Abstract
Mechanics of the extracellular matrix (ECM) play a pivotal role in governing cell behavior, such as cell spreading and differentiation. ECM mechanics have been recapitulated primarily in elastic hydrogels, including with dynamic properties to mimic complex behaviors (e.g., fibrosis); however, these dynamic hydrogels fail to introduce the viscoelastic nature of many tissues. Here, we developed a two-step crosslinking strategy to first form (via platinum-catalyzed crosslinking) networks of polydimethylsiloxane (PDMS) and then to increase PDMS crosslinking (via thiol-ene click reaction) in a temporally-controlled manner. This photoinitiated reaction increased the compressive modulus of PDMS up to 10-fold within minutes and was conducted under cytocompatible conditions. With stiffening, cells displayed increased spreading, changing from ~1300 to 1900 μm2 and from ~2700 to 4600 μm2 for fibroblasts and mesenchymal stem cells, respectively. In addition, higher myofibroblast activation (from ~2 to 20%) for cardiac fibroblasts was observed with increasing PDMS substrate stiffness. These results indicate a cellular response to changes in PDMS substrate mechanics, along with a demonstration of a mechanically dynamic and photoresponsive PDMS substrate platform to model the dynamic behavior of ECM.
Keywords: Viscoelasticity, Photocrosslinking, Dynamic, Elastomer, Mechanics, Mechanotransduction
1. Introduction
The extracellular matrix (ECM) is a complex three-dimensional (3D) network composed of fibrous matrix proteins that influence cell behaviors and tissue organization. The physical features of the ECM (e.g., stiffness [1–6], viscoelasticity [7] and topography [8–10]) play pivotal roles in governing cell behavior. In particular, ECM stiffness is a key regulator of cellular morphology [11–13], migration [14–16], proliferation [17–19], gene expression [20–22] and differentiation [23–25]. The elastic modulus of native ECM varies among different tissue types, ranging from 0.5 to 1 kPa in brain to 10-20 GPa in bone. Therefore, recapitulating the native mechanical environment is a useful strategy to investigate cell interactions with their microenvironment.
Numerous hydrogel systems have been developed to mimic the mechanical properties of ECM [26–29], including natural polymers (e.g., hyaluronic acid (HA), alginate, collagen, gelatin and chitosan) and synthetic polymers (e.g., polyacrylamide and polyethylene glycol (PEG)). To recapitulate the dynamic mechanical evolution of native ECM during biological processes (e.g., fibrosis [30], tumorigenesis [31,32] and tissue remodeling during wound healing [33–35]), both stiffening [36–38] and softening [39–42] hydrogels have been applied to cell culture studies. However, these dynamic hydrogels behave like elastic materials, lacking the viscoelastic nature of most tissues.
Only a few hydrogels (e.g., polyacrylamide [43,44] and alginate [45–47]) have been designed to be viscoelastic to directly investigate the influence of substrate viscoelasticity on cell behavior. Mesenchymal stem cells (MSCs) displayed increased proliferation, spread area, Rac activation and differentiation towards multiple lineages on acrylamide hydrogels with high loss modulus [43,44]. In alginate hydrogels, increased substrate stress relaxation promoted cell spreading, proliferation and osteogenic differentiation, indicating that stress relaxation is a key characteristic of cell-ECM interactions [47]. In addition, cells presented altered behavior (e.g., more spreading, proliferation, actin stress fiber organization and focal adhesions) in viscoelastic hydrogels compared to elastic hydrogels [47]. However, these viscoelastic hydrogels have not yet been designed with changing mechanics. Thus, a biomaterial system featuring both dynamic mechanics and viscoelasticity is desired to model the natural ECM.
Polydimethylsiloxane (PDMS) is a viscoelastic elastomeric material that has been widely used in cell culture [6,23,48–52]. PDMS can be simply prepared by mixing base and curing agent, where a platinum (Pt) catalyst facilitates hydrosilylation to crosslink vinylterminated PDMS (V-PDMS) base polymer with methylhydrogen siloxane units (from the curing agent), turning short polymer chains into an elastomeric network (Fig. 1a). The moduli of PDMS (from kPa to MPa) fabricated through this mechanism can be controlled by changing the base-to-curing agent mixing ratio, the curing temperature, and the reaction time [6,23,48,49]. PDMS substrates with a range of moduli have been used to investigate cellular responses to changing mechanical environments. For instance, 3T3 fibroblasts manifest larger cell spread areas and focal adhesions on stiff PDMS substrates [53] and myocardial cells and cardiac fibroblasts show increased spreading as well as higher matrix metalloproteinase-2 gene expression and protein activity with increasing PDMS modulus [54]. In addition to changes in spreading and focal adhesion size, lung myofibroblasts also exhibit higher expression of α-smooth muscle actin (α-SMA) on stiff PDMS substrates [55]. These reports present PDMS mechanics in a static manner and current strategies used to modulate the modulus of PDMS can only be utilized in the absence of cells, as the PDMS substrates are normally cured at high temperatures.
Fig. 1. Crosslinking of PDMS.
