Abstract
Background
Synthesis of silver nano-compounds with enhanced antimicrobial effects is of great interest for the development of new antibacterial agents. Previous studies have reported the antibacterial properties of pegylated silver-coated carbon nanotubes (pSWCNT-Ag) showing less toxicity in human cell lines. However, the mechanism underlining the pSWCNT-Ag as a bactericidal agent remained unfolded. Here we assessed the pSWCNT-Ag effects against foodborne pathogenic bacteria growth and proteome profile changes.
Results
Measurements of bioluminescent imaging, optical density, and bacteria colony forming units revealed dose-dependent and stronger bactericidal activity of pSWCNT-Ag than their non-pegylated counterparts (SWCNT-Ag). In ovo administration of pSWCNT-Ag or phosphate-buffered saline resulted in comparable chicken embryo development and growth. The proteomic analysis, using two-dimensional electrophoresis combined with matrix assisted laser desorption/ionization time of flight/time of flight mass spectrometry, was performed on control and surviving Salmonella enterica serovar Typhimurium to pSWCNT-Ag. A total of 15 proteins (ten up-regulated and five down-regulated) differentially expressed proteins were identified. Functional analyses showed significant reduction of proteins associated with biofilm formation, nutrient and energy metabolism, quorum sensing and maintenance of cell structure and cell motility in surviving S. Typhimurium. In contrast, proteins associated with oxygen stress, DNA protection, starvation, membrane rebuilding, and alternative nutrient formation were induced as the compensatory reaction.
Conclusions
This study provides further evidence of the antibacterial effects of pSWCNT-Ag nanocomposites and knowledge of their mechanism of action through various protein changes. The findings may lead to the development of more effective and safe antimicrobial agents.
Keywords: Foodborne pathogens, Pegylated silver-coated carbon nanotubes, Bacterial growth kinetics, Bioluminescence imaging, Chicken embryo development, Proteomics
Background
The emergence of multidrug-resistant bacteria is a global issue in human and veterinary medicine. Recent epidemiological studies have indicated the high prevalence of multidrug-resistant bacteria as a consequence of the abuse utilization of antibiotics and sharing of antibiotic-resistant genes among bacteria [1]. Furthermore, increasing transportation of humans, animals, and foods across the world also contributes to the prevalence of multidrug-resistant bacteria [2, 3]. Studies reported that antibiotic-resistant bacteria could be detected in a population that has rarely or never been exposed to antimicrobial agents in the past [3–5]. Although various broad-spectrum antibiotics have been developed during last decades, bacteria still show multidrug-resistance [3, 4].
Studies demonstrated that foodborne pathogens exhibiting drug resistance against five or more antibiotics were mainly isolated from foodborne outbreaks involving ground meat, poultry and dairy products (31/55, 56%) [6]. It is believed that the overuse of antibiotics in the animal industry and biofilm formation during food processing contribute to the emergence of multidrug-resistant foodborne bacteria [3, 5, 7]. As a consequence, the consumption of food contaminated with pathogens such as Salmonella spp., Escherichia coli, and Campylobacter spp. causes foodborne disease outbreaks, and investigating new technologies or agents is becoming vital for efficient microbial growth control [8–10].
The recent development of nanotechnology tools and their combination with microbiology have shown promising applications in inhibition of bacterial growth [11]. Numerous studies have demonstrated the antimicrobial effects of nano-sized metals (i.e., silver, silica, and gold), metal oxides (silver oxide, titanium oxide, and copper oxide), and carbon-based compounds (i.e., fullerene, graphene oxide and single-walled carbon nanotubes or SWCNTs) against Gram-negative and -positive bacteria [12–14]. These nanoparticles are easy to synthesize and possess large surface area-to-volume ratio and high versatility that favor their uses for antimicrobial growth without affecting human eukaryote cells [11, 15]. Among the currently tested nanoparticles, silver and SWCNT have exhibited high potentials for the treatment of multidrug resistance in various bacteria, including foodborne strains [16–18].
Although the mechanism of actions of both silver (Ag) and SWCNT nanocomposites remain unclear, their direct interactions with bacterial cell membrane leading to death are believed to be the primary path for bacterial growth inhibition [13, 19]. Various studies have shown that silver compounds mainly induce ribosomal destabilization, DNA and mitochondrial damages, and free radical formation that results in cell wall destruction [11, 20–23], while SWCNTs preferentially fusion the bacterial cell membrane to destabilize its structure and perturb the metabolism process [18, 24]. A recent study combined both nanoparticles to form high-performance nanocomposites (SWCNT-Ag) for greater antimicrobial strengths, while their further polymerization with polyethylene glycol or pegylation (pSWCNT-Ag) to enhance dispersion (hydrophilicity) and reduce cytotoxicity has led to comparable or stronger antibacterial effects of pSWCNT-Ag [25]. Both nanocomposites maintained antibacterial properties and only pSWCNT-Ag exhibited non-toxicity to various human cell lines [25]. Current findings indicated the internalization of nanocomposites into foodborne pathogens, leading to cell rupture and expulsion of cytoplasmic contents while the molecular mechanisms underlying the antimicrobial property of pSWCNT-Ag remain unknown [25].
In this study, we used three major foodborne pathogens (E. coli, Salmonella enterica serovar Typhimurium and S. enterica serovar Anatum) to evaluate the antibacterial effects of pSWCNT-Ag. Bacteria were transformed for bioluminescence emission, followed by growth assessments through optical density (spectrophotometer) measurement, real-time bioluminescence imaging, and colony forming unit counts. The cytotoxicity of pSWCNT-Ag on a hypothetical host was determined through in ovo chicken embryo development. Proteomic changes induced by the pSWCNT-Ag exposure were examined on the S. Typhimurium strain, used as a model. The results (i) confirmed antibacterial and non-cytotoxicity effects of pSWCNT-Ag, (ii) revealed real-time bioluminescence imaging as a useful tool for early detection of bacteria, and (iii) indicated low expression levels of proteins associated with biosynthesis of amino acids and secondary metabolites, carbon metabolism and cell motility as principal targets of pSWCNT-Ag antimicrobial property.
Methods
Production of bioluminescent bacteria
Strains of Escherichia coli O157:H7 (ATCC 43888), Salmonella enterica serovar Typhimurium (ATCC 14028) and Salmonella enterica serovar Anatum (ATCC 9270) were cultured in Luria–Bertani broth (LB broth, BD, Franklin Lakes, NJ, USA) with shaking or on solid LB agar at 37 °C. Bioluminescent bacteria were constructed by electroporation of pXen5-luxCDABE (Caliper life sciences, Hopkinton, MA, USA) containing ampicillin resistance gene into the target strains. Colonies of successfully transformed bacteria exhibiting bioluminescence were positively selected on solid agar medium containing ampicillin (100 µg/ml). The bioluminescence imaging of bacteria colony was performed using the In Vivo Imaging System (IVIS, Lumina XRMS Series III system, Perkin Elmer, Waltham, MA, USA).
Properties of silver nanocomposites
Silver-coated single-walled carbon nanotubes (SWCNT-Ag) and pegylated SWCNT (pSWCNT-Ag) were prepared and characterized by Chaudhari et al. [25] as previously described. The prepared pSWCNT-Ag had positive zeta potential (8.99) and more positive charges than SWCNT-Ag (− 41.9). The pSWCNT-Ag exhibited bigger size (54 nm vs. 6 nm in diameter; TEM imaging) and high hydrophilicity than the SWCNT-Ag [25]. In the present study, both nanocomposites were further characterized using the X-ray diffraction (XRD—Rigaku’s Ultima III XRD; The Woodlands, TX, USA) operated under 40 kV and 44 mA (scan range_10°–90°, step size_0.02°, and scan speed_1°/min). The JEM 2100 TEM (Jeol USA, Inc.; Peabody, MA, USA) coupled with the energy dispersive X-ray spectroscopy (EDS—Oxford Instrument’s X-Max 80T EDS detector) was also used.
