JR, Needham R, Staver KW, Wershaw RL. Environmental fate of roxarsone in poultry litter. Part II. Mobility of arsenic in soils amended with poultry litter. Environ Sci Technol. 2003;37(8):1515–1520. doi: 10.1021/es026222+. [DOI] [PubMed] [Google Scholar]
- 47.Stolz JF, Perera E, Kilonzo B, Kail B, Crable B, Fisher E, Ranganathan M, Wormer L, Basu P. Biotransformation of 3-nitro-4-hydroxybenzene arsonic acid (roxarsone) and release of inorganic arsenic by Clostridium species. Environ Sci Technol. 2007;41(3):818–823. doi: 10.1021/es061802i. [DOI] [PubMed] [Google Scholar]
- 48.Cortinas I, Field JA, Kopplin M, Garbarino JR, Gandolfi AJ, Sierra-Alvarez R. Anaerobic biotransformation of roxarsone and related N-substituted phenylarsonic acids. Environ Sci Technol. 2006;40(9):2951–2957. doi: 10.1021/es051981o. [DOI] [PubMed] [Google Scholar]
- 49.Yan Y, Ye J, Xue XM, Zhu YG. Arsenic demethylation by a C-As lyase in cyanobacterium Nostoc sp. PCC 7120. Environ Sci Technol. 2015;49(24):14350–14358. doi: 10.1021/acs.est.5b03357. [DOI] [PubMed] [Google Scholar]
- 50.Cho HJ, Kim K, Sohn SY, Cho HY, Kim KJ, Kim MH, Kim D, Kim E, Kang BS. Substrate binding mechanism of a type I extradiol dioxygenase. J Biol Chem. 2010;285(45):34643–34652. doi: 10.1074/jbc.M110.130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis MI, Orville AM, Neese F, Zaleski JM, Lipscomb JD, Solomon EI. Spectroscopic and electronic structure studies of protocatechuate 3,4-dioxygenase: nature of tyrosinate-Fe(III) bonds and their contribution to reactivity. J Am Chem Soc. 2002;124(4):602–614. doi: 10.1021/ja011945z. [DOI] [PubMed] [Google Scholar]
- 52.Orville AM, Lipscomb JD, Ohlendorf DH. Crystal structures of substrate and substrate analog complexes of proto-catechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding. Biochemistry. 1997;36(33):10052–10066. doi: 10.1021/bi970469f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.