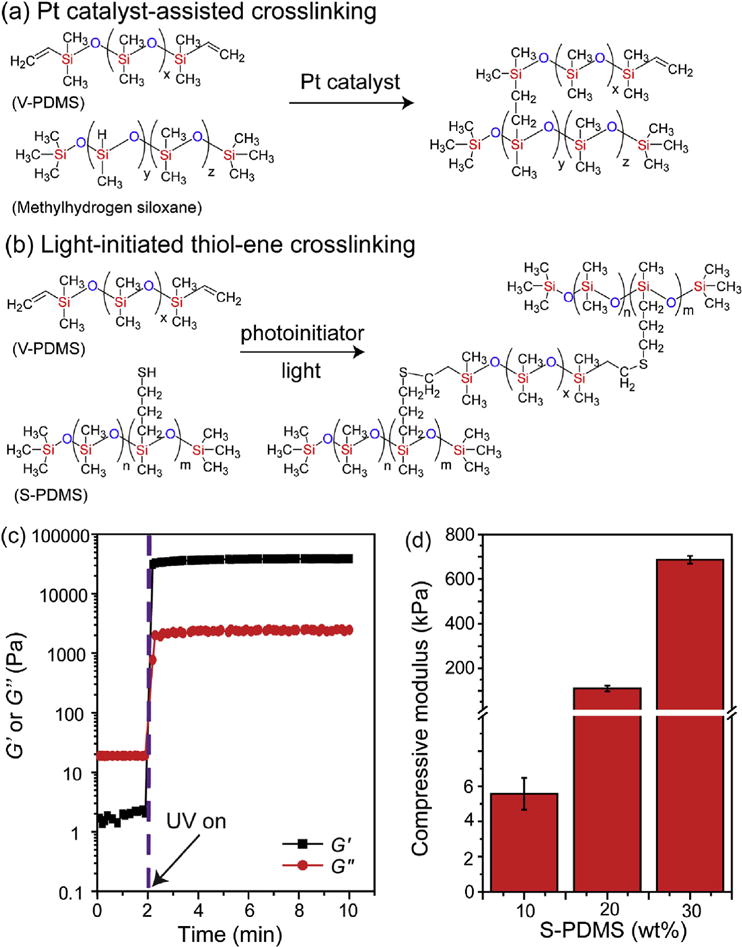
(a) Pt-catalyzed crosslinking and (b) light-initiated thiol-ene crosslinking. (c) Rheological characterization of the photocrosslinking of base agent (V-PDMS, 80 wt%) and thiolated crosslinker (S-PDMS, 20 wt%) with DMPA (0.5 wt%) under UV exposure (365 nm, 15 mW/cm2). (d) Compressive moduli of photocrosslinked V-PDMS and S-PDMS with varied concentrations of S-PDMS after UV exposure (365 nm, 15 mW/cm2) for 2 min. The mixing ratios between V-PDMS and S-PDMS were 90:10, 80:20 and 70:30 by weight. (n = 3. Data presented as mean ± s.d.).
Radiation curing of PDMS has been reported as an alternative and efficient way to convert viscous and reactive PDMS liquids into solids [56–60]. Thiol-ene “click” photochemistry [61,62] provides several advantages over the traditional platinum-catalyzed approach to crosslink PDMS, including: (i) no requirement for expensive heavy-metal catalysts, (ii) quantitative conversions in relatively short timeframes at room temperature, (iii) reduced oxygen inhibition, and (iv) no generation of intracellular reactive oxygen species (ROS). Here, we developed a two-step crosslinking strategy by combining platinum-catalyzed crosslinking and light-initiated thiol-ene crosslinking to temporally manipulate the modulus of PDMS (Fig. 1a and b). PDMS substrates were fabricated through a stepwise process to introduce thiolate crosslinker and photoinitiator to the network, allowing the use of secondary photocrosslinking to stiffen the substrate and to modulate cellular response. Thus, this work demonstrates a photo-controllable and stiffness-tunable PDMS substrate for use as a platform to dynamically manipulate cell responses.
2. Materials and methods
2.1. Materials
Sylgard 184 Elastomer Kit (Dow Corning) was used to fabricate PDMS substrates. 4-6% (mercaptopropyl)methylsiloxane-dimethylsiloxane copolymer (S-PDMS) was purchased from Gelest. Bovine collagen type 1 solution was purchased from Advanced BioMatrix. All other chemicals were purchased from Sigma-Aldrich unless otherwise indicated.
2.2. Mechanical testing of photocurable PDMS substrates
Photocurable PDMS substrates were prepared by mixing V-PDMS, S-PDMS and 2,2-dimethoxy-2-phenylacetophenone (DMPA) (0.5 wt%). Dynamic oscillatory time sweeps were performed using an AR2000ex rheometer (TA Instruments) with an ultraviolet light-guide accessory (SmartSwap™, TA Instruments) connected to an ultraviolet light source (Omnicure S1000, EXFO). The photocrosslinking of PDMS mixtures were carried out under exposure to UV light (365 nm, 15 mW/cm2). Storage (G′) and loss (G″) moduli with time were monitored under 0.5% strain and 1 Hz, using a cone and plate geometry (1°, 20-mm diameter, 27 μm gap) at 25°C.
Compressive moduli of photocured PDMS substrates were measured by dynamic mechanical analysis (DMA, Q800, TA Instruments) fitted with a compression clamp, running at a constant strain rate of 10%/min in air at room temperature. The modulus was calculated from the slope of the stress-strain curve between 10 and 20% strain. PDMS mixtures (80 μL, including V-PDMS, S-PDMS and DMPA (0.5 wt%)) were transferred to a mold and covered with a glass coverslip. The samples were irradiated with UV light (365 nm, 15 mW/cm2) for 2 min. Photocured PDMS substrates (~2 mm height, 5 mm diameter) were removed from molds for mechanical testing.
2.3. Fabrication of photo-stiffening PDMS substrates
The photo-stiffening PDMS substrates were all prepared through a two-step process. In the first step, PDMS mixtures were prepared by mixing base and curing agent at a ratio of 65:1, and placed under vacuum to remove any air bubbles. PDMS mixtures (80 μL) were transferred to molds, covered with cover slips, and cured at 37°C for 2 days. In the second step, PDMS samples were then removed from the molds and incubated in 1 mL toluene containing varying amounts of S-PDMS (0-30 wt%), V-PDMS (0-30 wt%) and DMPA (10 mM), where the total amount of S-PDMS and V-PDMS was 30 wt/% in toluene. After 3 h incubation at 37°C, PDMS samples were washed twice, once with toluene and once with hexane/acetone (1:1), and finally dried at 37°C overnight to evaporate the solvent. The secondary photocrosslinking of PDMS was carried out under exposure to UV light (365 nm, 15 mW/cm2) for 2 min. Compressive moduli of PDMS samples (~2 mm height, 5 mm diameter) were measured using DMA as described above.
