Abstract
Objectives
Low copy numbers and deletion of complement C4 genes are potent risk factors for systemic lupus erythematosus (SLE). However, it is not known whether this genetic association affects the clinical outcome. We investigated C4 copy number variation and its relationship to clinical and serological features in a Northern European lupus cohort.
Methods
We genotyped the C4 gene locus using polymerase chain reaction (PCR)-based TaqMan assays in 169 patients with SLE classified according to the 1997 revised American College of Rheumatology (ACR) criteria and in 520 matched controls. In the patient group the mean C4 serum protein concentrations nephelometrically measured during a 12-month period prior to genetic analysis were compared to C4 gene copy numbers. Severity of disease was classified according to the intensity of the immunosuppressive regimens applied and compared to C4 gene copy numbers, too. In addition, we performed a TaqMan based analysis of three lupus-associated single-nucleotide polymorphisms (SNPs) located inside the major histocompatibility complex (MHC) to investigate the independence of complement C4 in association with SLE.
Results
Homozygous deficiency of the C4A isotype was identified as the strongest risk factor for SLE (odds ratio (OR) = 5.329; p = 7.7 × 10−3) in the case-control comparison. Moreover, two copies of total C4 were associated with SLE (OR = 3.699; p = 6.8 × 10−3). C4 serum levels were strongly related to C4 gene copy numbers in patients, the mean concentration ranging from 0.110 g/l (two copies) to 0.256 g/l (five to six copies; p = 4.9 × 10−6). Two copies of total C4 and homozygous deletion of C4A were associated with a disease course requiring cyclophosphamide therapy (OR = 4.044; p = 0.040 and OR = 5.798; p = 0.034, respectively). Homozygous deletion of C4A was associated with earlier onset of SLE (median 24 vs. 34 years; p = 0.019) but not significant after correction for multiple testing. SNP analysis revealed a significant association of HLA-DRB1*0301 with SLE (OR = 2.231; p = 1.33 × 10−5).
Conclusions
Our findings confirm the important role of complement C4 genes in the development of SLE. Beyond the impact on the susceptibility for lupus, C4 copy numbers may be related to earlier onset and a more severe course of the disease. The association of homozygous deletion of C4A and SLE is accompanied by the presence of HLA-DRB1*0301 without a proven pathophysiological mechanism.
Keywords: Systemic lupus erythematosus, complement C4, copy number variation, HLA
Introduction
Systemic lupus erythematosus (SLE) is a complex multi-organ disease of unknown origin causing acute and chronic organ damage. A recent search for genetic factors revealed more than 30 associated genes and loci, almost all of which are attributable to functions of the immune system.1 As in most genome-wide association studies, the knowledge of the risk genes identified is limited to the association in general, whereas their causal role in the pathogenesis of the disease remains to be clarified.
Among these genes, the complement C4 gene with its isotypes C4A and C4B and the special feature of copy number variation has been repeatedly studied in lupus patients. Especially low copy numbers of C4 and deletion of C4A or C4B have consistently been reported as potent risk factors for SLE.2–4 However, the impact of specific genetic backgrounds on certain clinical variants of the disease or its clinical course is scarcely known. In our study, we investigated the contribution of genetic variations of complement C4 to the clinical course of the disease.
Since the C4 gene locus is part of the highly polymorphic major histocompatibility complex (MHC) gene region, the impact of other genetic factors located inside the MHC possibly confounding the association of C4 copy number variation and SLE has been discussed controversially. To assess the relative weight of C4 genes and potentially confounding genes within the MHC, we analyzed three single-nucleotide polymorphisms (SNPs) recently reported to have the strongest association with SLE in British and Spanish patient groups.3
Material and methods
Study participants
A total of 169 patients with the clinical diagnosis of SLE based on the 1997 revised American College of Rheumatology (ACR) criteria,5 who presented at the departments of Rheumatology and Nephrology of the University Hospital of Kiel between 1986 and 2013, were enrolled in this study. Inclusion criteria were a regular and structured follow-up and a treatment period of at least 12 months. The patient group consisted of 160 European individuals, five patients from the Middle East, three patients of Eastern Asian origin and one patient from Togo, Africa.
There were 520 age- and sex-matched unrelated controls drawn from the population-based German biobank POPGEN.6 The study was carried out according to the Declaration of Helsinki, written informed consent was obtained from all participants and the study has been approved by the local ethics committee. Ethylenediaminetetraacetate (EDTA) and serum blood samples were taken from all patients and controls.
Deoxyribonucleic acid (DNA) preparation and C4 genotyping
EDTA blood samples were stored at −80℃ up to genetic analysis. DNA extraction was performed automatically by the Autopure LS system with Gentra Puregene chemistry (Qiagen, Hilden, Germany). Whole genome amplification was performed using the Illustra GenomiPhi V2 DNA Amplification Kit according to the manufacturer’s guidelines (GE Healthcare, Little Chalfont, UK). C4 copy number genotyping was performed using the common method based on the TaqMan® real-time PCR technology as described elsewhere (assay description: C4A: Hs07226349; C4B: Hs07226350; Life Technologies Corporation, Foster City, CA, USA).7
In addition, we performed a replication analysis of three recently reported lupus-associated SNPs located inside the MHC locus that appear to be related to the C4 gene locus (assay description: rs2187668: hCV58662585; rs3135391: hCV2455638; rs558702: hCV940258; Life Technologies Corporation, Foster City, CA, USA).3 SNP genotyping was performed using the common TaqMan® SNP Genotyping Assays as described elsewhere.8
C4 concentrations from serum blood samples were measured by nephelometry using the BN II system (Siemens Healthcare, Erlangen, Germany).
