Abstract
Objective
The comprehensive assaying of low-molecular-weight compounds, for example, metabolomics, provides a unique tool to uncover novel biomarkers and understand pathways underlying myocardial infarction (MI). We used a targeted metabolomics approach to identify biomarkers for MI and evaluate their involvement in the pathogenesis of MI.
Methods and results
Using three independent, prospective cohorts (KORA S4, KORA S2 and AGES-REFINE), totalling 2257 participants without a history of MI at baseline, we identified metabolites associated with incident MI (266 cases). We also investigated the association between the metabolites and high-sensitivity C reactive protein (hsCRP) to understand the relation between these metabolites and systemic inflammation. Out of 140 metabolites, 16 were nominally associated (p<0.05) with incident MI in KORA S4. Three metabolites, arginine and two lysophosphatidylcholines (LPC 17:0 and LPC 18:2), were selected as biomarkers via a backward stepwise selection procedure in the KORA S4 and were significant (p<0.0003) in a meta-analysis comprising all three studies including KORA S2 and AGES-REFINE. Furthermore, these three metabolites increased the predictive value of the Framingham risk score, increasing the area under the receiver operating characteristic score in KORA S4 (from 0.70 to 0.78, p=0.001) and AGES-REFINE study (from 0.70 to 0.76, p=0.02), but was not observed in KORA S2. The metabolite biomarkers attenuated the association between hsCRP and MI, indicating a potential link to systemic inflammatory processes.
Conclusions
We identified three metabolite biomarkers, which in combination increase the predictive value of the Framingham risk score. The attenuation of the hsCRP–MI association by these three metabolites indicates a potential link to systemic inflammation.
INTRODUCTION
Myocardial infarction (MI) is the leading cause of death worldwide,1 and identifying individuals at an increased risk for MI represents a major opportunity and challenge for prevention.2 High-sensitivity C reactive protein (hsCRP) is a promising biomarker for MI,3 and shows modest added value to conventional risk scores.4,5 However, a Mendelian randomisation study provided no evidence for a causal association between hsCRP and MI.6 Genome-wide association studies have identified loci associated with MI, but genetic risk scores have not substantially improved the predictive value of established models.7 Thus, the research community remains in need of predictive and causal biomarkers for MI.8
Metabolomics, the study of intermediate and/or end products of physiological processes, provides a novel tool to reveal pathways associated with metabolism and uncover biomarkers for cardiovascular diseases (CVD).9 Metabolites and metabolic risk scores are associated with incident CVD.10–15 Ganna et al used a non-targeted Liquid hromatography–Mass spectrometry (LC-MS)-based method and reported four metabolites associated with coronary heart disease (CHD).16 Additionally, variations in metabolite levels are associated with risk factors for CVD, such as type 2 diabetes.17,18 These findings underscore the capacity of metabolomics as a tool to explore pathophysiological processes associated with CVD and MI.
Although these previous studies revealed metabolites associated with CVD, they often employed relatively small metabolite panels or focused on high-risk individuals. Our study assesses multiple classes of metabolites in prospective population-based cohorts to identify metabolite biomarkers for incident MI. We also investigate the association between the metabolites and hsCRP, to understand the relationship between MI-associated metabolites and inflammation.
METHODS
Study population
The Cooperative Health Research in the Region of Augsburg (KORA) surveys are population-based studies conducted in Augsburg, Germany, and initiated as part of the WHO Multinational Monitoring of Trends and Determinants in Cardiovascular Diseases (MONICA) project.19 The baseline KORA survey 4 (S4) consists of 4261 individuals (aged 25–74 years) examined between 1999 and 2001.20 Serum metabolite profiling was performed on 1610 individuals (1545 without prior MI) aged 55 to 74 years.
Our outcome was a combined endpoint of incident fatal and non-fatal MI. All MI events were identified via the KORA Augsburg coronary event registry or through questionnaires for subjects residing outside the study area. Through 31 December 2000, the diagnosis of a major non-fatal MI event was based on the MONICA algorithm taking into account symptoms, cardiac enzymes and ECG changes. Afterwards, MI events were diagnosed by the European Society of Cardiology and American College of Cardiology criteria.21 Deaths from MI were validated by chart reviews, death certificates, autopsy reports and information from the last treating physician.22 hsCRP was measured by latex-enhanced nephelometry (Siemens, Germany).23 Based on a follow-up conducted through 2009, 67 KORA S4 participants (4.5%) had an incident MI.
We replicated metabolite biomarkers found in KORA S4 in both non-fasting (KORA S2) and fasting (AGES and REFINE) cohorts. KORA S2 is a baseline survey of 4940 participants examined between 1989 and 1990.23 We constructed a case–cohort replication population from all incident MI cases aged <75 years, identified before 2003 (n=112) and a subcohort of randomly selected participants without prevalent MI at baseline (n=549).24 hsCRP was measured using a high-sensitivity immunordiometric assay for men aged 45–74 and a high-sensitivity latex-enhanced nephelometric assay for mean aged 35–44 and all women. Additional details can be found in the online supplementary methods.
The Age, Gene/Environment Susceptibility (AGES)-Reykjavik and the Risk Evaluation For Infarct Estimates (REFINE)-Reykjavik studies25 are two separate studies that were merged to create a nested case–control sample of fasting individuals. After merging, we selected an age and sex-matched nested case–control sample of 87 participants with incident MI and 167 participants without MI within 6 years of follow-up.25 hsCRP was measured on a Hitachi 912, using reagents from Roche Diagnostics (Mannheim, Germany).
All participants gave written informed consent, and all studies were approved by their respective ethics committees.
Population characteristics for all studies are given in table 1, and the timeline of baseline assessment and follow-up for all cohorts is given in online supplementary figure S1. Further details for all cohorts are given in the online supplementary materials.