2.4. Preparation of PDMS substrates on glass coverslips
The PDMS substrates on glass coverslips were all prepared through a two-step process.
In the first thermo-curing step, PDMS mixtures (base and curing agent at a ratio of 65:1, 100 mg) were spin-coated onto glass coverslips (22 mm × 22 mm) at a speed of 700 rpm for 30 s, and cured at 37°C for 2 days. The thickness of PDMS layer on glass coverslip was ~0.2 mm, which is also thick enough (>20 μm) for cells not to feel the underlying glass substrate [63]. In the second step, for unmodified PDMS substrates (UM), PDMS-coated glass coverslips were incubated in 4 mL toluene containing V-PDMS (30 wt%); for modified PDMS substrates (M), PDMS-coated glass coverslips were incubated in 4 ml toluene containing S-PDMS (10 wt%), V-PDMS (20 wt%) and DMPA (10 mM). V-PDMS (20 wt%) was used to prevent the uncrosslinked V-PDMS to be dissolved from the substrates by the pure toluene solution during the swelling process. The loss of V-PDMS in the substrate would weaken the mechanics of PDMS and decrease the efficiency of the secondary thiol-ene photocrosslinking. After 3 h incubation at 37°C, PDMS samples were washed twice, once with toluene and once with hexane/acetone (1:1), and finally dried at 37°C overnight to evaporate solvent.
2.5. Atomic force microscopy (AFM) characterization of topography and viscoelasticity
Topographic imaging of PDMS substrates was performed with Dimension Icon AFM (Bruker, Santa Barbara, CA) to determine the roughness of the modified PDMS substrates before and after UV exposure. Images were acquired in tapping mode using an integral cantilever with attached silicon tips (spring constant 30-40 N/m). The AFM scan size was chosen as 5.0 mm at a scan rate of 1.25 Hz, and the images were 256 by 256 pixels. AFM images were analyzed using NanoScope analysis software (Bruker, Santa Barbara, CA).
AFM-nanoindentation and ramp-and-hold force relaxation tests were performed using microspherical colloidal tips (radius R = 5 μm, nominal spring constant k = 0.6 N/m, HQ:NSC36/Tipless/Cr-Au, cantilever C, NanoAndMore, Watsonville, CA) and a Dimension Icon AFM (BrukerNano, Santa Barbara, CA) at 10 μm/s indentation rate up to a maximum load of ~120 nN [64]. The nanoindentation was performed via microspherical tip (R = 5 μm) up to ~ 1 μm maximum indentation depth, which is within the limit of linear deformation (strain ε < 0.2, Dmax < 0.4R) [65]. In our experiment, the thickness of PDMS substrate is h ~200 mm. According to the established contact mechanics theory [66], given h >> 12.8R in the linear deformation regime, the substrate constraint effect is negligible. Therefore, we calculated the effective indentation modulus by applying contact mechanics model within the Hertzian framework to the loading portion of indentation F-D curves by assuming a semi-infinite medium.
During the measurement, the PDMS substrates were immersed in phosphate buffered saline (PBS), and indentation was performed on at least 10 different locations for each sample to account for spatial heterogeneity.
2.6. Imaging of collagen and calculation of collagen distribution on PDMS substrates
Collagen was adsorbed to PDMS samples to enhance cell adhesion. To visualize collagen adsorption, PDMS substrates were incubated with a solution of FITC-labeled bovine type I collagen (Chondrex, 4001) at a concentration of 0.1 wt% at 37°C for 3 h. After collagen incubation, collagen-coated PDMS substrates were incubated in high glucose Dulbecco’s Modified Eagle’s Medium (DMEM, 4.5 g/L glucose) containing 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Invitrogen) at 37°C for 24 h before imaging. Adsorption was confirmed by imaging samples using an Olympus BX51 microscope (B&B Microscopes Limited) with a 10× magnification objective. To assess collagen distribution, binary masks of adsorbed collagen were generated using intensity-based thresholding from FITC-labeled collagen images, and the masks were then used to calculate the area covered by collagen. To assess any potential differences in collagen distribution resulting from PDMS stiffening, binary masks from collagen in the same PDMS regions were compared before and after UV exposure (365 nm, 15 mW/cm2, 2 min).
2.7. Cell culture of 3T3 fibroblasts and mesenchymal stem cells (MSCs)
NIH 3T3 fibroblasts and human bone marrow-derived MSCs were cultured at 37°C under a humidified-controlled environment with 5% CO2. 3T3 fibroblasts were cultured in high glucose DMEM containing 10% FBS and 1% penicillin/streptomycin. MSCs were purchased from Lonza, used at passage 3 for all experiments, and cultured in the media consisted of alpha-minimum essential media (α-MEM) supplemented with 16.7% FBS, 1% penicillin streptomycin and 1% :L-glutamine (Invitrogen). The cells were maintained in the above medium and media was replaced every three days. Before cell seeding, PDMS substrates on glass coverslips (22 mm × 22 mm) were placed in a non-tissue culture treated 6-well plate and incubated with ethanol (70%, 2.5 mL) for 1 h, collagen (0.1 mg/mL in PBS, 2.5 mL) for 3 h, and serum-contained media (10% FBS, 2.5 mL) for 30 min.
Cell viability was determined using an AlamarBlue assay according to the manufacturer’s protocol (Invitrogen). 3T3 fibroblasts were seeded at 1 × 105 cells/well (for short-term cell viability) or 1 × 104 cells/well (for long-term cell viability) on PDMS substrates (i.e., unmodified PDMS substrates (UM) and modified PDMS substrates (M)) placed in a non-tissue culture treated 6-well plate 24 h prior to the experiment. After cell seeding, PDMS substrates were exposed to UV light on day 1 to present another two samples: unmodified PDMS substrate with UV exposure (UM + UV) and modified PDMS substrate with UV exposure (M + UV). Cell-seeded PDMS substrates (UM, M, UM + UV and M + UV) were transferred to a new non-tissue culture treated 6-well plate for the AlamarBlue assay. Cells were treated with 1200 μL of 10% AlamarBlue reagent in serum-containing media. Subsequently, the cells were incubated at 37°C under a humidified-controlled environment with 5% CO2. After 4 h of incubation, 1000 μL (200 μL × 5) of solution from each well was transferred to a 96-well microplate and fluorescence measured (excitation/emission: 540 nm/590 nm, Infinite M200, Tecan) to determine the metabolic activity.