Quality control
Copy number results were assessed using the CopyCaller v1.0 Software (Life Technologies Corporation, Foster City, CA, USA) and were checked for quality according to the recommended procedure obtained from the CopyCaller user manual. In brief, quality control consisted of exclusion of all samples with fewer than three replicates, exclusion of all samples with confidence <0.95 and |CNpredicted – CNcalculated| >0.3, exclusion of all samples with Z score ≥2.65 and exclusion of all samples with 2.65 >Z score ≥1.75 and |CNpredicted –CNcalculated| >0.3.9
After quality control, 160 (total C4), 163 (C4A) and 164 (C4B) usable copy number values remained in the patient group and 460 (total C4), 479 (C4A) and 473 (C4B) usable copy number values in the control group.
SNP data were successfully collected from 168 (rs558702), 169 (rs2187668) and 165 (rs3135391) patients and 512 (rs558702), 514 (rs2187668) and 513 (rs3135391) controls.
Clinical features
Because a generally accepted scoring system for the severity of SLE is not available, we decided to use the most intensive treatment given to an individual patient as a surrogate parameter for the severity of the respective disease course. Therapeutic classification consisted of six categories, increasing from no therapy as the lowest to cyclophosphamide treatment as the highest category. In detail, treatment regimens were graded into the following categories: (1) no therapy, defined as the complete absence of any lupus-related medication; (2) treatment with hydroxychloroquine without accompanying prednisolone; (3) prednisolone therapy, either as monotherapy or combined with antimalarials and nonsteroidal antirheumatic drugs; (4) application of methotrexate or azathioprine; (5) administration of mycophenolate or cyclosporine; (6) treatment with cyclophosphamide.
Statistical analyses
Copy number groups to be compared are based on results found by Yang et al.4 Here, significant differences between cases and controls were found for C4 for 2 vs. >2 and ≥5 vs. <5 and for C4A for homozygous deficiency (0 vs. >0), homozygous and heterozygous deficiency (≤1 vs. >1) and for ≥3 vs. <3. In the study presented by Yang and colleagues, a frequency of two copies of C4 was found in 9.3% of the patients and in 1.5% of the controls (odds ratio (OR) 6.5). With a significance level of 0.05, this yields a power of 97% for the sample size of our study for a two-sided χ2 test. Even for a considerable lower OR of, for example, 3.0, the power of our study is a reasonable 58% (PS power v3.1.2 software).
Independence of C4 gene copy numbers between cases and controls as well as independence of C4 gene copy numbers and therapeutic status in patients (and their related ORs) were tested with Fisher’s exact test. Independence of C4 gene copy numbers and peripheral protein concentration/age at first diagnosis were tested with the Kruskal-Wallis test. Results were adjusted for multiple testing using Bonferroni’s correction. The Jonckheere-Terpstra test was performed to elucidate a trend in peripheral protein concentration across C4 gene copy numbers. Multiple logistic regression with backward selection was performed to investigate the joint influence of C4/C4A gene copy numbers (with various groupings) and SNP data on the case-control status. Inheritance models of SNP data were tested with the χ2 test.
Statistical analysis was performed using SPSS 19 software (IBM).
Results
Patients
The patient group consisted of 154 females and 15 males with a mean age of 47.3 ± standard deviation (SD) 13.8 years, ranging from 19 to 78 years. The control group consisted of 471 females and 49 males with a mean age of 49.1 ± 15.5 years, ranging from 19 to 75 years. A total of 163 patients fulfilled at least four of the 1997 revised ACR criteria and six patients were classified as incomplete SLE with typical lupus-related manifestations matching three ACR criteria. Demographic data and clinical characteristics of the patients are shown in Table 1.
Table 1.
Demographic data and clinical presentation of 169 patients according to the 1997 revised ACR criteria and the most intensive lupus-related treatment ever applied
|
(a)
|
Patients
(n = 169) |
Controls (n = 520) |
| Sex | 154 females and 15 males | 471 females and 49 males |
| Mean age | 47.3 ± 13.8 years | 49.1 ± 15.5 years |
| Age range | 19–78 years | 19–75 years |
| Age at diagnosis | 36.6 ± 14.1 years | – |
| Duration of disease | 11.1 ± 8.7 years | – |
| BMI | 25.3 ± 5.1 | – |
| Median ACR criteria | 5 | – |
|
(b)
|
ACR criteria
|
Number of patients (frequency)
|
| Malar rash | 83 (49.1%) | |
| Discoid rash | 66 (39.1%) | |
| Photosensitivity | 63 (37.3%) | |
| Oral ulcerations | 42 (24.9%) | |
| Arthritis | 136 (80.5%) | |
| Serositis | 40 (23.7%) | |
| Renal involvement | 37 (21.9%) | |
| CNS involvement | 13 (7.7%) | |
| Hematologic disorders: | ||
| Hemolytic anemia | 23 (13.6%) | |
| Leukocytopenia | 49 (29.0%) | |
| Lymphocytopenia | 35 (20.7%) | |
| Thrombocytopenia | 22 (13.0%) | |
| Immunologic disorders: | ||
| Antibodies against dsDNA | 132 (78.1%) | |
| Antibodies against Sm | 14 (8.3%) | |
| Antinuclear antibodies | 169 (100%) | |
|
(c)
|
Grading of therapy
|
Number of patients (frequency)
|
| No therapy (Grade 1) | 2 (1.2 %) | |
| Hydroxychloroquine (Grade 2) | 10 (5.9%) | |
| Prednisolone (Grade 3)a | 27 (16.0%) | |
| Methotrexate or azathioprine (Grade 4) | 24 (14.2%) | |
| Mycophenolate or cyclosporine (Grade 5) | 53 (31.4%) | |
| Cyclophosphamide (Grade 6) | 53 (31.4%) |
Characterization of 169 patients according to the ACR criteria and the most potent treatment ever applied. A: If not otherwise denoted, mean ± standard deviations are given.