Bacterial growth analyses
Escherichia coli, S. Typhimurium and S. Anatum were grown in LB broth until early stationary phase. Cell concentrations were then adjusted to 1 × 107 colony forming unit (CFU)/ml of fresh culture medium containing various concentrations of SWCNT-Ag or pSWCNT-Ag (0, 25, 31.25, 50 and 62.5 μg/ml). One set of each culture plate and treatment was incubated in the Cytation 5 spectrophotometer (Bioteck, Winooski, VT, USA) to monitor bacterial growth (at 600 nm OD) throughout a 48 h period. Another set of plates was placed in the IVIS chamber (at 37 °C) for culture, with the bioluminescence emission signal captured every hour, 48 times total, to monitor real-time bacterial growth kinetics. In a different set of culture plates, suspensions of bacteria were overnight-cultured in a solid phase (agar medium) to determine the number of live bacteria (CFU/ml) after 48 h cultivation under pSWCNT or SWCNT.
Evaluation of toxicity effect of pSWCNT-Ag in developing chicken embryo
Fertilized White Leghorn eggs (n = 6, 54 ± 3.5 g) were a gift of Professor Christopher McDaniel (Department of Poultry Science, Mississippi State University). Eggs were incubated at 37 °C with 60% humidity and automatically tilted to 30° angle every hour. At day 12 post-fertilization, the viability of egg was measured by candling, followed by the disinfection of the top of eggshell with 70% ethanol and the drilling of a hole of approximately 0.1 cm at the edge of air sac of each selected viable egg. The amount of pSWCNT-Ag were determined by a previous research utilizing 62.5 µg/ml as the highest concentration to evaluate in vitro cytotoxicity on A549, J774, and Hep-2 cell lines [25]. A volume of 100 µl of PBS containing or 62.5 µg/ml pSWCNT-Ag was injected into the allantoic cavity of each egg using a tuberculin syringe. The hole was sealed with parafilm and eggs were immediately placed in the incubator. At day 20 of post-fertilization, chick embryos were taken from shells and humanely sacrificed to weigh and evaluate growth according to Hamburger and Hamilton standards [26]. The embryos were submitted to X-ray imaging using IVIS.
Proteomic analysis
Protein extraction and 2D-gel electrophoresis
Salmonella Typhimurium bacteria were grown in the LB medium containing 0 or 25 µg/ml of pSWCNT-Ag. Cells were harvested at early stationary phase, washed three times with PBS, and frozen at − 80 °C until use. Total protein was extracted from frozen-thawed samples (Gene elute RNA/DNA/Protein purification plus kit; Sigma, MO, USA), precipitated (TCA/Acetone), and resulting pellets were dissolved in 220 µl rehydration sample buffer [7 M urea, 2 M thiourea, 4% CHAPS (w/v), and 20 mM dithiothreitol]. The isoelectric focusing (IEF) was carried out using IEF-strips (ReadyStrip IPG strips, 11 cm, pH 4–7, Bio-Rad; Hercules, CA, USA) and IEF conditions were 30 V for 12 h (rehydration), and 500 V for 15 min and 8000 V for 2.5 h with a total of 35 kVh. After IEF, the strips were then incubated for 15 min in equilibration buffer (6 M urea, 20% glycerol, 2% SDS, and 0.375 M Tris–HCL pH 6.8) containing 2% dithiothreitol (w/v), followed by additional incubation for 15 min in equilibration buffer containing 2.5% iodoacetamide (w/v). Electrophoresis was carried out by transferring the strips onto 4–20% gradient pre-casting gels (Criterion TGX Precast Gels 4–20%, Bio-Rad). Gels were then stained with Bio-safe Coomassie G-250 solution (Bio-Rad).
Three independent replicate gels per bacterial culture conditions (0 or 25 µg/ml pSWCNT-Ag) were obtained for scanning using Proteome Works Plus Spot Cutter (Bio-Rad). All images were aligned and protein spots were detected (PDQuest 2-D analysis software v7, Bio-Rad). Each spot was normalized based on the total valid spot intensity and protein quantitative analysis was performed by Student’s t test at P < 0.05. Significantly increased or decreased protein spots were extracted for protein identification using matrix-assisted laser desorption ionization time of flight/time of flight mass spectrometry (MALDI-TOF/TOF MS).
In-gel digestion, MALDI-TOF/TOF and functional analyses
In-gel tryptic digestion and subsequent MS spectra analysis were carried out at the Institute for Genomics, Biocomputing, and Biotechnology (IGBB), Mississippi State University. Briefly, the protein spots showing significant differences were automatically excised from the gels. Digested and dried peptides were dissolved in 2 μl of a saturated solution of α-Cyano-4-hydroxycinnamic acid in 50% acetonitrile and 0.1% of trifluoroacetic acid. As the internal standards, des-Arg-bradykinin (monoisotopic mass, 904.4681) and angiotensin I (1296.6853) were mixed with dissolved peptide samples which were loaded onto MALDI target plates.
Spotted peptide samples were analyzed by MALDI-TOF/TOF MS (4700 Proteomics Analyzer, Applied Biosystems, MA, USA). Monoisotopic peptide masses were selected in the mass range of 800–2500 Da with an acceleration voltage of 20 kV. The mass spectra were acquired by a cumulative average of 300 laser pulses, and protein identification was performed by peptide mass finger printing (MS-Fit program: http://prospector.ucsf.edu or MASCOT: http://www.matrixscience.com) with a mass tolerance of ± 50 ppm.
The classification of biological process and functional annotation of identified proteins were performed using the clusters of orthologous groups (COGs) analysis (https://www.ncbi.nlm.nih.gov/COG/). The subcellular localization was predicted using the pSORTb v3 (http://www.psort.org/psortb/), and protein interaction analysis was performed using the STRING v10.5 (https://string-db.org/).
Statistical analysis
Statistical analyses were carried out using Graphpad Prism version 7.0.1. Significant differences were determined by Student’s t test with two-tailed nonparametric analysis (P < 0.05) and data were presented as mean ± standard deviation (SD).
Results
Characterization of pSWCNT-Ag and SWCNT-Ag
Figure 1 (blue box) shows the EDS mapping images of the SWCNT-Ag (a–c) and pSWCNT-Ag (d–f) samples. The carbon ions representing the principal element of both nanocomposites are seen as yellow dots in micrographs b and e, while the deposition of silver ions are shown in micrographs c and f. Micrographs b/c and e/f are merged to form a and d, respectively. The XRD pattern analysis in Fig. 1 (green box) shows the presence of silver ions with four 2θ peaks at 32.2°, 38.2°, 44.3° and 46.4° in both nanocomposites, and an additional peak at 26.5° for SWCNT only. The successful pegylation is also observed through the detection of specific peaks (18.9° and 23.3°) in the pSWCNT-Ag alone, indicating the presence of the PL-PEG 5000-amine.
Fig. 1.