2.8. Cell staining, imaging, and analysis
Phase contrast images of 3T3 fibroblasts on PDMS substrates were acquired using a Zeiss Axiovert 200 inverted microscope (Hitech Instruments, Inc.). Cells on PDMS substrates were fixed in 10% formalin for 15 min at room temperature (RT). Samples were then permeabilized with 0.1% Triton X-100 for 3 min. To visualize cells and nuclei, samples were stained for actin (rhodamine phalloidin, 0.5% in 3% bovine serum albumin (BSA) solution in PBS) and double-stranded DNA (DAPI, 0.1% in PBS) for 20 and 5 min, respectively. After staining, fluorescent images were acquired using an Olympus BX51 microscope (B&B Microscopes Limited).
Time-lapse microscopy (EVOS FL Auto Imaging System equipped with motorized stage for multi-point acquisition and environmental chamber with gas mixer for live cell imaging) was used to track individual 3T3 fibroblasts and MSCs following substrate stiffening. Images of 3T3 fibroblasts and MSCs cultured on Soft and Stiff static PDMS substrates, as well as in situ stiffened PDMS (Soft-to-Stiff) were captured every 20 min for at least 16 h and used to assess cell spreading of individual cells in real-time.
All image analysis was performed using ImageJ NIH image processing software (Bethesda, MD).
2.9. Cardiac fibroblasts isolation and culture
Rat cardiac fibroblasts were harvested from Sprague Dawley rat pups post-natal day 0-1, as previously described [67]. Briefly, hearts were trimmed of atria and vessels, minced and subjected to sequential rounds of trypsin digestion, washing, filtering, and centrifugation steps to remove red blood cells and debris. The cells were cultured in 3:1 ratio of DMEM:M199 (1×), 10% FBS, 10 mM HEPES (4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid)), 1% Glutamine/Penicillin/Streptomycin (Gibco) at 37°C in a humidified incubator with 5% CO2.
A two-layer Percoll density gradient was used to separate the fibroblasts and myocytes, as previously described [68] utilizing Percoll solutions of density 1.07 g/mL and 1.09 g/mL. The cell suspension was layered on top of the gradient and centrifuged at 25°C at 2060 × g for 27 min using a low acceleration rate and no brake. After centrifugation, two distinct cell bands were visible: i) an upper band, located at the interface between the media and low density Percoll layer corresponding to the fibroblast layer, and ii) a lower band, located at the interface between the low and high Percoll densities corresponding to the myocyte layer. The fibroblast layer was collected and spun down at 700 μ g for 10 min at 25°C with mid-range acceleration and deceleration. Cells were then resuspended in fresh media and plated for 24 h on a Collagen IV-coated Bioflex® plate (Flexcell International Corporation) to keep the fibroblasts quiescent before plating on experimental substrates.
2.10. Immunostaining of cardiac fibroblasts and quantification of myofibroblast activation
Following the completion of the experiment, cells were rinsed with PBS, fixed in 4% formaldehyde for 10 min at RT, and simultaneously permeabilized and blocked with a solution containing 1% (w/v) BSA and 5% (v/v) goat serum (Cell Signaling Technology) in 0.1% Triton X-100 for 15 min at RT. Cells were then washed in PBS and first incubated with rabbit anti-vimentin (1:500, Abcam) primary antibody overnight at 4°C followed by a 2 h incubation with TRITC-conjugated goat anti-rabbit (1:2000, Jackson ImmunoR-esearch) IgG at room temperature in the dark. A second round of staining was performed by simultaneously re-permeabilizing and blocking as described above. The cells were then incubated with mouse anti-α-smooth muscle actin (Sigma-Aldrich) primary antibody overnight at 4°C. Cells were then washed with PBS and then incubated with FITC-conjugated goat anti-mouse (1:2000, Jackson ImmunoResearch) IgG for 2 h. Cells were counterstained with DAPI at 1 μg/mL in methanol for 10 min. Finally, the substrates were mounted on glass slides using Fluoromount G (Electron Microscopy Science) and imaged using a Nikon Eclipse 80i. Images were acquired under a 20× objective at a minimum of 8 locations per sample using NIS Elements software. The three fluorescent channels were merged using ImageJ and total number of cells was counted along with the number of activated cells, indicated by α-SMA stress fiber formation.
2.11. Statistical analysis
All images and data are representative of the results of at least two or more independent biological experiments. The box and binned data plots were generated using Origin software. Boxes present first and third quartiles with median lines; circles show the mean value; error bars indicate the range of 1.5 × interquartile. One-way analysis of variance (ANOVA) was performed for statistical analysis. Significance was set at p < 0.05 with *, **, or *** indicating p < 0.05, 0.01, or 0.001 respectively. Error is reported in figures as the standard deviation of the mean unless otherwise noted.
3. Results and discussion
3.1. Fabrication of mechanically dynamic PDMS substrates via a two-step crosslinking strategy
Standard crosslinking of PDMS occurs with a Pt-catalyzed reaction between V-PDMS and methylhydrogen siloxane (major component in curing agent) (Fig. 1a). To introduce photoinducible crosslinking, we utilized thiol-ene click chemistry where V-PDMS can be crosslinked with the (mercaptopropyl)methylsiloxane-dimethylsiloxane copolymer (S-PDMS), where the thiols from S-PDMS react with the terminal vinyl groups of V-PDMS in the presence of photoinitiator DMPA and UV light (365 nm, 15 mW/cm2) (Fig. 1b). Rheology was used to monitor the crosslinking of V-PDMS and S-PDMS, where the crossover of the storage (G′) and the loss (G″) moduli occurred within seconds and a plateau modulus was reached within ~1 min (Fig. 1c), indicating rapid crosslinking of V-PDMS and S-PDMS in response to UV light. The moduli of photo- initiated V-PDMS/S-PDMS networks increased in a crosslinker concentration-dependent manner (Fig. 1d). These results demonstrate that PDMS networks can be fabricated through a photo-induced thiol-ene reaction with short light exposure times and the network moduli can be tuned through the concentration of the crosslinker.