Table 1.
Population characteristics of the discovery and replication cohorts
| Clinical parameters |
KORA S4 | KORA S2 | AGES-REFINE | |||
|---|---|---|---|---|---|---|
| Incident MI | non-MI | Incident MI | non-MI | Incident MI | non-MI | |
| n=67 | n=1275 | n=112 | n=549 | n=87 | n=167 | |
| Age | 65.5±5.2 | 63.8±5.4 | 59.7±7.5 | 53.6±11.0 | 65.6±8.4 | 65.8±7.9 |
| Sex, male, n, % | 52 (77.9) | 623 (48.9) | 85 (75.9) | 271 (49.4) | 53 (61.0) | 103 (62.0) |
| BMI, kg/m2 | 29.6±4.8 | 28.3±4.2 | 28.3±3.9 | 27.1±4.0 | 28.1±4.3 | 27.7±5.4 |
| Waist-to-hip ratio | 1.0±0.1 | 0.9±0.1 | 0.9±0.1 | 0.9±0.1 | 1.0±0.1* | 1.0±0.1† |
| Type 2 diabetes, n (%) | 14 (20.9) | 104 (8.2) | 20 (17.9) | 18 (3.3) | 12 (13.8) | 17 (10.2) |
| Smoking | ||||||
| Non-smoker, n (%) | 26 (38.2) | 663 (49.7) | 32 (28.6) | 269 (49.0) | 21 (24.1) | 62 (37.1) |
| Former smoker, n (%) | 30 (44.1) | 463 (36.3) | 42 (37.5) | 151 (27.5) | 41 (47.1) | 67 (40.1) |
| Smoker, n (%) | 12 (17.7) | 117 (13.9) | 38 (33.9) | 129 (23.5) | 25 (28.7) | 39 (23.4) |
| Alcohol intake, n (%)‡ | 12 (17.7) | 262 (20.6) | 28 (25) | 153 (27.9) | 9 (18.0)§ | 24 (25.0)¶ |
| Physical activity, n (%)** | 16 (23.5) | 560 (43.9) | 30 (26.78) | 208 (37.7) | 26 (30.0) | 69 (42.0) |
| Systolic BP, mm Hg | 142.9±24.3 | 135.2±19.8 | 143.8±22.1 | 133.4±18.4 | 138.1±21.5 | 134.4±18.7 |
| Total cholesterol, mg/dL | 240.4±38.7 | 244.1±41.2 | 260.7±50.4 | 240.3±43.2 | 228.1±50.3 | 212.9±37.2 |
| HDL-C, mg/dL | 53.1±16.8 | 58.9±16.5 | 50.8±15.5 | 58.3±16.7 | 54.8±17.1 | 61.2±18.5 |
| hsCRP, mg/L‡‡ | 2.9±5.2 | 1.7±6.6 | 3.0±6.4 | 1.6±7.8 | 2.3±8.1 | 1.7±4.6 |
| Statin user, n (%) | 4 (6.0) | 109 (8.5) | 0 (0) | 3 (0.5) | 8 (9.0) | 25 (15.0) |
Data available in 37 participants with incident MI.
Data available in 71 participants.
≥20 g/day for women; ≥40 g/day for men.
Data available in 49 participants.
Data available in 95 participants.
>1 hour per week.
Geometric mean.
Mean ± SD is provided when appropriate.
BMI, body mass index; BP, blood pressure; HDL-C, high-density lipoprotein cholesterol; hsCRP, high-sensitivity C reactive protein; KORA, Cooperative Health Research in the Region of Augsburg; MI, myocardial infarction; AGES-REFINE, Age, Gene/Environment Susceptibility and the Risk Evaluation For Infarct Estimates-Reykjavik studies.
Metabolite quantification
Serum samples from KORA participants were assayed for 188 metabolites using the AbsoluteIDQ p180 Kit (BIOCRATES Life Sciences AG, Innsbruck, Austria, online supplementary methods). The targeted metabolomics approach allows simultaneous quantification of 188 metabolites using liquid chromatography and flow injection analysis–mass spectrometry. Serum samples from AGES and REFINE Reykjavik studies were assayed with the Biocrates AbsoluteIDQ p150 Kit. Identical quality control (QC) procedures were used for all studies.18 Each analysed metabolite met two criteria: (1) a coefficient of variance of <25% in the reference samples and (2) 50% of all measured sample concentrations above 3× the median of the zero samples. The metabolites that met QC included one hexose (H1), 21 acylcarnitines, 21 amino acids, 8 biogenic amines, 13 sphingomyelins, 33 diacyl (aa) phosphatidylcholines (PCs), 35 acyl-alkyl (ae) PCs and 8 lysophosphatidylcholines (LPCs) (see online supplementary table S1). Metabolite concentrations are given in micromolar and were natural-log transformed for all analyses.
Metabolite biomarker discovery
The study design is shown in figure 1, and online supplementary figure S2 contains an overview of the analytical strategy. Cox regression models were used to assess the association between metabolite concentration and incident MI. We used a basic model adjusting for age and sex, followed by a multivariable model additionally adjusting for body mass index, smoking status, alcohol intake, diabetes, systolic blood pressure, high-density lipoprotein cholesterol and total cholesterol. To examine the relation between the metabolites and systemic inflammation, we added log-transformed hsCRP to the multivariable model, and examined the change in the MI–hsCRP association when the metabolites were additionally added to this model. Sensitivity analyses were conducted to understand the effects of statin medication and diabetes.