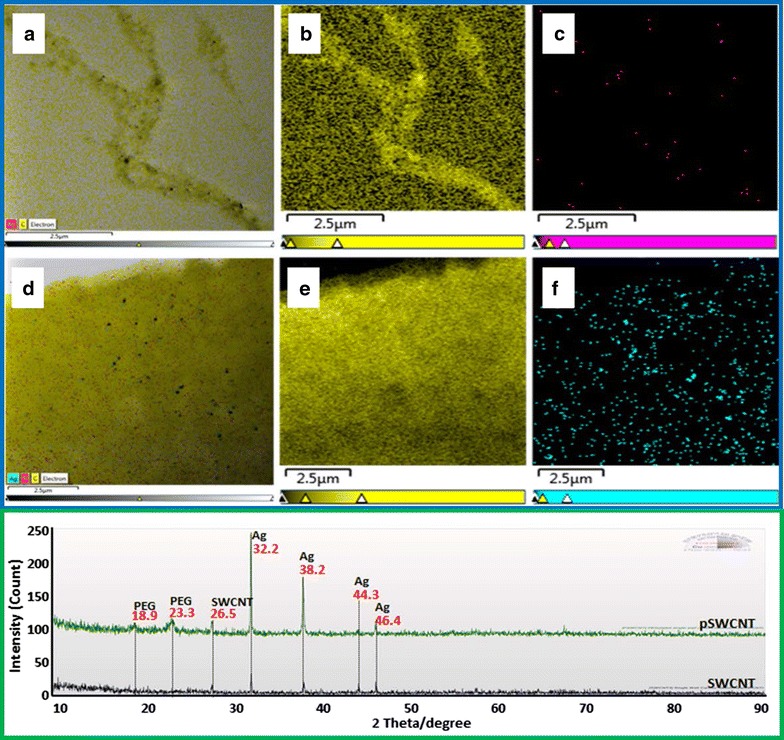
Energy dispersive X-ray spectroscopy (EDS) and X-ray diffraction (XRD) pattern analysis of pSWCNT-Ag and SWCNT-Ag. The upper panel (blue box) shows EDS mapping analysis of SWCNT-Ag (a–c) and pSWCNT-Ag (d–f). Micrographs a, d are merged images of carbon (b, e) and silver (c, f) atoms. The lower panel (green box) represents the XRD patterns depicting the presence of silver atoms (Ag) in both nanocomposites, and polyethylene glycol or PL-PEG 5000-amine (PEG) in pSWCNTs
Growth measurements of E. coli, S. Typhimurium and S. Anatum
The optical density results are summarized in Fig. 2. There were dose-dependent effects of pSWCNT-Ag and SWCNT-Ag on all bacteria growth, as referred to the significant delayed initiation of the exponential phases (or extended lag-phases) in comparison to their respective controls (Fig. 2). For instance, the exponential phase in E. coli started only after 18 and 36 h by the presence of 25 and 31.25 µg/ml pSWCNT-Ag, versus 3 h in the control. Similar patterns were observed with both Salmonellas, irrespective of the nanocomposite. The pSWCNT-Ag exhibited greater antibacterial potential than SWCNT-Ag, characterized by longer delayed exponential phase initiation (or longer lag-phases) in both E. coli and S. Typhimurium (Fig. 2). For example, E. coli in culture media containing 25 µg/ml pSWCNT-Ag had a delayed exponential initiation of 18 h (vs. 13 h for SWCNT), while the presence of 31.25 µg/ml provided 36 h (vs. 19 h) delayed. The S. Typhimurium growth curves clearly exemplify the greater effect of pSWCNT-Ag than that of SWCNT-Ag (delayed exponential phase initiation and complete growth inhibition at the highest pSWCNT-Ag). In addition, species-specificity responses of bacteria were observed at higher concentrations of either nanocomposite (50 and 62.5 µg/ml). The E. coli growth was completely inhibited by both concentrations of nanocomposites, while only 62.5 µg/ml pSWCNT-Ag provided similar effects on both Salmonellas. The concentration of 50 µg/ml pSWCNT-Ag and both 50 and 62.5 µg/ml SWCNT-Ag had limited to non-permanent inhibitory effects on S. Typhimurium and S. Anatum.
Fig. 2.
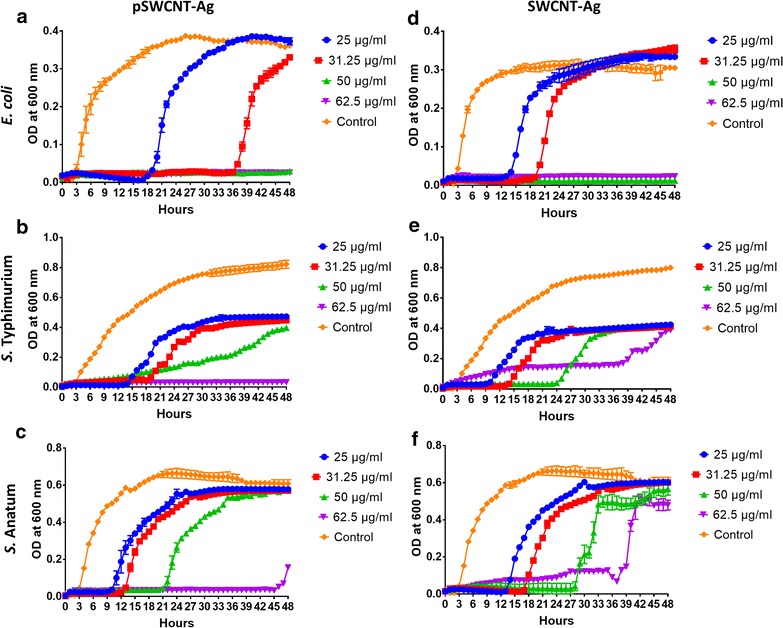
Effects of SWCNT-Ag and pSWCNT-Ag on bacterial growth. The graphs show optical density measurements (600 nm) of bacteria growth over 48-h exposure to various concentrations of pSWCNT-Ag (a–c) and SWCNT-Ag (d–f). Data are mean ± SD of three independent cultures of E. coli (a, d), S. Typhimurium (b, e) and S. Anatum (c, f)
Bacteria exposure to both nanocomposites revealed similar bioluminescence imaging (BLI) patterns, and Fig. 3 is a culture plate with bacteria exposed to pSWCNT-Ag. The bioluminescence intensities of E. coli, S. Typhimurium and S. Anatum appeared in a dose-dependent manner, regardless of the nanocomposite but the pSWCNT-Ag showed most potent effects. Bacteria suspensions in higher pSWCNT concentrations had no bioluminescence emission above background (E. coli in 50 and 62.5 µg/ml, and S. Typhimurium in 62.5 µg/ml), while S. Anatum exhibited lower bioluminescence signals only under 62.5 µg/ml pSWCNTs.
Fig. 3.
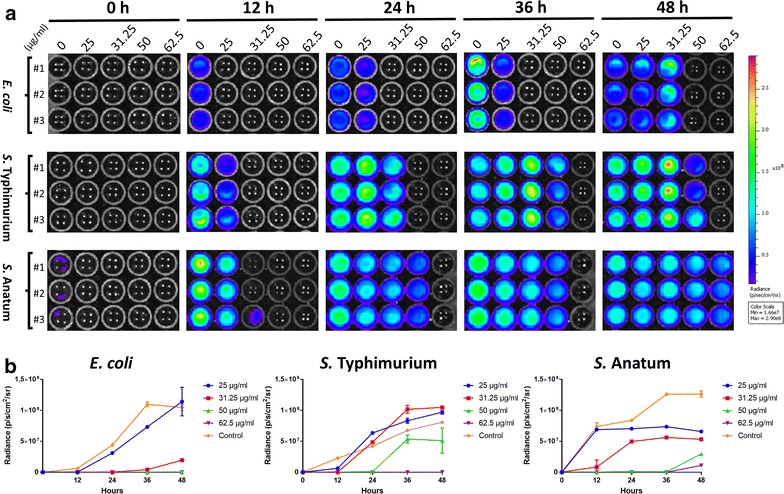
Real-time bioluminescence imaging of bacteria growth. Bioluminescence emission was imaged (a) and quantified (b) at 0, 12, 24, 36 and 48 h of culture. Bacteria (E. coli, S. Typhimurium and S. Anatum) exposed to SWCNT-Ag or pSWCNT-Ag showed similar patterns. Therefore, a representative culture plate containing bioluminescence-emitting bacteria under pSWCNT-Ag exposure is shown (a). Bioluminescence intensities (radiance) were plotted to generate the corresponding growth curves (b). Data (radiance) are mean ± SD of three independent cultures, with each bacteria strain plated in triplicate (#1, #2, and #3) per culture. The pseudo-color code scaling bar indicates the lowest (blue; 0.166 × 108) to the highest (red; 2.9 × 108) radiance
The CFU evaluations by the end of cultures (48 h) are summarized in Fig. 4. No CFU (living cells) were found after E. coli exposure to 50 or 62.5 µg/ml pSWCNT-Ag, or S. Typhimurium exposed to 62.5 µg/ml pSWCNT-Ag. However, CFU were counted in S. Anatum following exposure to 50 or 62.5 µg/ml pSWCNT-Ag. Additionally, bacteria exposed to lowest pSWCNT-Ag concentrations (≤ 50 µg/ml) had comparable CFU numbers with controls (P > 0.05).