The treatments in this table are listed only in case they represent the most intensive modality used in a given patient, i.e. those patients in categories 4 to 6 receiving concomitant prednisolone were not included in category 3.
ACR: American College of Rheumatology; BMI: body mass index; CNS: central nervous system; dsDNA: double-stranded DNA.
Gene copy numbers and ORs
The frequencies of C4 gene copies of total C4 and its isotypes C4A and C4B in cases and controls are shown in Table 2. Four copies of total C4 and two copies of C4A and C4B were the most common findings both in patients and controls. The distribution of total C4 and C4A differed significantly between cases and controls with an apparent shift to lower copy numbers in the patient group (total C4: p = 9.2 × 10−3; padjusted = 0.028; C4A: p = 2.1 × 10−4; padjusted = 6.3 × 10−4), whereas no significant effect was observed in C4B (p = 0.062, padjusted = 0.185). C4B was therefore not further investigated.
Table 2.
Distribution of C4 gene copy numbers in patients and controls
| Total C4 | Two copies | Three copies | Four copies | Five copies | Six copies |
|---|---|---|---|---|---|
| SLE (n = 160) | 11 (6.9%) | 59 (36.9%) | 74 (46.3%) | 15 (9.4%) | 1 (0.6%) |
| Controls (n = 460) | 9 (1.9%) | 135 (29.4%) | 254 (55.2%) | 58 (12.6%) | 4 (0.9%) |
| Fisher’s exact test: | p = 9.2 × 10−3 | ||||
| Bonferroni’s correction: | p = 0.028 | ||||
| C4A |
Zero copies
|
One copy
|
Two copies
|
Three copies
|
Four copies
|
| SLE (n = 163) | 7 (4.3%) | 54 (33.1%) | 79 (48.5%) | 22 (13.5%) | 1 (0.6%) |
| Controls (n = 479) | 4 (0.8%) | 101 (21.1%) | 260 (54.3%) | 101 (21.1%) | 13 (2.7%) |
| Fisher’s exact test: | p = 2.1 × 10−4 | ||||
| Bonferroni’s correction: | p = 6.3 × 10−4 | ||||
| C4B |
Zero copies
|
One copy
|
Two copies
|
Three copies
|
Four copies
|
| SLE (n = 164) | 0 (0.0%) | 28 (17.1%) | 129 (78.7%) | 7 (4.3%) | 0 (0.0%) |
| Controls (n = 473) | 11 (2.3%) | 109 (23.0%) | 335 (70.8%) | 18 (3.8%) | 0 (0.0%) |
| Overall testing (Fisher’s exact test): | p = 0.062 | ||||
| Bonferroni’s correction: | p = 0.185 | ||||
|
|
Copies |
Odds ratio
|
95% CI
|
p value |
Bonferroni’s correction
|
| Total C4 | 2 vs. >2 | 3.699 | 1.504–9.101 | 6.7 × 10−3 | 0.034 |
| ≥5 vs. <5 | 0.713 | 0.399–1.276 | 0.272 | 1.00 | |
| C4A | 0 vs. >0 | 5.329 | 1.539–18.445 | 7.7 × 10−3 | 0.039 |
| ≤1 vs. >1 | 2.130 | 1.451–3.128 | 1.8 × 10−4 | 8.8 × 10−4 | |
| ≥3 vs. <3 | 0.526 | 0.323–0.857 | 0.011 | 0.053 | |
The frequencies of gene copy numbers for total C4, C4A and C4B are shown in patients and controls. Testing for independence was performed using Fisher’s exact test. P values for overall testing were adjusted for multiple testing with Bonferroni’s correction (n = 3 tests). P values for odds ratios were adjusted for multiple testing with Bonferroni’s correction (n = 5 tests).
SLE: systemic lupus erythematosus; CI: confidence interval.
Individuals carrying only two copies for total C4 were at risk for SLE (OR = 3.699; 95% confidence interval (95% CI) = 1.504–9.101; p = 6.7 × 10−3, padjusted = 0.034). A protective effect for five and six copies could not be found in our data (OR = 0.713; 95% CI = 0.399–1.276; p = 0.272, padjusted = 1.00). Homozygous deficiency of C4A was identified as the strongest risk factor in our cohort (OR = 5.329; 95% CI = 1.539–18.445; p = 7.7 × 10−3, padjusted = 0.039), followed by the combined group including homozygous-deficient individuals and patients with one copy of C4A, i.e. heterozygous deficiency (OR = 2.13; 95% CI = 1.451–3.128; p = 1.8 × 10−4; padjusted = 8.8 × 10−4). No effect could be shown for more than three copies of C4A after adjusting for multiple testing (OR = 0.526; 95% CI = 0.323–0.857; p = 0.011; padjusted = 0.053).