After illustration of these two crosslinking mechanisms, we then combined them into a two-step crosslinking process of Pt-catalyzed crosslinking and then thiol-ene click reaction to fabricate photoresponsive and stiffness-tunable PDMS substrates. PDMS substrates were prepared via a stepwise process featuring 1) crosslinking of V-PDMS and methylhydrogen siloxane under Pt catalyst and 2) swelling of S-PDMS and DMPA into the cured PDMS substrate where V-PDMS was also added in the solution to inhibit the dissolution of uncrosslinked V-PDMS from the PDMS substrate by toluene (Fig. 2a). It was not possible to directly introduce S-PDMS during the initial crosslinking, as thiolates can react with the Pt catalyst and diminish its catalytic ability.
Fig. 2. Stiffening PDMS substrates through secondary thiol-ene photocrosslinking.
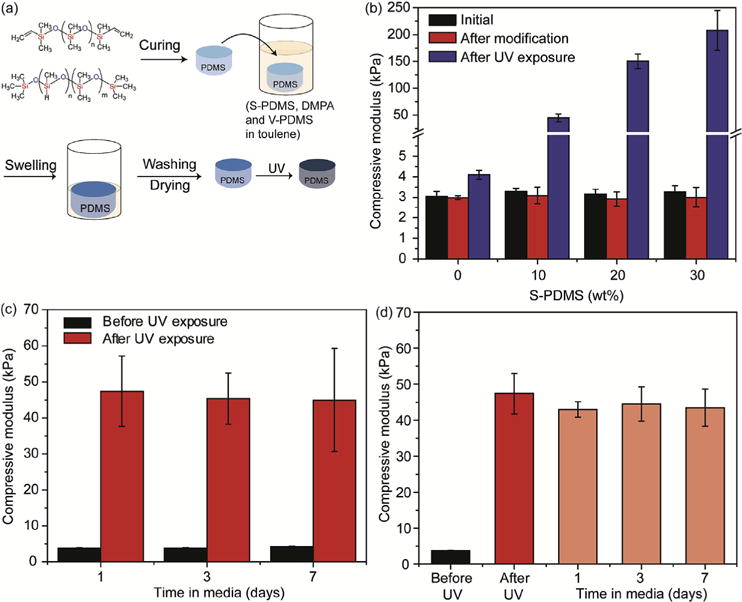
(a) Schematic of the stepwise preparation of photoresponsive PDMS substrates, where cured V-PDMS via Pt-catalyzed crosslinking was placed in toluene with S-PDMS, DMPA and V-PDMS, the toluene was allowed to evaporate, and the sample was exposed to light for crosslinking. (b) Moduli of photoresponsive PDMS substrates with different concentrations of S-PDMS as a crosslinker in the presence of DMPA with UV exposure (365 nm, 15 mW/cm2) for 2 min. PDMS substrates were prepared through two steps: 1) V-PDMS and curing agent were mixed at a ratio of 65:1 to form soft substrates after curing at 37°C for 2 days; 2) PDMS substrates were incubated in toluene containing varying amounts of S-PDMS (0-30 wt%), V-PDMS (0-30 wt%) and DMPA (10 mM), with the total amount of S-PDMS and V-PDMS at 30 wt/% in toluene. (c) Moduli of photoresponsive PDMS substrates (10wt% S-PDMS) after incubation in serum-containing media (10% FBS) for different time points (1, 3, 7 days) and then stiffened. (d) Moduli of photo-stiffened PDMS (10wt% S-PDMS) incubated in serum-containing media (10% FBS) for up to 7 days. (n = 3. Data presented as mean ± s.d.). For PDMS substrates used in (c) and (d), PDMS substrates were also prepared through the two-step process as used in (a), while S-PDMS (10 wt%) and V-PDMS (20 wt %) and DMPA (10 mM) were used in the swelling step. The presence of V-PDMS (20 wt%) prevents any uncrosslinked V-PDMS from diffusing from the substrates during the swelling process.
To generate a soft PDMS substrate while maintaining excess V-PDMS for secondary crosslinking, V-PDMS and methylhydrogen siloxane were mixed at a 65:1 ratio and cured at 37°C for 2 days, presenting an initial modulus of ~3 kPa. The moduli of PDMS substrates did not change after the S-PDMS and DMPA swelling step alone; however, stiffening was introduced with UV light exposure in a S-PDMS concentration-dependent manner (Fig. 2b), up to a modulus of ~200 kPa with 30 wt% S-PDMS. Furthermore, the modified PDMS substrates could be stiffened after prolonged incubation in serum-containing media, indicating that the crosslinker (S-PDMS) and photoinitiator (DMPA) were quite stable within the PDMS substrates (Fig. 2c). Also, post-stiffening, the mechanics of PDMS substrates were maintained in serum-containing media for up to 7 days (Fig. 2d). These results confirm that our two-step crosslinking approach can elicit substrate stiffening in a user-defined manner for up to 7 days in a biological environment.
These photo-responsive PDMS substrates can also be stiffened gradually with intermittent light exposure. Three light intensities (5, 10 and 15 mw/cm2) were applied to demonstrate the gradual stiffening as well as the tunability of the stiffening rate in changing the PDMS modulus (Fig. S1). PDMS was stiffened at a slower rate using lower light intensity, generating modulus changes in the range of 3-45 kPa over multiple time points. In addition, this two-step PDMS crosslinking can be applied to PDMS prepared with various base-to-curing agent ratios. PDMS modulus varied with changes in the initial base-to-curing agent ratio and the extent of modulus change was dependent on this initial ratio (Fig. S2).