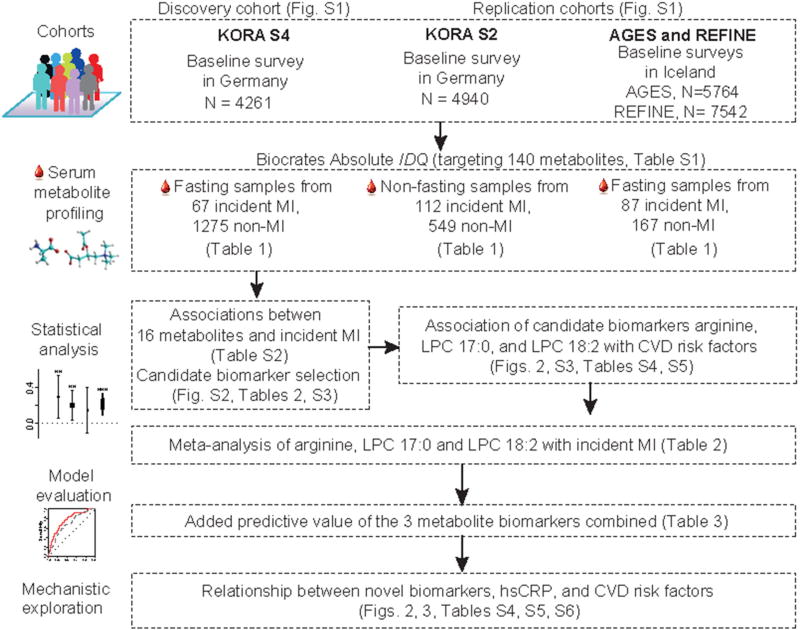
Figure 1.
Cohort, metabolite profiling and analysis outline. Description of the cohorts, metabolite profiling, statistical analysis, model evaluation and mechanistic exploration for this study. CVD, cardiovascular disease; hsCRP, high-sensitivity C reactive protein; KORA, Cooperative Health Research in the Region of Augsburg; LPC, lysophosphatidylcholine; MI, myocardial infarction.
Potential biomarkers were selected from metabolites with p<0.05 in all three Cox regression models via a two-step process in KORA S4. We used a nominal p value cut-off to retain metabolites that might be significant predictors in a multivariate model despite not independently having a multiple-test significant p value. First, metabolites with non-zero β estimations after L1-regularised estimation were retained. A priori we chose to filter these metabolites via Akaike information criterion (AIC)-guided backward stepwise regression, following commonly used practices. The metabolites remaining after this step were referred to as the potential biomarkers.
We obtained estimates of the predictive capabilities of the metabolite biomarkers by including them in models containing the Framingham risk score (FRS)26 in KORA S4 and also adding them to the multivariable model in KORA S4. Increased predictive power was replicated in KORA S2 and the AGES and REFINE Reykjavik studies (see online supplementary figure S2). In KORA, the oversampling of cases was accounted for by weighting the subcohort samples by the inverse of the sampling probability.24 The AGES and REFINE Reykjavik studies were analysed using conditional logistic regression models. We designated our potential biomarkers from KORA S4 as metabolite biomarkers if they significantly increased incident MI prediction when added to the FRS as measured by the area under the receiver operating characteristic curve (AUC). Sensitivity analyses were performed to evaluate the influence of potential confounders on the metabolite biomarkers.
Further details of statistical methods and sensitivity analysis are described in the supplemental methods. All statistical analyses were performed in R,27 using the packages penalised,28 pROC29 and PredictABEL.30
RESULTS
Identification of metabolite biomarkers for incident MI
Sixteen metabolites were associated (p<0.05) with incident MI in the basic and multivariable models in KORA S4 (see online supplementary table S2). These 16 metabolites belong to three classes (amino acids, LPCs, PCs), and displayed high intraclass correlations and low interclass correlations. The PCs were modestly correlated with total cholesterol in KORA S4 and S2, while the LPCs were negatively correlated with hsCRP levels (see online supplementary figure S3). Arginine, LPC 17:0 and LPC 18:2 were identified as potential biomarkers via AIC-guided backwards stepwise selection in KORA S4. In the KORA S4 multivariable model, an SD increase in log-transformed arginine, LPC 17:0 and LPC 18:2 was associated with a +35%, −31% and −35% risk of incident MI, respectively (table 2).
Table 2.
Risk of incident myocardial infarction via Cox regression models according to individual metabolite biomarkers
| Metabolites | KORA S4 | KORA S2 | AGES-REFINE | Meta-analysis | ||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Basic model | ||||||||
| Arginine | 1.40 (1.10 to 1.78) | 0.006 | 1.26 (1.05 to 1.52) | 0.01 | 1.16 (0.90 to 1.50) | 0.25 | 1.27 (1.12 to 1.43) | 5.2×10−4 |
| LPC 17:0 | 0.65 (0.52 to 0.82) | 2.5×10−4 | 0.77 (0.64 to 0.93) | 0.01 | 0.91 (0.69 to 1.19) | 0.48 | 0.77 (0.65 to 0.91) | 0.002 |
| LPC 18:2 | 0.71 (0.56 to 0.91) | 5.3×10−5 | 0.67 (0.56 to 0.82) | 4.8×10−5 | 0.65 (0.48 to 0.88) | 0.005 | 0.65 (0.57 to 0.74) | 1.7×10−5 |
| Multivariable model | ||||||||
| Arginine | 1.35 (1.06 to 1.72) | 0.016 | 1.23 (1.02 to 1.47) | 0.03 | 1.23 (0.93 to 1.64) | 0.15 | 1.26 (1.11 to 1.43) | 3.0×10−4 |
| LPC 17:0 | 0.69 (0.53 to 0.89) | 0.005 | 0.72(0.52 to 0.090) | 0.004 | 0.95 (0.68 to 1.34) | 0.78 | 0.76 (0.66 to 0.88) | 3.0×10−4 |
| LPC 18:2 | 0.65 (0.49 to 0.87) | 0.004 | 0.79 (0.64 to 0.96) | 0.02 | 0.66 (0.46 to 0.95) | 0.02 | 0.73 (0.63 to 0.85) | 2.7×10−5 |
The basic model was adjusted for age and sex. The multivariable model was additionally adjusted for body mass index diabetes, systolic blood pressure, smoking status, alcohol intake, total cholesterol, high-density lipoprotein cholesterol. Values are provided as estimated HR (95% CI). CI given for a 1 SD increase/decrease in the respective metabolite.