Fig. 4.
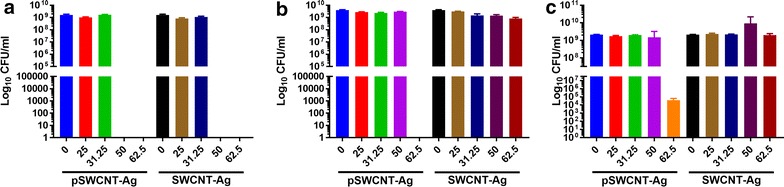
Bacterial survival after exposure to pSWCNT-Ag and SWCNT-Ag. Colony forming units (CFU/ml) were evaluated from each E. coli (a), S. Typhimurium (b), and S. Anatum (c) harvested after 48 h cultures in the presence of various concentration of pSWCNT-Ag and SWCNT-Ag (0, 25, 31.25, 50, and 62.5 µg/ml). Data are mean ± SD of three independent cultures
Evaluation of pSWCNT-Ag toxicity on developing chicken embryo
As seen in Fig. 5, in ovo injections of fertilized eggs with PBS or pSWCNT-Ag (62.5 µg/ml) on day 12 post-fertilization did not perturb embryo development. Both PBS (control) and pSWCNT-injected eggs were comparable, and chicks were alive at the collection on day 20. The weight of both control and pSWCNT-Ag chicks were not different (24.03 ± 1.57 and 23.13 ± 0.77 g, respectively; P > 0.05). Embryos of both groups exhibited similar skeletal structures and bone mass, with no structural abnormality observed through X-ray imaging (Fig. 5). The weight of various internal organs such as brain, hearts, liver, spleen and bursa of Fabricius harvested from both control and treated chicks showed no significant differences (P > 0.05; data not shown).
Fig. 5.
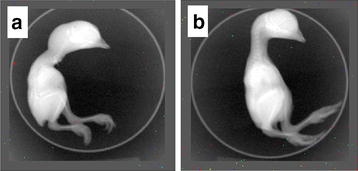
Toxicity evaluation of pSWCNT-Ag on developing chicken embryos. Fertilized chicken eggs were inoculated at 12th day of development with either PBS (Control) or pSWCNT-Ag within the allantoid. Eggs were broken at 20th day of development and fetuses were sacrificed for evaluation. Photographs are X-ray images (IVIS XRMS series III) of chicken controls (a) and exposed (b)
Proteomic analyses and identification of differentially expressed proteins
Figure 6 shows representative gel electrophoreses of control or treated S. Typhimurium with 25 µg/ml pSWCNT-Ag. The matching of three independent and highly reproducible gel replicates per treatment group was performed, followed by quantitative spot intensity analyses that revealed a total of 114 protein spots being significantly differentially expressed between control and treated samples (P < 0.05). Among them, 12 spots (2506, 3603, 3605, 4405, 4508, 4601, 5401, 5408, 5503, 6406, 8003, and 9001) were down-regulated (Fig. 6a), with ten identified as flagellin FliC, aspartate ammonia-lyase, outer membrane protein A, adenylosuccinate synthetase, arginine deiminase, ornithine carbamoyltransferase, carbamate kinase, l-asparaginase 2, universal stress protein F, and ethanolamine utilization protein EutM. In contrast, 6 spots (2101, 2204, 3202, 5303, 7105, and 8401) were up-regulated (Fig. 6b) and five were identified as outer membrane protease, alkyl hydroperoxide reductase subunit C, propanediol utilization microcompartment protein PtuB, DNA starvation/stationary phase protection protein, and aldehyde dehydrogenase. Quantitative analyses of protein spots revealed downregulation of 1.4- to 100-fold and upregulation of 2.56- to 7.53-fold changes (Fig. 7).
Fig. 6.
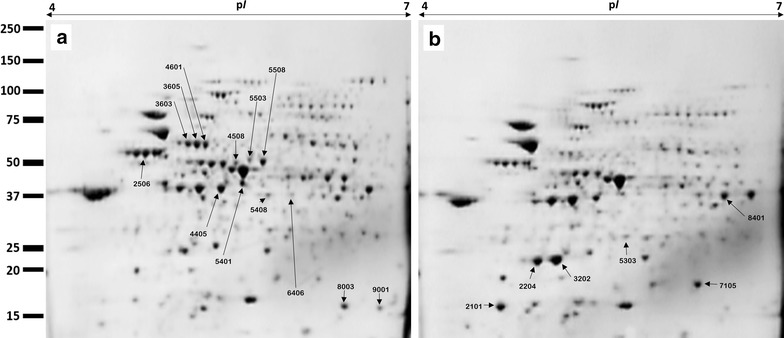
Representative two-dimensional electrophoresis gels of control (a) and pSWCNT-Ag-exposed (b) S. Typhimurium. Down- and up-regulated protein spots are shown in (a) and (b), respectively
Fig. 7.
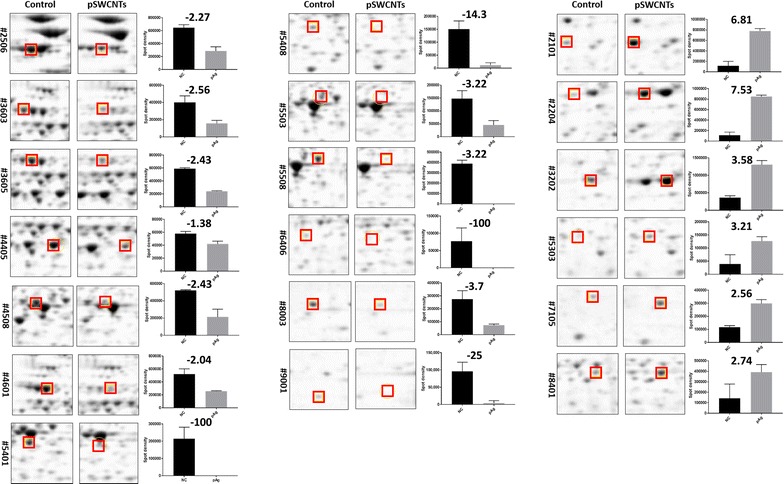
Comparative analysis of differentially detected protein spot intensities. A total of 19 representative altered or spiked protein spots are shown with negative or positive fold change values, respectively. Quantification of spot intensities were analyzed by PDQuest 2-D software and ratios of spot intensity (exposed/control) are shown in each graph. Data are mean ± SD of three gels derived from three independent bacteria cultures. Differential regulation was called with P < 0.05 (Student’s t test)
Table 1 summarizes the characteristics of identified proteins. All proteins showed 100% protein score confidence interval (CI) % and total ion CI %. Functional analyses of identified proteins showed association with different sub-cellular localization such as cytoplasm (53%), outer membrane (13%), extracellular (7%), periplasm (7%) and unknown (20%). Functional prediction using COGs analysis indicated that proteins were involved in amino acid transport and metabolism (E, 40%), cell wall/membrane/envelope biogenesis (M, 13%), translation, ribosomal structure and biogenesis (J, 13%), cell motility (N, 7%), nucleotide transport and metabolism (F, 7%), defense mechanisms (V, 7%), secondary metabolites biosynthesis, transport and catabolism (Q, 7%) and energy production and conversion (C, 7%).
Table 1.