SNP analysis
Results from SNP analysis are shown in Table 3. All SNPs showed no deviation from Hardy-Weinberg equilibrium. The SNPs rs558702 (intronic C2 region) and rs2187668 (human leukocyte antigen (HLA)-DRB1*0301/HLA-DR3 tag SNP) were associated with SLE in the allelic and dominant model (OR = 2.290; 95% CI = 1.579–3.322; p = 9.63 × 10−6 and OR = 2.231; 95% CI = 1.548–3.216; p = 1.33 × 10−5, respectively), whereas rs3135391 (HLA-DRB1*1501/HLA-DR2 tag SNP) was not associated with SLE overall. The SNPs rs558702 (intronic C2 region) and rs2187668 (HLA-DRB1*0301/HLA-DR3 tag SNP) were in moderate to strong linkage disequilibrium (D’ = 0.87; r2 = 0.7).
Table 3.
SNP results: Minor allele frequencies and inheritance models
| (a) Minor allele frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients | Controls | Minor allele | Location | ||||||
| rs558702 | 21.43% | 11.82% | A | Intronic C2 region | |||||
| rs2187668 | 22.19% | 13.13% | A | HLA-DRB1*0301 (DR3) tag SNP | |||||
| rs3135391 | 16.16% | 13.84% | T | HLA-DRB1*1501 (DR2) tag SNP | |||||
|
(b) Inheritance models
| |||||||||
|
Allelic model
|
Dominant model | Recessive model | |||||||
|
SNP
|
OR
|
95% CI
|
p (χ2) |
OR
|
95% CI
|
p (χ2) |
OR
|
95% CI
|
p (χ2) |
| rs558702 | 2.035 | 1.474–2.809 | 1.18 × 10−5 | 2.290 | 1.579–3.322 | 9.63 × 10−6 | 2.587 | 0.779–8.588 | 0.108 |
| rs2187668 | 1.886 | 1.378–2.582 | 6.21 × 10−5 | 2.231 | 1.548–3.216 | 1.33 × 10−5 | 1.36 | 0.413–4.475 | 0.611 |
| rs3135391 | 1.2 | 0.851–1.691 | 0.298 | 1.309 | 0.886–1.933 | 0.176 | 0.777 | 0.256–2.356 | 0.654 |
Results from SNP analysis with minor allele frequencies and inheritance models are shown. P values were calculated with χ2 test.
SNP: single-nucleotide polymorphism; HLA: human leukocyte antigen; OR: odds ratio; CI: confidence interval.
When considering jointly C4 or C4A copy number and SNPs as influence variables for SLE status, only SNP rs558702 (intronic C2 region) remained in the model and the copy numbers were no longer significant (data not shown).
C4 serum levels
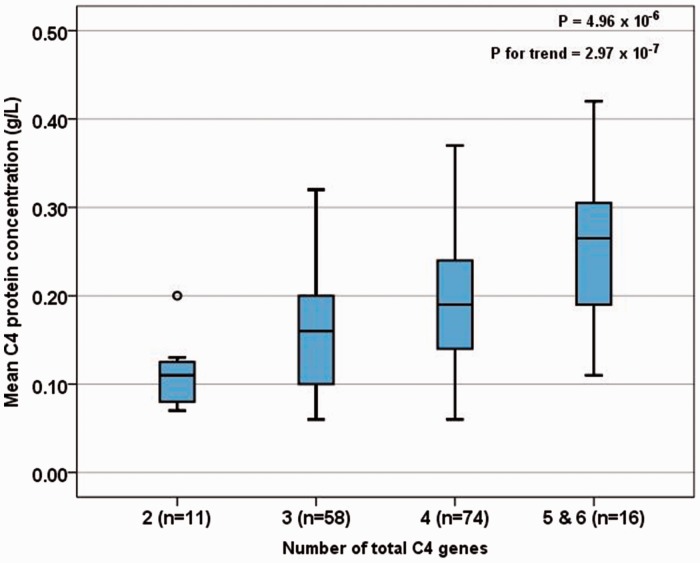
To determine the C4 serum concentration, we calculated the mean of at least five serum samples obtained from every patient during the 12-month period prior to genetic analysis. The mean C4 serum concentration was highly related to the C4 gene copy numbers and increased from 0.110 g/l (two copies) over 0.157 g/l (three copies) and 0.189 g/l (four copies) to 0.256 g/l (five and six copies). The Kruskal-Wallis test revealed a significant association (p = 4.96 × 10−6) with a substantial trend (pfor trend = 2.97 × 10−7). Detailed results are shown in Figure 1.
Figure 1.
Association of C4 gene copy numbers and C4 protein levels.
The mean serum C4 concentration measured during a 12-month period prior to genetic analysis and its relationship to C4 gene copy numbers. P values from the Kruskal-Wallis test and Jonckheere-Terpstra test (p for trend). The length of the box shows the interquartile range. The band within the box indicates the median of the group. The ends of the whiskers identify the upper and the lower 1.5-fold interquartile range, respectively. The circle in group 2 denotes an outlier.
Therapeutic grading
Because of the highly unequal sizes of therapeutic groups (see Table 1), all patients from grade 1 to 5 were summarized and compared to the group of patients who had received cyclophosphamide treatment. Results are shown in Table 4.
Table 4.