LPC, lysophosphatidylcholine; KORA, Cooperative Health Research in the Region of Augsburg.
Arginine, LPC (17:0) and LPC (18:2) were significant (p<0.0003) in the meta-analysis of all three cohorts (table 2). The direction of the association was consistent in all cohorts where increased arginine concentrations and decreased concentrations of LPC (17:0) and LPC (18:2) were associated with increased risk of incident MI.
Increased predictive capability of metabolite biomarkers
When added to the FRS in KORA S4, the three potential metabolite biomarkers significantly improved MI risk prediction as the AUC increased from 0.70 to 0.78 (p=0.001, table 3). When calibrated the FRS components’ coefficients to KORA S4, the KORA S4 AUC remained significantly improved (AUC: 0.68 to 0.73, p=0.007). We replicated associations in AGES-REFINE (AUC: 0.70 to 0.76, p=0.02, table 3), but not in the non-fasting KORA S2 cohort (AUC: 0.74 to 0.77, p=0.14). The AUC, net reclassification index and integrated discrimination improvement when the metabolite biomarkers were added to both the multivariable model and FRS are given in table 3. Individually, none of the metabolite biomarkers significantly increased the AUC in any cohort (data not shown), and only LPC 18:2 was independently associated with MI in all cohorts (table 2). The metabolite–MI associations are independent of diabetes status and statin users, and remain when the limited non-fasting samples were included in KORA S4 (see online supplementary table S3). Based on these results term, these three metabolites ‘metabolite biomarkers’ (figure 1, see online supplementary figure S2).
Table 3.
Added value of three candidate metabolites in prediction of incident myocardial infarction in the three studies
| AUC (95 % CI) | p Value | NRI (95 % CI) | IDI (95 % CI) | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| Categorical* | p Value | Continuous | p Value | |||||
| KORA S4 | ||||||||
| Multivariable model | 0.68 (0.62 to 0.74) | |||||||
| +3 metabolites | 0.73 (0.67 to 0.79) | 0.007 | 0.17 (0.04 to 0.30) | 0.01 | 0.61 (0.40 to 0.84) | 1.24×10−4 | 0.10 (0.06 to 0.14) | 9.58×10−7 |
| Framingham score | 0.70 (0.64 to 0.76) | |||||||
| +3 metabolites | 0.78 (0.73 to 0.84) | 0.001 | 0.22 (0.09 to 0.36) | 9.1×10−4 | 0.62 (0.42 to 0.81) | 1.23×10−9 | 0.15 (0.10 to 0.20) | 4.1×10−9 |
| KORA S2 | ||||||||
| Multivariable model | 0.76 (0.72 to 0.81) | |||||||
| +3 metabolites | 0.77 (0.73 to 0.82) | 0.33 | 0.07 (−0.04 to 0.18) | 0.20 | 0.32 (0.12 to 0.52) | 0.002 | 0.02 (0.00 to 0.04) | 0.02 |
| Framingham score | 0.74 (0.70 to 0.79) | |||||||
| +3 metabolites | 0.77 (0.72 to 0.80) | 0.14 | 0.10 (−0.01 to 0.20) | 0.06 | 0.35 (0.15 to 0.55) | 6.03×10−4 | 0.01 (−0.01 to 0.02) | 0.21 |
| AGES-REFINE | ||||||||
| Multivariable model | 0.76 (0.71 to 0.82) | |||||||
| +3 metabolites | 0.80 (0.75 to 0.86) | 0.02 | 0.07 (−0.04 to 0.17) | 0.20 | 0.50 (0.25 to 0.74) | 8.85×10−5 | 0.06 (0.03 to 0.09) | 8.85×10−5 |
| Framingham score | 0.70 (0.63 to 0.77) | |||||||
| +3 metabolites | 0.76 (0.69 to 0.82) | 0.02 | 0.15 (0.07 to 0.24) | 2.37×10−4 | 0.40 (0.16 to 0.65) | 0.001 | 0.06 (0.03 to 0.09) | 8.85×10−5 |
Categories were set at 0%–10%, 11%–20% and more than 21%. AUC, NRI and IDI with 95% CI are provided in the table. The multivariable model adjusted for age, sex, body mass index, diabetes, systolic blood pressure, smoking, alcohol consumption, total cholesterol and high-density lipoprotein cholesterol.
AUC, area under the receiver operating characteristic curve; IDI, integrated discrimination improvement; KORA, Cooperative Health Research in the Region of Augsburg; NRI, net reclassification index; AGES-REFINE, Age, Gene/Environment Susceptibility and the Risk Evaluation For Infarct Estimates-Reykjavik studies.
Association of metabolite biomarkers with hsCRP
To examine links with inflammation, we evaluated the association between the metabolite biomarkers and hsCRP and their attenuation of MI–hsCRP associations. The three metabolites were significantly correlated (Spearman) with each other and with hsCRP with the exception of arginine which was strongly correlated with hsCRP but weakly correlated with LPC 17:0 and LPC 18:2 (see online supplementary table S4). hsCRP was associated with each metabolite biomarker in all three cohorts (see online supplementary table S5).