Differentially expressed proteins in S. Typhimurium exposed to pSWCNT-Ag
| Spot ID | Protein name | NCBI no. | Abbr. | Func.a | Local.b | MW (kDa)c | pId | No. of peptide matched | Protein scoree | Total ion scoref | Protein score CI %g | Total ion CI %h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Down-regulated | ||||||||||||
| 2506 | Flagellin FliC | gi|446001950 | hag | N | Extracellular | 51.5 | 4.79 | 16 | 930 | 830 | 100 | 100 |
| 3603 3605 4601 |
Aspartate ammonia-lyase | gi|445991585 | aspA | E | Cytoplasmic | 52.2 | 5.15 | 17 | 450 | 335 | 100 | 100 |
| 4405 | Outer membrane protein A | gi|487406894 | ompA | M | Outer membrane | 37.4 | 5.6 | 16 | 571 | 449 | 100 | 100 |
| 4508 | Adenylosuccinate synthetase | gi|446450117 | purA | F | Cytoplasmic | 47.3 | 5.31 | 21 | 265 | 116 | 100 | 100 |
| 5503 5508 |
arginine deiminase | gi|446332991 | STM4467 | E | Cytoplasmic | 45.5 | 5.47 | 22 | 774 | 586 | 100 | 100 |
| 5401 | Ornithine carbamoyltransferase | gi|446159176 | STM4465 | E | Cytoplasmic | 36.7 | 5.28 | 11 | 585 | 514 | 100 | 100 |
| 5408 | Carbamate kinase | gi|446350896 | STM4466 | E | Cytoplasmic | 33.3 | 5.45 | 13 | 399 | 307 | 100 | 100 |
| 6406 | l-Asparaginase 2 | gi|446316334 | ansB | E | Periplasmic | 36.9 | 5.84 | 13 | 347 | 262 | 100 | 100 |
| 8003 | Universal stress protein F | gi|447005040 | uspF | J | Unknown | 15.7 | 5.93 | 11 | 406 | 296 | 100 | 100 |
| 9001 | Ethanolamine utilization protein EutM | gi|446309861 | eutM | E | Unknown | 9.8 | 6.06 | 6 | 164 | 110 | 100 | 100 |
| Up-regulated | ||||||||||||
| 2101 | Outer membrane protease | gi|446639417 | ompX | M | Outer membrane | 18.4 | 5.74 | 9 | 500 | 425 | 100 | 100 |
| 2204 3202 |
Alkyl hydroperoxide reductase subunit C | gi|445974947 | ahpC | V | Cytoplasmic | 20.7 | 5.03 | 10 | 437 | 354 | 100 | 100 |
| 5303 | Propanediol utilization microcompartment protein PduB | gi|446019642 | pduB | Q | Unknown | 27.9 | 5.21 | 9 | 378 | 318 | 100 | 100 |
| 7105 | DNA starvation/stationary phase protection protein | gi|446022950 | dps | J | Cytoplasmic | 18.7 | 5.73 | 14 | 476 | 336 | 100 | 100 |
| 8401 | Aldehyde dehydrogenase | gi|446075650 | gapA | C | Cytoplasmic | 35.5 | 6.33 | 20 | 572 | 393 | 100 | 100 |
aFunctional classification was performed using COGs (Clusters of Orthologous Groups) functional annotation. C energy production and conversion, E amino acid transport and metabolism, F nucleotide transport and metabolism, J translation, ribosomal structure and biogenesis, M cell wall/membrane/envelope biogenesis, N cell motility, Q secondary metabolites biosynthesis, transport and catabolism, V defense mechanisms
bSubcellular localization was predicted using pSORTb version 3.0
cPredicted molecular weight (MW) and d isoelectric point (pI)
eProtein scores are derived from ion scores as a non-probabilistic basis for the ranking protein hits. Ions score is − 10 log(P), where P is the probability that the observed peptide match is a random event (http://www.matrixscience.com/help/interpretation_help.html)
fTotal ion score is calculated by weighting ion scores for all individual peptides matched to the protein that is associated with this peptide and MS/MS spectrum (http://www.matrixscience.com/help/interpretation_help.html)
gConfidence interval % (CI %) rates the confidence level of the protein score [MS] or ion score [MS/MS]. The CI % is the statistical calculation based on the distribution of the probability that enables to compare the searched data base with the number of submitted mass spectra for the database searches
The KEGG pathway analysis revealed high relationships between up- or down-regulated proteins, with four major pathways identified (P < 0.05; Fig. 8). The glycolysis/gluconeogenesis (A: gapA, pgk, and tpiA) pathway was increased, while the flagella assembly (B: flbC, fliS and hag), alanine, aspartate and glutamate metabolism (C: ansB, aspA, and purA), arginine and proline metabolism (D: STM4465, STM4466, and STM4467) pathways were decreased after S. Typhimurium culture in the presence of pSWCNT-Ag.
Fig. 8.
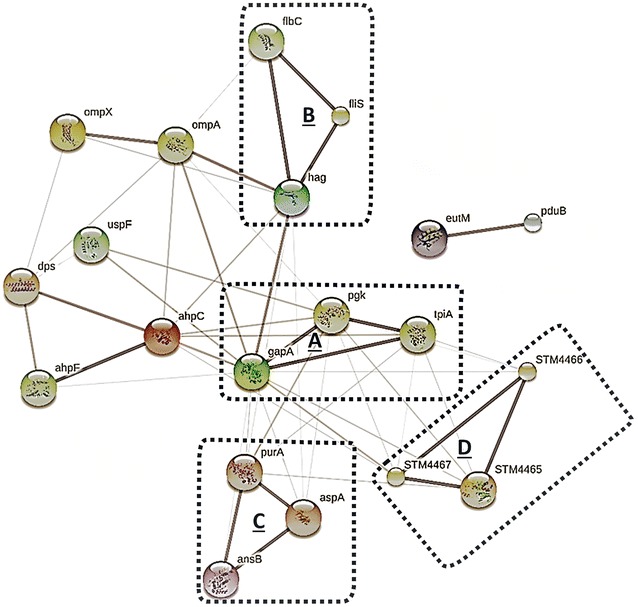
Interaction network analysis of differentially regulated proteins under pSWCNT-Ag. Four representative networks (a–d P = 0.0198) were predicted using STRING software (http://string-db.org). a glycolysis/gluconeogenesis, b Flagellar assembly pathway, c alanine, aspartate and glutamate metabolism, and d Arginine and proline metabolism
Discussion
Numerous studies have reported single-walled carbon nanotubes (SWCNTs) and silver nanoparticles (AgNP) as excellent candidates for alternative therapeutic agents to antibiotics [14, 15, 18]. Additionally, the formation of hydrophilic and functionalized nanocomposites such as pSWCNT-Ag and PLGA-SWCNT (poly-lactic-co-glycolic acid) can enhance antimicrobial strengths as compared to single unique nanoparticles [14, 27]. In the present study, the presence of SWCNT-Ag or pSWCNT-Ag dose-dependently inhibited growth kinetics of tested foodborne bacteria (E. coli, S. Typhimurium and S. Anatum), as observed through optical density (at 600 nm) and CFU measurements, both standard approaches to evaluate bacterial growth. However, the dose-dependent increased darkness of culture media together with the nanocomposite concentrations interfered with OD analyses, making bioluminescence imaging an excellent approach to precisely evaluate bacteria growth due to intrinsic real-time expression of the lux gene. Consequently, bioluminescence imaging allowed for early detection of bacteria growth than the OD method, regardless of the turbidity of the culture medium.