Frequencies of C4 gene copy numbers in two groups of patients with different treatment
| Total C4 | Two copies | Three copies | Four copies | Five copies | Six copies |
|---|---|---|---|---|---|
| Patients with cyclophosphamide treatment (n = 52) | 7 (13.5%) | 17 (32.7%) | 22 (42.3%) | 5 (9.6%) | 1 (1.9%) |
| Patients without cyclophosphamide treatment (n = 108) | 4 (3.7%) | 42 (38.9%) | 52 (48.1%) | 10 (9.3%) | 0 (0.0%) |
| Overall testing (Fisher’s exact test): | p = 0.108 | ||||
| Bonferroni’s correction: | p = 0.216 | ||||
|
C4A
|
Zero copies
|
One copy
|
Two copies
|
Three copies
|
Four copies
|
| Patients with cyclophosphamide treatment (n = 52) | 5 (9.6%) | 17 (32.7%) | 23 (44.2%) | 7 (13.5%) | 0 (0.0%) |
| Patients without cyclophosphamide treatment (n = 111) | 2 (1.8%) | 37 (33.3%) | 56 (50.5%) | 15 (13.5%) | 1 (0.9%) |
| Overall testing (Fisher’s exact test): | p = 0.226 | ||||
| Bonferroni’s correction: | p = 0.452 | ||||
|
|
Copies
|
Odds ratio
|
95% CI
|
p value |
Bonferroni’s correction
|
| Total C4 | 2 vs. >2 | 4.044 | 1.128–14.507 | 0.040 | 0.120 |
| C4A | 0 vs. >0 ≤1 vs. >1 | 5.789 1.345 | 1.086–30.957 0.690–2.657 | 0.034 0.391 | 0.102 1.00 |
The distribution of C4 gene copy numbers in patients who had received cyclophosphamide treatment and all other patients who received less aggressive treatment are shown. P values for overall testing were calculated with Fisher’s exact test and adjusted for multiple testing with Bonferroni’s correction (n = 2 tests). P values for odds ratios were calculated with Fisher’s exact test and adjusted for multiple testing with Bonferroni’s correction (n = 3 tests).
CI: confidence interval.
Similar to the association in general, four copies of total C4 and two copies of C4A were the most frequently detected copy numbers in both groups. The group receiving cyclophosphamide treatment included slightly more patients with only two copies of total C4 and 0 copies of C4A, but in overall testing, there was no significant difference in the general distribution of gene copy numbers between the patients receiving cyclophosphamide and the patients with less aggressive therapy (total C4: p = 0.108; padjusted = 0.216; C4A: p = 0.226; padjusted = 0.452). Comparing the subgroups of patients, individuals carrying only two copies for total C4 had more often been exposed to cyclophosphamide therapy (OR = 4.044; 95% CI = 1.128–14.507; p = 0.040, padjusted = 0.120) and individuals with homozygous deficiency of C4A had an even higher exposure to cyclophosphamide treatment (OR = 5.798; 95% CI = 1.086–30.957; p = 0.034, padjusted = 0.102) even though the results remained no longer significant after adjustment for multiple testing. Summarizing the patients carrying homozygous and heterozygous deficiency of C4A, no significant exposure to cyclophosphamide therapy could be detected (OR = 1.345; 95% CI = 0.690–2.657; p = 0.391; padjusted = 1.00).
Age at diagnosis
Age at diagnosis was significantly associated with total C4 and C4A gene copy numbers even after adjustment for multiple testing (total C4: p = 7.2 × 10−3, padjusted = 0.014; C4A: p = 2.8 × 10−3, padjusted = 5.6 × 10−3) without a trend. Detailed results are shown in Table 5.
Table 5.
Age at first diagnosis and C4 gene copy numbers
| Age at first diagnosis | Gene copy numbers of total C4 |
|||
|---|---|---|---|---|
| Two (n = 11) | Three (n = 58) | Four (n = 74) | Five and six (n = 16) | |
| Median | 25 | 39 | 31 | 35 |
| Mean | 29.18 | 40.81 | 33.07 | 39.44 |
| Range | 19–45 | 15–71 | 14–60 | 16–69 |
| 25% quartile | 20 | 29.25 | 23.75 | 25.25 |
| 75% quartile | 41 | 53 | 42.25 | 52.50 |
| Kruskal-Wallis test: Bonferroni’s correction: | p = 7.2 × 10−3 p = 0.014 | |||
|
Gene copy numbers of C4A |
||||
|
Age at first diagnosis
|
Zero
(n = 7) |
One
(n = 53) |
Two (n = 79) |
Three and four (n = 23) |
| Median | 24 | 41 | 31 | 41 |
| Mean | 25.43 | 41.21 | 33.52 | 38.26 |
| Range | 19–42 | 17–71 | 14–69 | 16–67 |
| 25% quartile | 20 | 28.50 | 24 | 25 |
| 75% quartile | 28 | 53 | 41 | 48 |
| Kruskal-Wallis test: | p = 2.8 × 10−3 | |||
| Bonferroni’s correction: | p = 5.6 × 10−3 | |||
|
|
Copies
|
Median age at diagnosis
|
p value |
Bonferroni’s correction
|
| Total C4 | 2 vs. >2 | 25 vs. 34 | 0.069 | 0.207 |
| C4A | 0 vs. >0 ≤1 vs. >1 | 24 vs. 34 37 vs. 32.5 | 0.019 0.073 | 0.057 0.219 |
The age at first diagnosis of SLE in relation to the C4 gene copy numbers are shown. P values were calculated using Kruskal-Wallis test and adjusted for multiple testing with Bonferroni’s correction (n = 2 tests). P values for grouped copy numbers were calculated with Mann-Whitney-U test and adjusted for multiple testing with Bonferroni’s correction (n = 3 tests).