3.2. Characterization of PDMS substrates using atomic force microscopy
Atomic force microscopy (AFM) was used to characterize the topography and mechanical properties of modified PDMS substrates. For AFM measurements, PDMS substrates were spin-coated on glass coverslips, cured, and modified through the stepwise process to introduce crosslinker (S-PDMS) and photoinitiator (DMPA). The roughness of modified PDMS substrates did not change after UV exposure as the root mean square roughness (Rq) values were 0.67 ± 0.01 nm and 0.60 ± 0.02 nm before and after UV exposure, respectively (Fig. S3).
AFM-nanoindentation and force relaxation were performed to quantify the elastic and viscoelastic properties of the PDMS substrates at the microscale. Exposure to UV light resulted in significant stiffening of the PDMS substrates, showing ~10-fold increase in both the equilibrium modulus (E∞, increased from ~3 to 37 kPa) and the effective indentation modulus (Eind, increased from ~6 to 60 kPa) (Fig. 3a). The equilibrium modulus (changed from 3 to 37 kPa after UV exposure) measured by AFM in nanoscale was quite similar to the compressive modulus (changed from 3 to 45 kPa after UV exposure) measured by DMA in bulk. In addition, the degree of elasticity, as indicated by the ratio E∞/Eind became significantly higher (Fig. 3b), indicating a more elastic polymeric network due to the presence of higher covalent crosslink density. In addition, E∞ and two viscoelastic relaxation modes, (E1, τ1) and (E2, τ2), were extracted from the five-element standard linear solid (SLS) model shown in Fig. 3c [69]. We also observed an increase in the longterm relaxation time, τ2, after UV exposure (Fig. 3d), indicating the increased crosslinking density in the polymer network as well as the increased hindrance of polymer chain movement following UV-mediated secondary crosslinking. These AFM results confirm that secondary crosslinking increased the modulus as well as the degree of elasticity and viscoelasticity of PDMS network after UV treatment.
Fig. 3. AFM-based nanoindentation and force relaxation measurements of PDMS substrates.
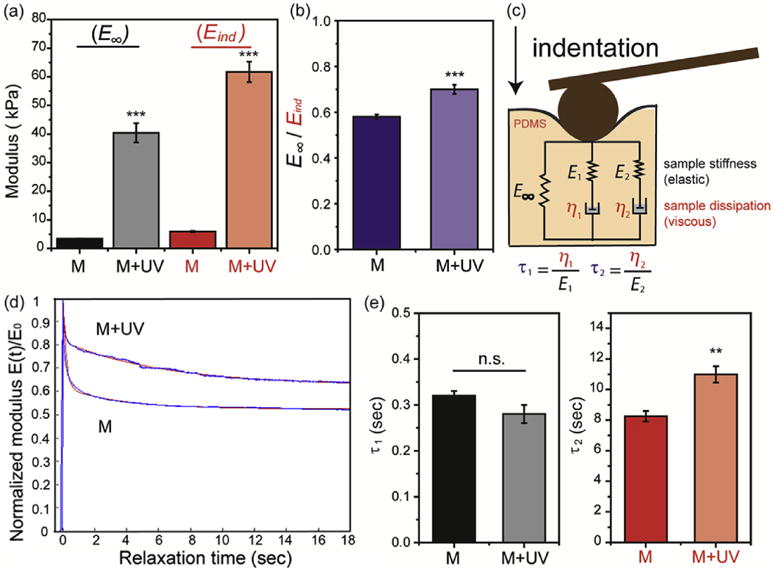
(a) Equilibrium modulus (E∞), indentation modulus (Eind), and (b) the ratio of E∞/Eind of modified PDMS substrates before (M) and after UV exposure (M + UV). (c) Schematic of the five-element SLS model implemented with two viscoelastic relaxation time constants, τ1 and τ2. Temporal modulus, E(t) = E∞ + E1 exp(−t/τ2) + E2 exp(−t/τ2), where E1 and E2 are the moduli, η1 and η2 are the viscosities corresponding to the two relaxation time constants, τ1 and τ2, respectively. (d) Representative relaxation curves of PDMS substrates. Normalized E(t) versus initial, instantaneous indentation modulus (E0), curves showed distinctive relaxation behaviors of modified PDMS substrates before and after UV exposure. Red lines are the SLS model fits by non-linear least squares regression via the Levenberg-Marquardt algorithm (LMA). (e) Relaxation time constants of PDMS substrates. M = modified PDMS, M + UV = modified PDMS with UV exposure (n = 10 per group. Data presented as mean ± s.d.) ***: p < 0.001, **: p < 0.01, n.s.: not significant. Spin-coated PDMS substrates were prepared through a two-step process: 1) V-PDMS and curing agent were mixed at a ratio of 65:1 to form soft substrates after curing at 37°C for 2 days; 2) PDMS substrates were incubated in toluene containing S-PDMS (10 wt%), V-PDMS (20 wt%) and DMPA (10 mM). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Cell spreading in response to in situ stiffening of PDMS substrates
Cells were seeded on PDMS substrates that were spin-coated on glass coverslips. To facilitate cell adhesion on PDMS substrates, collagen was used as a coating material through physical adsorption. Fluorescent dye-labeled collagen was used to visualize collagen distribution on the PDMS substrates. The morphology of collagen did not change after UV exposure and the collagen- covered area on the PDMS substrates with different conditions was quite similar (Fig. S4), indicating that the modification and UV treatment did not affect collagen distribution on the PDMS substrates.