In this study, all analytical approaches confirmed the greater bactericidal effects of pSWCNT-Ag than SWCNT-Ag, which could be attributed to reduced agglomerates, higher dispersed and available pSWCNT-Ag to target bacteria [25, 28]. The observed antibacterial effects on E. coli and S. Typhimurium were in agreement with previous studies [25, 29], and the growth patterns differed across bacteria strains, depending on the pSWCNT-Ag concentrations. Despite S. Anatum showing the highest resistance against both nanocomposites, duration of the lag phases or initiation of the exponential phases of all bacteria strains were significantly and dose-dependently increased, in comparison to their respective controls. In all bacteria strains, pSWCNT-Ag consistently maintained the stronger effects than SWCNT-Ag, suggesting the possibility for decreased antibiotic concentrations when combining them pSWCNT-Ag nanocomposites for disease control or disinfection process. Yet, there are conflicting reports regarding the antibacterial effects of the silver nanomaterial itself that may affect the efficacy of derived carbon nanocomposites. These discrepancies are attributable to bacteria species and/or nanomaterial physico-chemistry characteristics such as surface/volume, shape, size and zeta potential that may vary among studies [30–32]. The recent studies suggested 50 and 62.5 µg/ml as the most effective inhibitory concentrations for all tested bacteria strains, not affecting eukaryotic cells, such as human cell lines [25, 33, 34],
In the current study, the potential toxic effects of pSWCNT-Ag nanocomposites were evaluated on developing chicken embryos, reported as an outstanding animal model to assess the toxicity of organic and inorganic compounds [35], due to their ability to overcome the blood–brain barrier and cause developmental toxicity during embryogenesis [36]. Here, the in ovo injection of pSWCNT-Ag at 62.5 µg/ml (the highest concentration in bacterial evaluations) did not affect embryo development prior to hatching. Chicks harvested from control and injected eggs were still alive and comparable based upon the developmental stage 45 of the Hamburger and Hamilton standards, referring to eggs with half of the yolk sac enclosed in the body cavity and chorio-allantoic membrane glued in embryo with less blood [26]. There were no differences in terms of bone mass observation through X-ray imaging, embryo weight, embryo size and weight of various internal organs of both control and injected chicken. Most importantly, our results reveal the non-toxicity of pSWCNT-Ag to during in ovo embryo-fetal development.
The selective targeting of prokaryote cells implies specific mechanisms of action of pSWCNT-Ag nanocomposites. Carbon-based nanotubes are known to induce growth inhibition through direct contact with bacteria, leading to various cellular perturbations and cell death [14]. A genomic DNA microarray study has revealed the downregulation of a large number of genes encoding flagella components of Bacillus cereus after exposure to silver nanoparticles [37]. A recent study has reported the inhibition of mRNA expression levels of genes involved in the regulation of normal physiology (ybeF), quorum sensing (sdiA), outer membrane structure (safC), invasion (ychP) and virulence (safC, ychP, sseA and sseG) of S. Typhimurium exposed to pSWCNT-Ag [25]. These later findings prompted us to use the foodborne pathogenic and multidrug resistant S. Typhimurium as a bacterium model to investigate the protein changes. As the functional unit of the cell, the study of the bacteria global proteomic analysis provides a unique and important knowledge of the molecular mechanisms of having direct effects on S. Typhimurium, as observed in this study. A total of 15 proteins playing crucial roles in bacterial growth, survival, and division were differentially regulated by the presence of pSWCNT-Ag.
These proteins were predicted to significantly alter the glycolysis/gluconeogenesis, alanine, aspartate and glutamate metabolism, arginine and proline metabolism, and flagella assembly. Previous studies have demonstrated the adverse effect of l-asparaginase 2 (AnsB) and aspartate ammonia-lyase (AspA) down-regulation in S. Typhimurium. In support of our findings, the deletion of l-asparaginase 2 (AnsB) enables bacteria to avoid recognition and clearance by the host immune system [38], while lower aspartate ammonia-lyase (aspA) levels in cells reduce communications between neighboring bacteria, due to the conversion of aspartate into fumarate with decreased production of ammonia used as quorum sensing molecule in bacteria [39, 40]. In the same line, the downregulation of adenylosuccinate synthetase (purA), having a crucial role in the purine biosynthesis, reduces bacterial growth and attenuates pathogenicity at all stages of infection such as colonization, invasion, and internalization [41, 42]. Likewise, the down-regulation of fermentative enzymes appears in favor of bacterial growth inhibition observed under the pSWCNT-Ag exposure. These enzymes corresponded to arginine deiminase (STM4467), ornithine carbamoyl-transferase (STM4465) and carbamate kinase (STM4466), participating into arginine and proline metabolism, as well as the arginine deiminase (ADI or STM4467) system that is important for bacterial protection to acidic extracellular damage [43]. Indeed, the ADI system converts l-arginine into ornithine, ammonia, carbon dioxide, and energy (ATP), all products contributing to acidic pH neutralization within the phagosome and therefore the survival of S. Typhimurium in the host cell (i.e., macrophage) [44, 45]. Other down-regulated proteins corresponded to flagellin FliC (hag, spot2506, 2.3×), outer membrane protein A (ompA, spot4405, 1.4×), universal stress protein F (uspF, spot8803, 1.4×), ethanolamine utilization protein EutM (eutM, spot9001, 25×), and outer membrane protein A (ompA, spot4405, 1.38×). These proteins are involved in various bacterial function such as locomotion (flagellin FliC—[37]), bacteria growth and colonization (eutM—[46]), and survival in hostile environments such as pH, osmolality and temperature (protein F—[47]), bacterial structure, survival and communications in hostile environments, and nutrient transportation (ompA—[48, 49]). Altogether, these results support the antibacterial effects of pSWCNT-Ag through the suppression of various metabolic pathway regulations, cell motility, and biofilm formation, resulting in the attenuation of virulence and reduction of bacterial growth. In support of these predictions, both ultrastructure (scanning electron microscopy) and nanoscale (atomic force microscope) imaging have shown silver nanoparticle attachments from the basal body to the distal end of the flagellum of bacteria, leading to serious damages and reduced motility [50, 51].
As a compensatory bacterial response to pSWCNT-Ag exposure, upregulations of outer membrane protease (ompX, spot2101, 6.81×), alkyl hydroperoxide reductase subunit C (ahpC, spot2204, 7.53×), propanediol utilization microcompartment protein pduB (pduB, spot5303, 3.21×), DNA starvation/stationary phase protection protein (dps, spot7105, 2.56×) and aldehyde dehydrogenase (gapA, spot8401, 2.74×) were observed. These increases help to protect bacteria through membrane rearrangements by removal or destruction of damaged proteins (ompX—[52]), elimination of reactive oxygen species that can be induced by silver nanomaterials (ahpC—[53, 54]), optimization of metabolic pathways (pduB—[55]), DNA protection from oxidative damages and survival to starvation (dps—[56]), improved energetic metabolism (gapA—[57]). Overall, it appears that under pSWCNT-Ag exposure, S. Typhimurium develops resistance through protection against oxidative respiratory stress and supplementary gain of energy and nutrients via other sources such as nitrogen, carbon and energy from ethanol, 1-propanol and 2-propanol.
Conclusions
The present study confirms the dispersed SWCNT-Ag (or pSWCNT-Ag) has an excellent antimicrobial property against foodborne pathogens with undetectable toxic effects during in ovo embryo-fetal development of chicken. Most importantly, the study provides, in our knowledge, the first and comprehensive proteomic analysis revealing significant protein changes and predicted biological functions in S. Typhimurium bacteria following exposure to pSWCNT-Ag. With all tested foodborne bacteria showing extended lag phases or initiation of exponential growth phases and lack of eukaryotic cell toxicity, the current study contributes to support the use of dispersed nanocomposites, such as the pSWCNT-Ag, for disinfection and therapeutic procedures.
Authors’ contributions
SBP and JMF conceived and designed the study—AAC, SRP and SRS designed and synthesized the nanocomposites—SBP performed all experiments, assisted by CSS and JMF—SBP and JMF analyzed the data and drafted the manuscript—PLR and STW assisted in the study design and data interpretation. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Dr. Tibor Pechan and Dr. Olga Pechanova (IGBB, MSU) for their assistance in proteomics analyses.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
All authors agree to publish the manuscript.