SLE: systemic lupus erythematosus.
Patients with homozygous deficiency of C4A were a median of 10 years younger in age at diagnosis compared to patients without homozygous C4A null alleles (median 24 vs. 34 years; p = 0.019, padjusted = 0.057). Patients with two copies of total C4 showed a similar trend that was, however, not clear enough to reach statistical significance (median 25 vs. 34 years; p = 0.069, padjusted = 0.207). The group containing patients with 0 and one copy of C4A did not significantly differ from all other patients (median 37 vs. 32.5 years; p = 0.073; padjusted = 0.219).
Discussion
We investigated the complement C4 gene locus to evaluate the impact of the individual genetic variations on the clinical course of SLE. Our findings confirm the association of low C4 gene copy numbers and of homozygous C4A deficiency with SLE. Although complete deficiency of C4A was detected in a minority of both groups, its impressive impact on the risk of SLE emphasizes the important role of C4 in the susceptibility of SLE. The more than fivefold increased risk of SLE in C4A-deficient individuals is well in line with similar findings in other SLE cohorts.3,4,10
As a new finding, low copy numbers of total C4 and homozygous deficiency of C4A were associated with a distinctly higher rate of exposure to cyclophosphamide therapy (although not significant after adjustment for multiple testing), implicating that these genetic states may predispose to a more severe and aggressive course of the disease.
As an additional aspect, the role of complement C4 is underscored by our finding of an association between complete C4A deficiency and an earlier onset of SLE. To our knowledge, this is the first genetic study demonstrating a median 10 years’ difference in the age of first diagnosis between individuals completely lacking C4A and the remaining patients.
Taken together, our results support hypotheses suggesting a strong immunological effect of complement C4 both on the general risk of systemic lupus as well as on the clinical course of the disease.
Moreover, the role of C4 copy numbers is substantiated by our finding of a clear and highly significant relationship between complement C4 copy numbers and C4 serum levels. This correlation has repeatedly been described in healthy individuals,11–13 but has rarely been reported in lupus patients.14 In the study presented here, the C4 protein levels represent a mean of an average of five samples taken over a period of one year. Therefore, the chance of misinterpreting individual disease flares associated with complement consumption is low.
The importance of complement C4 in the immunobiology of systemic lupus has been widely studied.15,16 As a substantial component of the classical complement pathway, C4 is involved in the clearance of immune complexes as well as in the clearance of apoptotic cell bodies.17,18 This has been repeatedly demonstrated in complement-deficient individuals, as well as by in vitro experiments, showing that C4 promotes the opsonization and the clearance of immune complexes in concert with complement C3 in a dose-dependent manner.19 C4A appears to play a more important role than C4B among the C4 isotypes, based on its acidic chemical properties and higher affinity to immunoglobulins.20,21 Defects in those processes have been observed in patients suffering from SLE as well as in other disorders classified as immune complex-mediated diseases.22,23 In addition, a state of C4 deficiency has been shown to affect the regulation and suppression of B-cell tolerance, leading to an increased survival of autoreactive B-cells.24,25
However, recent findings suggest that low copy numbers of C4 are not an independent risk factor for SLE, but that they are related to other SLE risk loci such as HLA-DRB1*0301 (DR3 isotype) or HLA-DRB1*1501 (DR2 isotype) located inside the MHC.6 Indeed, the association of SLE and HLA-DRB1*0301 was observed in our patients and this finding is consistent with common genetic features of other Northern European lupus samples.26,27 In contrast, HLA-DRB1*1501 did not increase the risk for SLE in our cohort. These findings are in accord with studies indicating that the association with HLA-DRB1*1501/HLA-DR2 is either less strong28,29 or even not significantly increased30 in Caucasian or Hispanic cohorts, whereas the strongest evidence of the role of HLA-DR2 in SLE appears to be found in patients of Asian origin.31
In a logistic regression considering the joint effect of C4 and C4A copy number variation and of the HLA genotypes and rs558702 (intronic region of the complement C2 gene) as influence variables on SLE, neither C4 status nor HLA genotypes remained significant but only rs558702 did. However, note the substantial linkage disequilibrium between rs558702 and HLA-DRB1*0301 (r2 = 0.7).
Thus, our findings confirm the linkage between the gene loci of C4 and of HLA-DRB1*0301, an association which primarily appears to reflect a highly conserved ancestral haplotype in European lupus patients.32 Despite the reproducibility of these findings, the relative weight of either component appears to be less clear. Indeed, there are several findings of genetic or functional C4A null alleles in patients with SLE not carrying the HLA-DRB1*0301 genotype.2,33 The predominant HLA alleles appear to differ among global patient groups with various ethnic backgrounds, especially in nonwhite populations and in patients of non-European ancestry.30,34 In contrast, the C4A null allele appears not to be restricted to certain ethnicities in general.2,35
As an additional aspect, the implication of HLA-DR3 for the susceptibility to SLE is not substantiated by a known or proven pathophysiological mechanism supporting the role of this gene locus. Whereas the HLA genes identified in rheumatoid arthritis or celiac disease have been shown to be involved in direct or indirect molecular interactions with the presumable disease-associated antigen,36,37 similar findings have not been described for HLA-DR3 and the known autoantigens of SLE.