Fibroblasts are the most abundant cell type in connective tissue, where they produce structural proteins that comprise the ECM. Here, 3T3 fibroblasts were used to investigate the cytocompatibility of modified PDMS substrates as well as the cell response to the change in substrate stiffness. Cells were seeded on unmodified (UM) and modified (M) PDMS substrates, and UV light (365 nm, 15 mW/cm2, 2 min) was applied after cell seeding for 24 h to form two UV-treated PDMS samples: unmodified PDMS substrates with UV exposure (UM + UV) and modified PDMS substrates with UV exposure (M + UV). An AlamarBlue assay was performed to quantify the cellular metabolic activity and used as a proxy for cell viability. The metabolic activity of cells was examined right after UV irradiation, indicating that UV irradiation did not change the cells as the UM + UV and M + UV samples presented similar metabolic activity compared to the UM and M samples, respectively (Fig. S5). In addition, no toxicity was observed for the cells cultured on these four PDMS substrates and cells continued to proliferate for up to 7 days (Fig. 4), demonstrating that modified PDMS substrates were cytocompatible.
Fig. 4. Cytocompatibility testing of PDMS substrates.
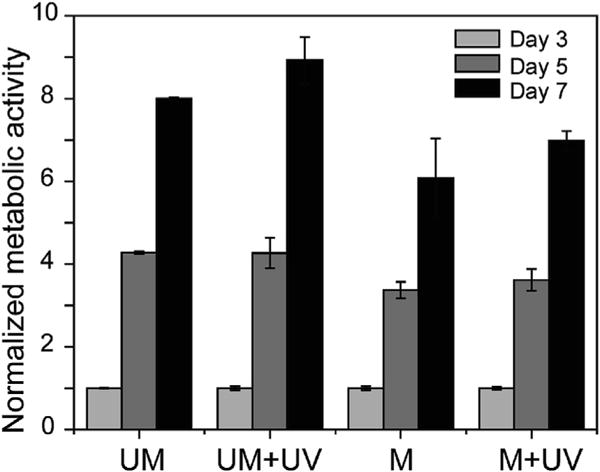
Metabolic activity of 3T3 fibroblasts on PDMS substrates, with normalization to UM at day 3. UV exposure was applied to form UM + UV and M + UV samples after cell seeding for 24 h (day 1), and the AlamarBlue assay was performed on day 3, 5 and 7. UM = unmodified PDMS, UM + UV = unmodified PDMS with UV exposure, M = modified PDMS, M + UV = modified PDMS with UV exposure. (n = 3 per group. Data presented as mean ± s.d.). The values of metabolic activity at all time points on each PDMS substrate were statistically significant from each other (p < 0.001 for all comparisons).
Recent studies have shown that the mechanical properties of culture substrates impact fibroblast spreading and adhesion morphology [53,70]. Here, 3T3 fibroblasts seeded on soft modified PDMS substrates manifested a smaller spread area (~1300 μm2), while 3T3 fibroblasts cultured on the stiff UV-exposed modified PDMS substrate spread to larger area (~1700 μm2) (Fig. 5a top and 5b). These results indicate that the increased PDMS substrate stiffness increased spreading of fibroblasts. Meanwhile, 3T3 fibroblasts initially cultured on soft substrates were responsive to secondary crosslinking-induced stiffness changes of the substrate, presenting similar spread area (~1900 μm2) to the cells cultured on stiff static substrates (~2100 μm2) (Fig. 5a bottom and 5c). 3T3 fibroblasts cultured on unmodified PDMS substrates remained small (~1300 μm2) even after UV exposure (Figs. S6a and S6b). This demonstrates that the modified PDMS substrates are mechanically dynamic and their tunable stiffness can direct cell response. Similar area changes of cardiac fibroblasts and MSCs were observed on these dynamic PDMS substrates (Fig. 6 and Figs. S6c-S6f), where cardiac fibroblast spreading increased from 1100 μm2 to 1500 μm2 and MSC spreading increased from 2700 μm2 to 4600 μm2. The change in cell spreading over time was monitored by tracking single cells immediately after stiffening of the PDMS substrates. Cells initially cultured on soft substrates responded to the in situ stiffening of the substrates, presenting an increase in spread area over time (Fig. S7).
Fig. 5. 3T3 fibroblast spreading in response to in situ stiffening of PDMS substrates.
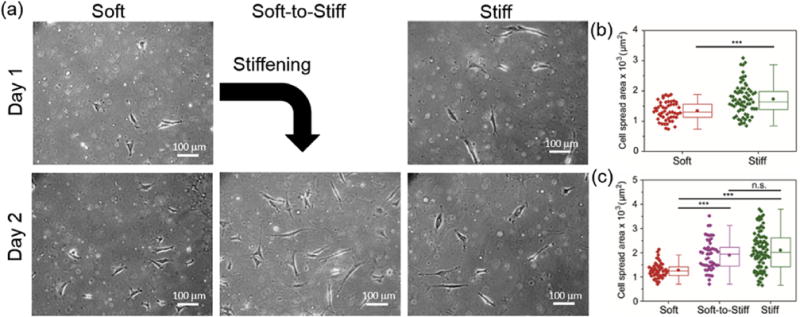
(a) Representative images of 3T3 fibroblasts on soft and stiff static substrates, as well as a dynamically stiffened soft-to-stiff substrate (stiffening performed on day 1). 3T3 fibroblast spreading quantification on (b) day 1 and (c) day 2. Stiff substrates were used as a positive control as photocrosslinking was applied before cell seeding and static soft substrates did not receive light exposure. The moduli for the Soft and Stiff substrates were ~3 and ~37 kPa (measured by AFM indentation), respectively. The modulus of the Soft-to-Stiff substrate was changed from ~3 to 37 kPa after stiffening. Thus, the final moduli of the Soft, Soft-to-Stiff and Stiff substrates were ~3, ~37 and ~37 kPa, respectively. n > 40 cells per group. ***p < 0.001, n.s.: not significant.
Fig. 6. Cardiac fibroblast and MSC spread areas are dynamically tuned to PDMS stiffness.