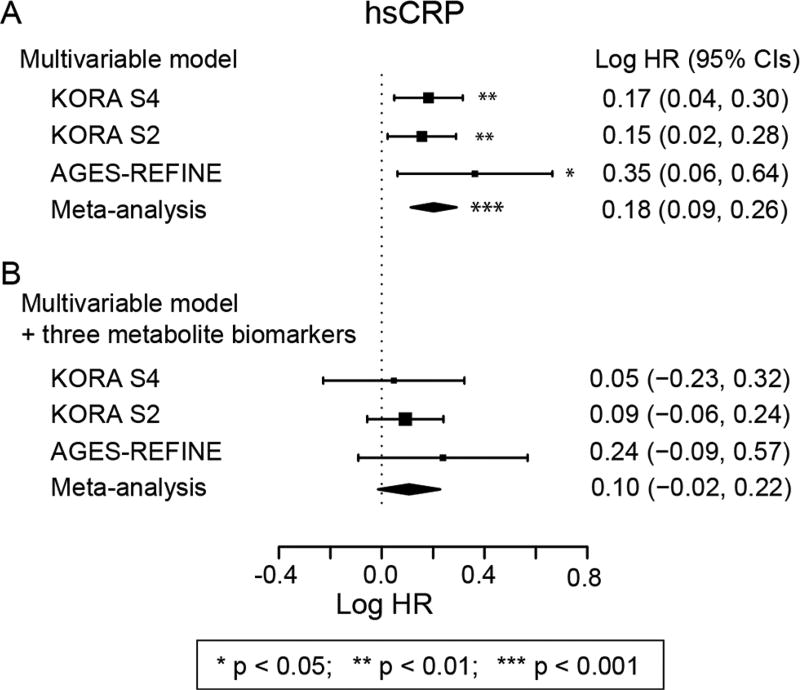
hsCRP was significantly associated with incident MI in the multivariable model (figure 2A). Addition of the metabolite biomarkers removed this association (figure 2B), but did not attenuate the association with MI for other clinical covariates (see online supplementary table S6). This attenuation implies that a substantial proportion of the hsCRP–MI association was mediated by these three metabolite biomarkers. Conversely, the metabolites were only slightly attenuated by the addition of hsCRP to the multivariable model (see online supplementary table S2).
Figure 2.
Attenuation of high-sensitivity C reactive protein–myocardial infarction (hsCRP–MI) association by metabolite biomarkers. Association of hsCRP with incident MI in the multivariable model (A) and in the multivariable model after including the three metabolite biomarkers (B). The multivariable model adjusted for age, sex, body mass index, alcohol consumption, systolic blood pressure, diabetes status, total cholesterol and high-density lipoprotein cholesterol. *=p<0.05, **=p<0.01, ***=p<0.001. KORA, Cooperative Health Research in the Region of Augsburg.
DISCUSSION
Using a targeted metabolomics approach across three cohorts, we identified arginine, LPC 17:0 and LPC 18:2 as metabolite biomarkers for incident MI. Adding arginine, LPC 17:0 and LPC 18:2 to the FRS significantly increased the predictive value in fasting cohorts and in a meta-analysis of the three cohorts. These metabolite biomarkers were independent of conventional risk factors for MI, but strongly attenuated the hsCRP–MI associations indicating a potential role in systemic inflammation. Although this is the first time that these three metabolites in combination have been suggested as biomarkers for incident MI, each has been linked to CVD.31–35
Arginine
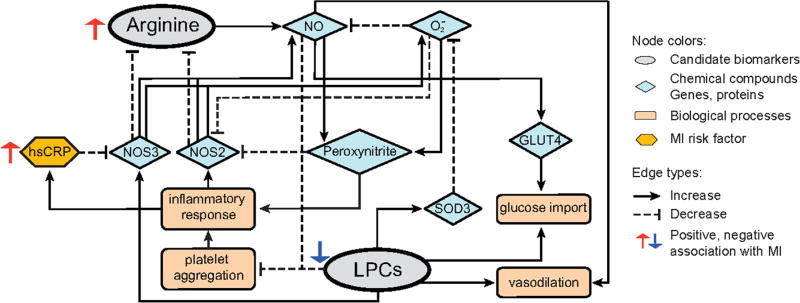
An SD increase in serum arginine concentrations was associated with a 26% higher risk of incident MI risk in the meta-analysis. Ornithine, a product of splitting urea from arginine, is associated with CHD.36 A possible mechanism linking arginine and inflammation is production of oxygen radicals (O2−) and peroxynitrite. Arginine is metabolised to NO via the NO synthases NOS3 and NOS2 in vascular cells.37 In the presence of high concentrations of O2−, NO reacts with O2− to generate peroxynitrite, reducing the bioavailability of NO and contributing to inflammatory processes.38,39 Additionally, peroxynitrite and O2− trigger the uncoupling of NO synthases, shifting the production of NO to O2−,40 creating a feedback loop (figure 3).
Figure 3.
Pathway annotation of metabolite biomarkers. Based on the literature references, we diagram the NO-O2−–peroxynitrite biochemical process which links the three candidate biomarkers. GLUT4, glucose transporter type 4; hsCRP, high-sensitivity C reactive protein; LPC, lysophosphatidylcholine; MI, myocardial infarction; NO, nitric oxide; NOS2, inducible nitric oxide synthase; NOS3, endothelial nitric oxide synthase; SOD3, extracellular superoxide dismutase.