The association of rs558702 and SLE has recently been reported38 but, because of its localization inside an intronic region of the complement C2 gene, its pathogenetic role is completely unclear. The demonstrated linkage disequilibrium in our findings suggests that rs558702 might be a concomitant feature of HLA-DRB1*0301 or it might be an indicator SNP of another risk locus inside the MHC that has not been detected yet.
In conclusion, the strong association of SLE in individuals with low copy numbers of C4 and in particular in patients with complete deficiency of C4A was confirmed by our data. In this lupus cohort, patients carrying the homozygous C4A null allele were exposed to an earlier onset of the disease and to the requirement of cyclophosphamide treatment, suggesting a more aggressive course of the disease. However, some of these associations were not significant after correction for multiple testing, probably arising from the sample size of only 169 patients in this single-center study. Replication studies with larger sample sizes are recommended to verify these novel results. Furthermore, it is difficult to estimate the severity of SLE. Given the heterogeneous patient group with several individuals suffering from SLE for more than 30 years, the commonly used assessment tools like the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) or Systemic Lupus International Collaborating Clinics Damage Index (SLICC) were not available for all patients. In the absence of other parameters, we used the most intensive treatment procedures ever applied as an approach to assess the severity of the disease in our study because the choice of the drug is determined largely by the severity of the disease and the major organ involvement.39 With respect to this approach, the highest grade of severity was attributed to the patients with a disease course requiring cyclophosphamide treatment since cyclophosphamide was regarded as the most intensive treatment option for severe lupus for two decades before the introduction of mycophenolate and is still of high importance in the appropriate indications.40,41 Further studies with commonly used damage indices are required to prove the associations shown here.
Summarizing these findings, we suggest that the biologic effects of low C4 copy numbers and homozygous deficiency of C4A may influence the clinical course of SLE and should be considered in the management of patients with SLE.
Acknowledgment
We thank Meike Zahnen for her editorial assistance.
Declaration of conflicting interests
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: MJ, FF, AC, WL, SS, AF and JOS have nothing to disclose. Dr Zeuner reports personal fees from Pfizer, UCB, Roche, CSL Behring, and Novartis, outside the submitted work.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: POPGEN is supported by a grant from the German Ministry for Education and Research (01EY1103).
References
- 1.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol 2010; 6: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y, Chung EK, Zhou B, et al. The intricate role of complement component C4 in human systemic lupus erythematosus. Curr Dir Autoimmun 2004; 7: 98–132. [DOI] [PubMed] [Google Scholar]
- 3.Boteva L, Morris DL, Cortés-Hernández J, Martin J, Vyse TJ, Fernando MM. Genetically determined partial complement C4 deficiency states are not independent risk factors for SLE in UK and Spanish populations. Am J Hum Genet 2012; 90: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, Chung EK, Wu YL, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): Low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet 2007; 80: 1037–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997; 40: 1725–1725. [DOI] [PubMed] [Google Scholar]
- 6.Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: Population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet 2006; 9: 55–61. [DOI] [PubMed] [Google Scholar]
- 7.Wu YL, Savelli SL, Yang Y, et al. Sensitive and specific real-time polymerase chain reaction assays to accurately determine copy number variations (CNVs) of human complement C4A, C4B, C4-long, C4-short, and RCCX modules: Elucidation of C4 CNVs in 50 consanguineous subjects with defined HLA genotypes. J Immunol 2007; 179: 3012–3025. [DOI] [PubMed] [Google Scholar]
- 8.De la Vega FM, Lazaruk KD, Rhodes MD, Wenz MH. Assessment of two flexible and compatible SNP genotyping platforms: TaqMan SNP Genotyping Assays and the SNPlex Genotyping System. Mutat Res 2005; 573: 111–135. [DOI] [PubMed] [Google Scholar]
- 9.Flachsbart F, Caliebe A, Heinsen FA, et al. Investigation of complement component C4 copy number variation in human longevity. PLoS One 2014; 9: e86188–e86188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Man XY, Luo HR, Li XP, Yao YG, Mao CZ, Zhang YP. Polymerase chain reaction based C4AQ0 and C4BQ0 genotyping: Association with systemic lupus erythematosus in southwest Han Chinese. Ann Rheum Dis 2003; 62: 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena K, Kitzmiller KJ, Wu YL, et al. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: A comparison of Asian-Indian and European American populations. Mol Immunol 2009; 46: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Chung EK, Zhou B, et al. Diversity in intrinsic strengths of the human complement system: Serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J Immunol 2003; 171: 2734–2745. [DOI] [PubMed] [Google Scholar]
- 13.Hammond A, Ollier W, Walport MJ. Effects of C4 null alleles and homoduplications on quantitative expression of C4A and C4B. Clin Exp Immunol 1992; 88: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragon-Durey MA, Rougier N, Clauvel JP, et al. Lack of evidence of a specific role for C4A gene deficiency in determining disease susceptibility among C4-deficient patients with systemic lupus erythematosus (SLE). Clin Exp Immunol 2001; 123: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickering MC, Walport MJ. Links between complement abnormalities and systemic lupus erythematosus. Rheumatology (Oxford) 2000; 39: 133–141. [DOI] [PubMed] [Google Scholar]