Ethics approval and consent to participate
Not applicable.
Funding
This research was supported by the USDA-ARS Biophotonics (Grant # 58-6402-3-018).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Ag
silver
- COG
clusters of orthologous groups
- ATCC
American type culture collection
- E. coli
Escherichia coli
- CFU
colony forming unit
- FT-IR
Fourier transform infrared spectroscopy
- IVIS
in vivo imaging system
- LB
Luria–Bertani broth
- MALDI-TOF/TOF MS
matrix assisted laser desorption/ionization time of flight/time of flight mass spectrometry
- PBS
phosphate-buffered saline
- S. Typhimurium
Salmonella enterica serovar Typhimurium
- SWCNT
single walled-carbon nanotube
- SWCNT-Ag
silver coated single walled-carbon nanotube
- pSWCNT-Ag
pegylated (polyethylene glycol) silver coated single-walled carbon nanotube
- PL-PEG
phospholipid-poly ethylene glycol-amine
- TEM
transmission electron microscopy
- EDS
energy dispersive X-ray spectroscopy
- XRD
X-ray diffraction
Contributor Information
Seong B. Park, Email: sp1679@msstate.edu
Christy S. Steadman, Email: csteadman@ads.msstate.edu
Atul A. Chaudhari, Email: atulvet@gmail.com
Shreekumar R. Pillai, Email: spillai@alasu.edu
Shree R. Singh, Email: ssingh@alasu.edu
Peter L. Ryan, Email: ryan@provost.msstate.edu
Scott T. Willard, Email: swillard@cals.msstate.edu
Jean M. Feugang, Phone: +1662-325-7567, Email: jn181@ads.msstate.edu
References
- 1.da Costa PM, Loureiro L, Matos AJ. Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int J Environ Res Public Health. 2013;10:278–294. doi: 10.3390/ijerph10010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Memish ZA, Venkatesh S, Shibl AM. Impact of travel on international spread of antimicrobial resistance. Int J Antimicrob Agents. 2003;21:135–142. doi: 10.1016/S0924-8579(02)00363-1. [DOI] [PubMed] [Google Scholar]
- 3.Turnidge J, Christiansen K. Antibiotic use and resistance—proving the obvious. Lancet. 2005;365:548–549. doi: 10.1016/S0140-6736(05)70770-4. [DOI] [PubMed] [Google Scholar]
- 4.Norrby SR, Nord CE, Finch R. Lack of development of new antimicrobial drugs: a potential serious threat to public health. Lancet Infect Dis. 2005;5:115–119. doi: 10.1016/S1473-3099(05)70086-4. [DOI] [PubMed] [Google Scholar]
- 5.Singer RS, Finch R, Wegener HC, Bywater R, Walters J, Lipsitch M. Antibiotic resistance—the interplay between antibiotic use in animals and human beings. Lancet Infect Dis. 2003;3:47–51. doi: 10.1016/S1473-3099(03)00490-0. [DOI] [PubMed] [Google Scholar]
- 6.DeWaal CS, Grooters S. Antibiotic resistance in foodborne pathogens. Washington, D.C.: Center for Science in the Public Interest; 2013. pp. 1–22. [Google Scholar]
- 7.Sofos JN, Geornaras I. Overview of current meat hygiene and safety risks and summary of recent studies on biofilms, and control of Escherichia coli O157: H7 in nonintact, and Listeria monocytogenes in ready-to-eat, meat products. Meat Sci. 2010;86:2–14. doi: 10.1016/j.meatsci.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong GL, Hollingsworth J, Morris JG., Jr Emerging foodborne pathogens: Escherichia coli O157: H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol Rev. 1996;18:29–51. doi: 10.1093/oxfordjournals.epirev.a017914. [DOI] [PubMed] [Google Scholar]
- 10.Møretrø T, Heir E, Nesse LL, Vestby LK, Langsrud S. Control of Salmonella in food related environments by chemical disinfection. Food Res Int. 2012;45:532–544. doi: 10.1016/j.foodres.2011.02.002. [DOI] [Google Scholar]
- 11.Hajipour MJ, Fromm KM, Ashkarran AA, de Aberasturi DJ, de Larramendi IR, Rojo T, Serpooshan V, Parak WJ, Mahmoudi M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012;30:499–511. doi: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Schabes-Retchkiman P, Canizal G, Herrera-Becerra R, Zorrilla C, Liu H, Ascencio J. Biosynthesis and characterization of Ti/Ni bimetallic nanoparticles. Opt Mater. 2006;29:95–99. doi: 10.1016/j.optmat.2006.03.014. [DOI] [Google Scholar]
- 13.Kim JS, Kuk E, Yu KN, Kim JH, Park SJ, Lee HJ, Kim SH, Park YK, Park YH, Hwang CY. Antimicrobial effects of silver nanoparticles. Nanomed Nanotechnol Biol Med. 2007;3:95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Al-Jumaili A, Alancherry S, Bazaka K, Jacob MV. Review on the antimicrobial properties of carbon nanostructures. Materials. 1066;2017:10. doi: 10.3390/ma10091066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franci G, Falanga A, Galdiero S, Palomba L, Rai M, Morelli G, Galdiero M. Silver nanoparticles as potential antibacterial agents. Molecules. 2015;20:8856–8874. doi: 10.3390/molecules20058856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galdiero S, Falanga A, Vitiello M, Cantisani M, Marra V, Galdiero M. Silver nanoparticles as potential antiviral agents. Molecules. 2011;16:8894–8918. doi: 10.3390/molecules16108894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C, Mamouni J, Tang Y, Yang L. Antimicrobial activity of single-walled carbon nanotubes: length effect. Langmuir. 2010;26:16013–16019. doi: 10.1021/la103110g. [DOI] [PubMed] [Google Scholar]
- 18.Kang S, Pinault M, Pfefferle LD, Elimelech M. Single-walled carbon nanotubes exhibit strong antimicrobial activity. Langmuir. 2007;23:8670–8673. doi: 10.1021/la701067r. [DOI] [PubMed] [Google Scholar]
- 19.Radzig M, Nadtochenko V, Koksharova O, Kiwi J, Lipasova V, Khmel I. Antibacterial effects of silver nanoparticles on Gram-negative bacteria: influence on the growth and biofilms formation, mechanisms of action. Colloids Surf B. 2013;102:300–306. doi: 10.1016/j.colsurfb.2012.07.039. [DOI] [PubMed] [Google Scholar]
- 20.Su HL, Chou CC, Hung DJ, Lin SH, Pao IC, Lin JH, Huang FL, Dong RX, Lin JJ. The disruption of bacterial membrane integrity through ROS generation induced by nanohybrids of silver and clay. Biomaterials. 2009;30:5979–5987. doi: 10.1016/j.biomaterials.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 21.Feng Q, Wu J, Chen G, Cui F, Kim T, Kim J. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J Biomed Mater Res. 2000;52:662–668. doi: 10.1002/1097-4636(20001215)52:4<662::AID-JBM10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Holt KB, Bard AJ. Interaction of silver (I) ions with the respiratory chain of Escherichia coli: an electrochemical and scanning electrochemical microscopy study of the antimicrobial mechanism of micromolar Ag+ Biochemistry. 2005;44:13214–13223. doi: 10.1021/bi0508542. [DOI] [PubMed] [Google Scholar]
- 23.Prabhu S, Poulose EK. Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int Nano Lett. 2012;2:32. doi: 10.1186/2228-5326-2-32. [DOI] [Google Scholar]
- 24.Mocan T, Matea CT, Pop T, Mosteanu O, Buzoianu AD, Puia C, Iancu C, Mocan L. Development of nanoparticle-based optical sensors for pathogenic bacterial detection. J Nanobiotechnol. 2017;15:25. doi: 10.1186/s12951-017-0260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaudhari AA, Jasper SL, Dosunmu E, Miller ME, Arnold RD, Singh SR, Pillai S. Novel pegylated silver coated carbon nanotubes kill Salmonella but they are non-toxic to eukaryotic cells. J Nanobiotechnol. 2015;13:23. doi: 10.1186/s12951-015-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. doi: 10.1002/jmor.1050880104. [DOI] [PubMed] [Google Scholar]