Lysophosphatidylcholines
LPCs are associated with lipid metabolism, inflammation and MI.31–34,41,42 In our study, participants with higher serum LPC 17:0 and LPC 18:2 concentrations had a lower risk of MI. LPCs also have a negative association with a combined CVD outcome of MI, ischaemic stroke and sudden cardiac death,14 further supporting our findings, and LPC 18:2 is associated with CHD.16
LPCs are associated with inflammatory processes via a variety of mechanisms (figure 3). LPCs can increase expression of NOS3, which is involved in NO synthesis.31 LPCs can also drive production of antioxidant enzymes, for example, SOD3, which reduce superoxide anion concentrations (figure 3).32 As a consequence, lower LPC concentrations may increase oxidative stress and promote inflammation as indicated by the negative association with hsCRP. Finally, LPCs increase the synthesis of prostaglandin I2 resulting in improved vasodilation.34,41 Thus, lower LPC concentrations may contribute to endothelial dysfunction, increasing the risk for MI.
High-sensitivity C reactive protein
hsCRP is a measure of systemic inflammation. Our three metabolite biomarkers explained 10% of the variation in hsCRP concentrations and attenuated the hsCRP–MI association coefficient by 34%–74%, making it no longer significant (see online supplementary table S6). Therefore, inflammation likely represents a pathway through which our novel biomarkers are associated with MI.
MI risk prediction and previous CVD studies
Metabolomics can potentially reveal causal biomarkers with strong predictive value for CVD,9 and baseline metabolomics profiles are associated with risk of death or incident MI.11 Recently, there have been several metabolomic studies using general population-based cohorts.14–16,36 Stegemann et al used a shotgun lipidomics approach to identify lipid species associated with incident CVD.14 Würtz et al used a Nuclear magnetic resonance (NMR)-based approach to identify CVD-associated metabolites and calculate a risk score for CVD.15 Vaarhorst et al also used an NMR-based approach and identified metabolites associated with incident CHD; however, their score did not significantly increase CHD risk prediction.36 Ganna et al used an LC-MS/MS-based lipidomics approach and associated LPC 18:2 with CHD.16 Covering 140 metabolites, our MS approach is more comprehensive than that taken by Stegemann et al (135 metabolites), Vaarhorst et al (100 metabolites) or Würtz et al (68 metabolites), and with 266 incident MI events, our study is the largest by the number of MI events. Additionally, while previous studies assessed a combined cardiovascular endpoint,11,14,36 we exclusively analysed incident MI.
Strengths and limitations of our study
A strength of our study is its prospective design using general population-based cohorts. Study participants were initially free of MI, and thus we analysed only incident events. All samples were handled in a similar, unbiased manner. We controlled for a variety of demographic characteristics, lifestyle factors, medication and clinical variables. All data were collected and verified by trained personnel16,23,25 to reduce measurement error. We applied stringent QC procedures to our metabolite assays,18 and observed no sources of systematic error. Though nested case–cohort and case–control designs are less efficient than assaying the entire cohort,43 we still observed associations with our approach. Also, associations for the LPC 18:2 and the metabolite score were similar for KORA and AGES-REFINE despite reports of potential bias with stratified case–control designs.43
A limitation of using backward stepwise selection to reduce the set of predictors is that the pruning is done in an agnostic manner. Thus, the predictors selected may not represent the most meaningful or causal metabolites, but may be markers for unmeasured or unselected metabolites. We were unable to control for dietary intake, which is associated with incident MI.44 This may have affected our inability to identify associations with metabolites influenced by diet, for example, tryptophan.45,46 Additionally, the non-fasting status of the KORA S2 cohort may have introduced variability that obscured associations in this cohort. We suggest that future studies of MI and metabolic profiles collect detailed dietary information and use fasting samples.
A key point of all biomarker discovery studies is generalisability. We incorporated samples from Germany and Iceland indicating that our associations generalise across European populations. Although we provide substantial evidence for improved baseline discrimination for future incident MI for our novel biomarkers, future studies are needed to formulate and validate an incident MI risk scoring function based on these metabolite biomarkers.
CONCLUSION
We identified three metabolite biomarkers associated with incident MI which improved MI prediction when added to the FRS. We replicated this MI prediction improvement in independent fasting samples. These three metabolites were associated with hsCRP and attenuated the hsCRP–MI association while being independent of other traditional CVD risk factors, indicating a potential link to systemic inflammation.
Supplementary Material
Key messages.
What is already known on this subject?
Metabolites have been associated with cardiovascular disease (CVD); however, the majority of published studies have focused on combined events (eg, myocardial infarction (MI), stroke, and sudden cardiac death) or limited, non-targeted metabolite panels. Large population-based studies using targeted metabolomics data are limited. There are also limited studies showing improvement in CVD risk assessment when metabolites are added to CVD risk scores.
What might this study add?
This study used a targeted metabolomics assay to uncover metabolites associated with incident MI. When assessed in fasting samples, three of these metabolites (arginine, lysophosphatidylcholines (LPC 17:0 and LPC 18:2)) significantly improved risk prediction as compared with the Framingham risk score. LPC 18:2 has been previously associated with a combined CVD outcome, and this study adds to these results by demonstrating the LPC 18:2 association specifically in MI in the largest study to date. Conditional analyses showed that the metabolites substantially attenuated the C reactive protein–MI association, and thus may be biochemical mediators of inflammation.
How might this impact clinical practice?
This study establishes that the addition of metabolite biomarkers to the Framingham risk score gives an improved assessment of risk in fasting samples. The association of the metabolite biomarkers with inflammation and mediation of C reactive protein associations suggests that they are new targets for studying the inflammation-driven pathophysiology of MI.