- 16.Giles BM, Boackle SA. Linking complement and anti-dsDNA antibodies in the pathogenesis of systemic lupus erythematosus. Immunol Res 2013; 55: 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schifferli JA, Ng YC, Peters DK. The role of complement and its receptor in the elimination of immune complexes. N Engl J Med 1986; 315: 488–495. [DOI] [PubMed] [Google Scholar]
- 18.Gullstrand B, Mårtensson U, Sturfelt G, Bengtsson AA, Truedsson L. Complement classical pathway components are all important in clearance of apoptotic and secondary necrotic cells. Clin Exp Immunol 2009; 156: 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Traustadottir KH, Sigfusson A, Steinsson K, Erlendsson K. C4A deficiency and elevated level of immune complexes: The mechanism behind increased susceptibility to systemic lupus erythematosus. J Rheumatol 2002; 29: 2359–2366. [PubMed] [Google Scholar]
- 20.Law SK, Dodds AW, Porter RR. A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J 1984; 3: 1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatenby PA, Barbosa JE, Lachmann PJ. Differences between C4A and C4B in the handling of immune complexes: The enhancement of CR1 binding is more important than the inhibition of immunoprecipitation. Clin Exp Immunol 1990; 79: 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arason GJ, Steinsson K, Kolka R, Víkingsdóttir T, D’Ambrogio MS, Valdimarsson H. Patients with systemic lupus erythematosus are deficient in complement-dependent prevention of immune precipitation. Rheumatology (Oxford) 2004; 43: 783–789. [DOI] [PubMed] [Google Scholar]
- 23.Ng YC, Peters DK, Walport MJ. Monoclonal rheumatoid factor-IgG immune complexes. Poor fixation of opsonic C4 and C3 despite efficient complement activation. Arthritis Rheum 1988; 31: 99–107. [DOI] [PubMed] [Google Scholar]
- 24.Prodeus AP, Goerg S, Shen LM, et al. A critical role for complement in maintenance of self-tolerance. Immunity 1998; 9: 721–731. [DOI] [PubMed] [Google Scholar]
- 25.Chatterjee P, Agyemang AF, Alimzhanov MB, et al. Complement C4 maintains peripheral B-cell tolerance in a myeloid cell dependent manner. Eur J Immunol 2013; 43: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skarsvåg S. The importance of C4A null genes in Norwegian patients with systemic lupus erythematosus. Scand J Immunol 1995; 42: 572–576. [DOI] [PubMed] [Google Scholar]
- 27.Steinsson K, Jónsdóttir S, Arason GJ, et al. A study of the association of HLA DR, DQ, and complement C4 alleles with systemic lupus erythematosus in Iceland. Ann Rheum Dis 1998; 57: 503–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galeazzi M, Sebastiani GD, Morozzi G, et al. HLA class II DNA typing in a large series of European patients with systemic lupus erythematosus: Correlations with clinical and autoantibody subsets. Medicine (Baltimore) 2002; 81: 169–178. [DOI] [PubMed] [Google Scholar]
- 29.Fernando MM, Stevens CR, Walsh EC, et al. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLoS Genet 2008; 4: e1000024–e1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reveille JD, Moulds JM, Ahn C, et al. Systemic lupus erythematosus in three ethnic groups: I. The effects of HLA class II, C4, and CR1 alleles, socioeconomic factors, and ethnicity at disease onset. LUMINA Study Group. Lupus in minority populations, nature versus nurture. Arthritis Rheum 1998; 41: 1161–1172. [DOI] [PubMed] [Google Scholar]
- 31.Yuan YJ, Luo XB, Shen N. Current advances in lupus genetic and genomic studies in Asia. Lupus 2010; 19: 1374–1383. [DOI] [PubMed] [Google Scholar]
- 32.Price P, Witt C, Allcock R, et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev 1999; 167: 257–274. [DOI] [PubMed] [Google Scholar]
- 33.Wu YL, Hauptmann G, Viguier M, Yu CY. Molecular basis of complete complement C4 deficiency in two North-African families with systemic lupus erythematosus. Genes Immun 2009; 10: 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim I, Kim YJ, Kim K, et al. Genetic studies of systemic lupus erythematosus in Asia: Where are we now? Genes Immun 2009; 10: 421–432. [DOI] [PubMed] [Google Scholar]
- 35.Dunckley H, Gatenby PA, Hawkins B, Naito S, Serjeantson SW. Deficiency of C4A is a genetic determinant of systemic lupus erythematosus in three ethnic groups. J Immunogenet 1987; 14: 209–218. [DOI] [PubMed] [Google Scholar]
- 36.Sollid LM. Coeliac disease: Dissecting a complex inflammatory disorder. Nat Rev Immunol 2002; 2: 647–655. [DOI] [PubMed] [Google Scholar]
- 37.James EA, Moustakas AK, Bui J, et al. HLA-DR1001 presents “altered-self” peptides derived from joint-associated proteins by accepting citrulline in three of its binding pockets. Arthritis Rheum 2010; 62: 2909–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armstrong DL, Zidovetzki R, Alarcón-Riquelme ME, et al. GWAS identifies novel SLE susceptibility genes and explains the association of the HLA region. Genes Immun 2014; 15: 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsokos GC. Systemic lupus erythematosus. N Engl J Med 2011; 365: 2110–2121. [DOI] [PubMed] [Google Scholar]
- 40.Boumpas DT, Austin HA, 3rd, Vaughn EM, et al. Controlled trial of pulse methylprednisolone versus two regimens of pulse cyclophosphamide in severe lupus nephritis. Lancet 1992; 340: 741–745. [DOI] [PubMed] [Google Scholar]
- 41.Zampeli E, Klinman DM, Gershwin ME, Moutsopoulos HM. A comprehensive evaluation for the treatment of lupus nephritis. J Autoimmun 2017; 78: 1–10. [DOI] [PubMed] [Google Scholar]