- 27.Xiu ZM, Zhang QB, Puppala HL, Colvin VL, Alvarez PJ. Negligible particle-specific antibacterial activity of silver nanoparticles. Nano Lett. 2012;12:4271–4275. doi: 10.1021/nl301934w. [DOI] [PubMed] [Google Scholar]
- 28.Dumortier H, Lacotte S, Pastorin G, Marega R, Wu W, Bonifazi D, Briand JP, Prato M, Muller S, Bianco A. Functionalized carbon nanotubes are non-cytotoxic and preserve the functionality of primary immune cells. Nano Lett. 2006;6:1522–1528. doi: 10.1021/nl061160x. [DOI] [PubMed] [Google Scholar]
- 29.Guzman M, Dille J, Godet S. Synthesis and antibacterial activity of silver nanoparticles against Gram-positive and Gram-negative bacteria. Nanomed Nanotechnol Biol Med. 2012;8:37–45. doi: 10.1016/j.nano.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Chernousova S, Epple M. Silver as antibacterial agent: ion, nanoparticle, and metal. Angew Chem Int Ed. 2013;52:1636–1653. doi: 10.1002/anie.201205923. [DOI] [PubMed] [Google Scholar]
- 31.Wu D, Fan W, Kishen A, Gutmann JL, Fan B. Evaluation of the antibacterial efficacy of silver nanoparticles against Enterococcus faecalis biofilm. J Endod. 2014;40:285–290. doi: 10.1016/j.joen.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 32.Zhang M, Zhang K, De Gusseme B, Verstraete W, Field R. The antibacterial and anti-biofouling performance of biogenic silver nanoparticles by Lactobacillus fermentum. Biofouling. 2014;30:347–357. doi: 10.1080/08927014.2013.873419. [DOI] [PubMed] [Google Scholar]
- 33.Travan A, Pelillo C, Donati I, Marsich E, Benincasa M, Scarpa T, Semeraro S, Turco G, Gennaro R, Paoletti S. Non-cytotoxic silver nanoparticle-polysaccharide nanocomposites with antimicrobial activity. Biomacromol. 2009;10:1429–1435. doi: 10.1021/bm900039x. [DOI] [PubMed] [Google Scholar]
- 34.Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 2009;27:76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Rashidi H, Sottile V. The chick embryo: hatching a model for contemporary biomedical research. BioEssays. 2009;31:459–465. doi: 10.1002/bies.200800168. [DOI] [PubMed] [Google Scholar]
- 36.Prasek M, Sawosz E, Jaworski S, Grodzik M, Ostaszewska T, Kamaszewski M, Wierzbicki M, Chwalibog A. Influence of nanoparticles of platinum on chicken embryo development and brain morphology. Nanoscale Res Lett. 2013;8:251. doi: 10.1186/1556-276X-8-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babu MMG, Sridhar J, Gunasekaran P. Global transcriptome analysis of Bacillus cereus ATCC 14579 in response to silver nitrate stress. J Nanobiotechnol. 2011;9:49. doi: 10.1186/1477-3155-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kullas AL, McClelland M, Yang HJ, Tam JW, Torres A, Porwollik S, Mena P, McPhee JB, Bogomolnaya L, Andrews-Polymenis H. l-Asparaginase II produced by Salmonella typhimurium inhibits T cell responses and mediates virulence. Cell Host Microbe. 2012;12:791–798. doi: 10.1016/j.chom.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 40.Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr Opin Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Van Immerseel F. The Salmonella Enteritidis lipopolysaccharide biosynthesis gene rfbH is required for survival in egg albumen. Zoonoses Public Health. 2009;56:145–149. doi: 10.1111/j.1863-2378.2008.01195.x. [DOI] [PubMed] [Google Scholar]
- 42.Sigwart D, Stocker B, Clements J. Effect of a purA mutation on efficacy of Salmonella live-vaccine vectors. Infect Immun. 1989;57:1858–1861. doi: 10.1128/iai.57.6.1858-1861.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Cheng C, Xia Y, Zhao H, Fang C, Shan Y, Wu B, Fang W. Lmo0036, an ornithine and putrescine carbamoyltransferase in Listeria monocytogenes, participates in arginine deiminase and agmatine deiminase pathways and mediates acid tolerance. Microbiology. 2011;157:3150–3161. doi: 10.1099/mic.0.049619-0. [DOI] [PubMed] [Google Scholar]
- 44.Marina A, Uriarte M, Barcelona B, Fresquet V, Cervera J, Rubio V. Carbamate kinase from Enterococcus faecalis and Enterococcus faecium. FEBS J. 1998;253:280–291. doi: 10.1046/j.1432-1327.1998.2530280.x. [DOI] [PubMed] [Google Scholar]
- 45.Choi Y, Choi J, Groisman EA, Kang DH, Shin D, Ryu S. Expression of STM4467-encoded arginine deiminase controlled by the STM4463 regulator contributes to Salmonella enterica serovar Typhimurium virulence. Infect Immun. 2012;80:4291–4297. doi: 10.1128/IAI.00880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Souza CS, Torres AG, Caravelli A, Silva A, Polatto JM, Piazza RM. Characterization of the universal stress protein F from atypical enteropathogenic Escherichia coli and its prevalence in Enterobacteriaceae. Protein Sci. 2016;25:2142–2151. doi: 10.1002/pro.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wexler HM. Outer-membrane pore-forming proteins in Gram-negative anaerobic bacteria. Clin Infect Dis. 2002;35:S65–S71. doi: 10.1086/341923. [DOI] [PubMed] [Google Scholar]
- 49.Kint G, De Coster D, Marchal K, Vanderleyden J, De Keersmaecker SC. The small regulatory RNA molecule MicA is involved in Salmonella enterica serovar Typhimurium biofilm formation. BMC Microbiol. 2010;10:276. doi: 10.1186/1471-2180-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barwal I, Ranjan P, Yadav SC, Kateriya S. Cellular oxido-reductive proteins of Chlamydomonas reinhardtii control the biosynthesis of silver nanoparticles. J Nanobiotechnol. 2011;9:56. doi: 10.1186/1477-3155-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suresh AK, Pelletier DA, Wang W, Moon JW, Gu B, Mortensen NP, Allison DP, Joy DC, Phelps TJ, Doktycz MJ. Silver nanocrystallites: biofabrication using Shewanella oneidensis, and an evaluation of their comparative toxicity on Gram-negative and Gram-positive bacteria. Environ Sci Technol. 2010;44:5210–5215. doi: 10.1021/es903684r. [DOI] [PubMed] [Google Scholar]
- 52.Dalbey RE, Wang P, van Dijl JM. Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol Mol Biol Rev. 2012;76:311–330. doi: 10.1128/MMBR.05019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L, Xie QW, Nathan C. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol Cell. 1998;1:795–805. doi: 10.1016/S1097-2765(00)80079-9. [DOI] [PubMed] [Google Scholar]
- 54.He D, Jones AM, Garg S, Pham AN, Waite TD. Silver nanoparticle—reactive oxygen species interactions: application of a charging—discharging model. J Phys Chem C. 2011;115:5461–5468. doi: 10.1021/jp111275a. [DOI] [Google Scholar]
- 55.Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1, 2-propanediol degradation by Salmonella. J Bacteriol. 2011;193:1385–1392. doi: 10.1128/JB.01473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect Immun. 2004;72:1155–1158. doi: 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dailly Y, Mat-Jan F, Clark DP. Novel alcohol dehydrogenase activity in a mutant of Salmonella able to use ethanol as sole carbon source. FEMS Microbiol Lett. 2001;201:41–45. doi: 10.1111/j.1574-6968.2001.tb10730.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