Acknowledgments
The authors express appreciation to all Cooperative Health Research in the Region of Augsburg (KORA) study participants for donating their blood and time. The authors thank the field staff in Augsburg conducting the KORA studies. The authors thank the staff from the Institute of Epidemiology at the Helmholtz Zentrum München, and the Metabolomics Platform of the Genome Analysis Center, who helped in the sample logistics, data collection and metabolomic measurements, particularly J. Scarpa, K. Sckell, F. Scharl, N. Lindemann, H. Chavez, A. Sabunchi, A. Schneider, A. Ludolph, S. Jelic and B. Langer.
Funding The KORA study was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of in part by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant 01ZX1313A-2014). Part of this project was supported by EU FP7 grants HEALTH-2009-2.2.1-3/242114 (Project OPTiMiSE) and HEALTH-2013-2.4.2-1/602936 (Project CarTarDis). Part of the work was funded by the German Ministry of Education and Research (Agreement No 01ZX1313C; DZHK MH5.1; MH5.2). KS is supported by ‘Biomedical Research Program’ funds at Weill Cornell Medical College in Qatar, a programme funded by the Qatar Foundation; The Age, Gene/Environment Susceptibility Reykjavik Study is funded by NIH contract N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association) and the Althingi (the Icelandic Parliament). IRH was supported by the PhD research fund at the University of Iceland. VG, TA and VE were supported by Icelandic Research Foundation grant (141101-051) at RANNIS.
Footnotes
Contributors Conception: CKW-C, TX, RW-S; study design: CKW-C, TX, RW-S, AP; data acquisition: TX, TA, BT, CM, ID-K, AZ, ZY, IRH, TBH, LJL, AG, LL, GE, MW, CP, KS, TI, JA, AR, WK, VG, VE; data analysis: TX, TA, AG, RW-S; data interpretation: CKW-C, TX, RW-S, VG, VE, AP, ID-K, CM, AR, WK; manuscript drafting: CKW-C, TX, RW-S; critical revision: CKW-C, TX, VG, WK, VE, RW-S, AP.
Competing interests None declared.
Ethics approval This study involved data collected from human subjects. The collection and analysis of data from KORA was approved by the ethics committee of the Bavarian Medical Association, Germany. The usage of data for Age, Gene/Environment Susceptibility (AGES)-Reykjavik and the Risk Evaluation For Infarct Estimates (REFINE)-Reykjavik studies are approved by the Icelandic National Bioethics Committee (VSN: 00-063, VSN:05-112 respectively) and the Data Protection Authority.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Yeh RW, Sidney S, Chandra M, et al. Population trends in the incidence and outcomes of acute myocardial infarction. N Engl J Med. 2010;362:2155–65. doi: 10.1056/NEJMoa0908610. [DOI] [PubMed] [Google Scholar]
- 2.Ajani UA, Ford ES. Has the risk for coronary heart disease changed among U.S. adults? J Am Coll Cardiol. 2006;48:1177–82. doi: 10.1016/j.jacc.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 3.Kaptoge S, Di Angelantonio E, Pennells L, et al. Emerging Risk Factors Collaboration. C-reactive protein, fibrinogen, and cardiovascular disease prediction. N Engl J Med. 2012;367:1310–20. doi: 10.1056/NEJMoa1107477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ioannidis JP, Tzoulaki I. Minimal and null predictive effects for the most popular blood biomarkers of cardiovascular disease. Circ Res. 2012;110:658–62. doi: 10.1161/RES.0b013e31824da8ad. [DOI] [PubMed] [Google Scholar]
- 5.D’Agostino RB, Vasan RS, Pencina MJ, et al. General cardiovascular risk profile for use in primary care: the framingham heart study. Circulation. 2008;117:743–53. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 6.Wensley F, Burgess S, Burgess S, et al. C Reactive Protein Coronary Heart Disease Genetics Collaboration (CCGC) Association between C reactive protein and coronary heart disease: mendelian randomisation analysis based on individual participant data. Bmj. 2011;342:d548. doi: 10.1136/bmj.d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nabel EG, Braunwald E. A tale of coronary artery disease and myocardial infarction. N Engl J Med. 2012;366:54–63. doi: 10.1056/NEJMra1112570. [DOI] [PubMed] [Google Scholar]
- 9.Hoefer IE, Steffens S, Ala-Korpela M, et al. ESC Working Group Atherosclerosis and Vascular Biology. Novel methodologies for biomarker discovery in atherosclerosis. Eur Heart J. 2015;36:2635–42. doi: 10.1093/eurheartj/ehv236. [DOI] [PubMed] [Google Scholar]
- 10.Cheng S, Rhee EP, Larson MG, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125:2222–31. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah SH, Bain JR, Muehlbauer MJ, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3:207–14. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- 12.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–20. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Würtz P, Raiko JR, Magnussen CG, et al. High-throughput quantification of circulating metabolites improves prediction of subclinical atherosclerosis. Eur Heart J. 2012;33:2307–16. doi: 10.1093/eurheartj/ehs020. [DOI] [PubMed] [Google Scholar]
- 14.Stegemann C, Pechlaner R, Willeit P, et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based bruneck study. Circulation. 2014;129:1821–31. doi: 10.1161/CIRCULATIONAHA.113.002500. [DOI] [PubMed] [Google Scholar]
- 15.Würtz P, Havulinna AS, Soininen P, et al. Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation. 2015;131:774–85. doi: 10.1161/CIRCULATIONAHA.114.013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganna A, Salihovic S, Sundström J, et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014;10:e1004801. doi: 10.1371/journal.pgen.1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang-Sattler R, Yu Z, Herder C, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keil U, Liese AD, Hense HW, et al. Classical risk factors and their impact on incident non-fatal and fatal myocardial infarction and all-cause mortality in southern Germany. results from the MONICA augsburg cohort study 1984–1992. monitoring trends and determinants in cardiovascular diseases. Eur Heart J. 1998;19:1197–207. doi: 10.1053/euhj.1998.1089. [DOI] [PubMed] [Google Scholar]
- 20.Holle R, Happich M, Löwel H, et al. KORA - A research platform for population based health research. Gesundheitswesen. 2005;67:19–25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 21.Alpert JS, Thygesen K, Antman E, et al. Myocardial infarction redefined--a consensus document of the joint european society of cardiology/American college of cardiology committee for the redefinition of myocardial infarction. J Am Coll Cardiol. 2000;36:959–69. doi: 10.1016/s0735-1097(00)00804-4. [DOI] [PubMed] [Google Scholar]
- 22.Karakas M, Zierer A, Herder C, et al. Leptin, adiponectin, their ratio and risk of coronary heart disease: results from the MONICA/KORA augsburg study 1984–2002. Atherosclerosis. 2010;209:220–5. doi: 10.1016/j.atherosclerosis.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Koenig W, Khuseyinova N, Baumert J, et al. Prospective study of high-sensitivity C-reactive protein as a determinant of mortality: results from the MONICA/KORA augsburg cohort study, 1984–1998. Clin Chem. 2008;54:335–42. doi: 10.1373/clinchem.2007.100271. [DOI] [PubMed] [Google Scholar]
- 24.Barlow WE, Ichikawa L, Rosner D, et al. Analysis of case-cohort designs. J Clin Epidemiol. 1999;52:1165–72. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 25.Harris TB, Launer LJ, Eiriksdottir G, et al. Age, gene/Environment Susceptibility-Reykjavik study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165:1076–87. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Agostino RB, Vasan RS, Pencina MJ, et al. General cardiovascular risk profile for use in primary care: the framingham heart study. Circulation. 2008;117:743–53. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 27.Core Team R. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. http://www.R-project.org/ [Google Scholar]
- 28.Goeman JJ. L1 penalized estimation in the cox proportional hazards model. Biom J. 2010;52:70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- 29.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kundu S, Aulchenko YS, van Duijn CM, et al. PredictABEL: an R package for the assessment of risk prediction models. Eur J Epidemiol. 2011;26:261–4. doi: 10.1007/s10654-011-9567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cieslik K, Zembowicz A, Tang JL, et al. Transcriptional regulation of endothelial nitric-oxide synthase by lysophosphatidylcholine. J Biol Chem. 1998;273:14885–90. doi: 10.1074/jbc.273.24.14885. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto M, Hara H, Adachi T. The expression of extracellular-superoxide dismutase is increased by Lysophosphatidylcholine in human monocytic U937 cells. Atherosclerosis. 2002;163:223–8. doi: 10.1016/s0021-9150(02)00007-2. [DOI] [PubMed] [Google Scholar]
- 33.Fukai T, Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal. 2011;15:1583–606. doi: 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zembowicz A, Jones SL, Wu KK, KK W. Induction of cyclooxygenase-2 in human umbilical vein endothelial cells by lysophosphatidylcholine. J Clin Invest. 1995;96:1688–92. doi: 10.1172/JCI118211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lechner M, Höhn V, Brauner B, et al. CIDeR: multifactorial interaction networks in human diseases. Genome Biol. 2012;13:R62. doi: 10.1186/gb-2012-13-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaarhorst AA, Verhoeven A, Weller CM, et al. A metabolomic profile is associated with the risk of incident coronary heart disease. Am Heart J. 2014;168:e7, 45–52. doi: 10.1016/j.ahj.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Knowles RG, Moncada S. Nitric oxide synthases in mammals. Biochem J. 1994;298:249–58. doi: 10.1042/bj2980249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mungrue IN, Husain M, Stewart DJ. The role of NOS in heart failure: lessons from murine genetic models. Heart Fail Rev. 2002;7:407–22. doi: 10.1023/a:1020762401408. [DOI] [PubMed] [Google Scholar]
- 39.Reiter CD, Teng RJ, Beckman JS. Superoxide reacts with nitric oxide to nitrate tyrosine at physiological pH via peroxynitrite. J Biol Chem. 2000;275:32460–6. doi: 10.1074/jbc.M910433199. [DOI] [PubMed] [Google Scholar]
- 40.Sun J, Druhan LJ, Zweier JL. Reactive oxygen and nitrogen species regulate inducible nitric oxide synthase function shifting the balance of nitric oxide and superoxide production. Arch Biochem Biophys. 2010;494:130–7. doi: 10.1016/j.abb.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riederer M, Ojala PJ, Hrzenjak A, et al. Acyl chain-dependent effect of lysophosphatidylcholine on endothelial prostacyclin production. J Lipid Res. 2010;51:2957–66. doi: 10.1194/jlr.M006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Gaal LF, Mertens IL, De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006;444:875–80. doi: 10.1038/nature05487. [DOI] [PubMed] [Google Scholar]
- 43.Ganna A, Reilly M, de Faire U, et al. Risk prediction measures for case-cohort and nested case-control designs: an application to cardiovascular disease. Am J Epidemiol. 2012;175:715–24. doi: 10.1093/aje/kwr374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estruch R, Ros E, Salas-Salvadó J, et al. PREDIMED Study Investigators. Primary prevention of cardiovascular disease with a mediterranean diet. N Engl J Med. 2013;368:1279–90. doi: 10.1056/NEJMc1806491. [DOI] [PubMed] [Google Scholar]
- 45.Krug S, Kastenmüller G, Stückler F, et al. The dynamic range of the human metabolome revealed by challenges. Faseb J. 2012;26:2607–19. doi: 10.1096/fj.11-198093. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa M, Maekawa K, Saito K, et al. Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects’ gender and age. PLoS One. 2014;9:e91806. doi: 10.1371/journal.pone.0091806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.