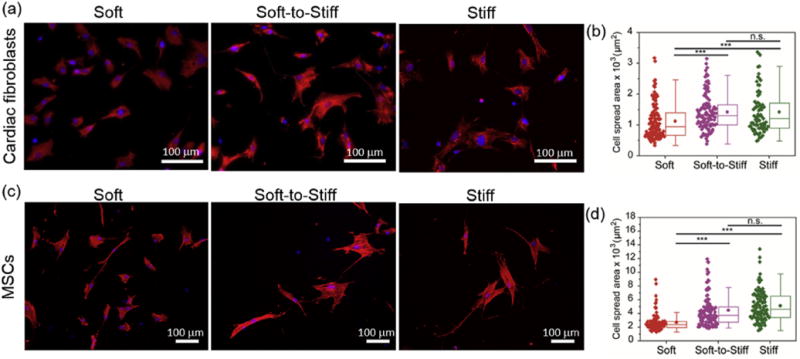
Representative images (a,c) and spread area quantification (b,d) of (top) cardiac fibroblasts and (bottom) MSCs on soft and stiff static substrates, as well as on a soft-to-stiff dynamic substrate on day 2 (stiffening performed on day 1). Stiff substrates were used as a positive control as photocrosslinking was applied before cell seeding and static soft substrates did not receive light exposure. The moduli for the Soft and Stiff substrates were ~3 and ~37 kPa, respectively. The modulus of Soft-to-Stiff substrate was changed from ~3 to 37 kPa after stiffening. Thus, the final moduli of the Soft, Soft-to-Stiff and Stiff substrates were ~3, ~37 and ~37 kPa, respectively. Cardiac fibroblasts were stained with vimentin (red), nuclei (blue); MSCs were stained with F-actin (red), nuclei (blue). n > 40 cells per group. ***: p < 0.001, n.s.: not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Stiffening PDMS substrates promote myofibroblast activation
Cardiac fibroblasts play essential roles in myocardial function, where they are involved in ECM deposition and remodeling as well as in chemical and mechanical signaling. The differentiation of cardiac fibroblasts to myofibroblasts is currently recognized as a key step during maladaptive cardiac remodeling [71,72]. Sustained activation of myofibroblasts results in excess production and deposition of ECM proteins in the myocardium, known as fibrosis, which leads to distorted architecture and cardiac dysfunction [73–75]. Here, we applied photoresponsive PDMS substrates to investigate the contribution of stiffness changes to myofibroblast differentiation. After stiffening of the PDMS substrates, we observed an increase in the percentage of activated myofibroblasts, using α-SMA as a marker, to levels comparable to glass substrates (Fig. 7a top and 7b). Additionally, activation of fibroblasts was found to be time-dependent: the highest activation (~20%) was observed after 4 days in vitro on the dynamic soft-to-stiff substrates (Fig. 7a bottom and 7b). As a control, cardiac fibroblasts cultured on unmodified PDMS substrates presented low activation even after UV exposure (Fig. S8). These results demonstrate that photoresponsive PDMS substrates can model the mechanically dynamic features of ECM during cardiac fibrosis, where the activation of cardiac fibroblasts is promoted by substrate stiffening.
Fig. 7. Stiffening of PDMS substrates promotes myofibroblast activation.
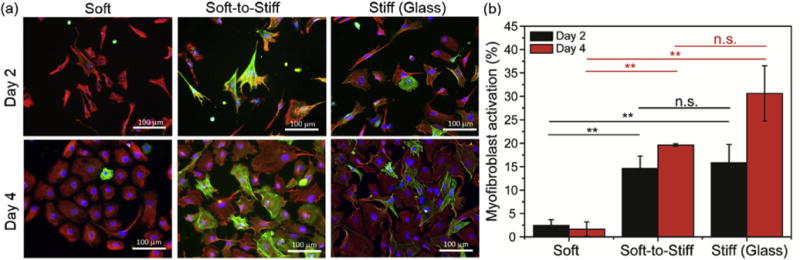
(a) Representative images for cardiac fibroblasts on soft and stiff static substrates, as well as on soft-to- stiff dynamic substrates on day 2 and day 4 (stiffening performed on day 1). (b) Myofibroblast activation quantification of cardiac fibroblasts at these same time points. The moduli for the Soft and Stiff (Glass) substrates were ~3 and ≥ 1 GPa, respectively. The modulus of Soft-to-Stiff substrates changed from ~3 to 37 kPa after stiffening. Thus, the final moduli of the Soft, Soft-to-Stiff and Stiff (Glass) substrates were ~3 kPa, ~37 kPa and ≥1 GPa, respectively. Cells were stained with vimentin (red), nuclei (blue) and α-SMA (green). n ≥ 2 measurements per group. **: p < 0.01, n.s.: not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusion
Here, we present a two-step crosslinking strategy to fabricate a mechanically dynamic PDMS substrate, where Pt-catalyzed crosslinking was used to cure the PDMS and thiol-ene photocrosslinking was subsequently applied to stiffen the PDMS. The viscoelasticity and stiffness of the PDMS substrates were temporally controlled using light. The photoresponsive PDMS substrates were cytocompatible and capable of modulating cell behavior, showing a strong correlation between substrate stiffness and cell spreading. When substrates were stiffened using light, adhered cells increased their spreading, and cardiac fibroblasts were activated to myofibroblasts. Taken together, this photo-stimulated crosslinking approach is a fast and cytocompatible way to control the stiffness and viscoelasticity of PDMS, and the photoresponsive PDMS substrates provide a promising platform to study cell-matrix interactions in a dynamic manner.
Supplementary Material
Acknowledgments
The authors would like to thank Matthew Brukman (Singh Center for Nanotechnology, University of Pennsylvania) for help with AFM for topography, Anbin Mu and Vivek Kumar (Cardiovascular Institute, University of Pennsylvania) for help with cardiac fibroblast isolation and culture. This work was supported by an American Heart Association Established Investigator Award and the Center for Engineering MechanoBiology through a grant from the National Science Foundation’s STC Program (CMMI: 15-48571).
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.biomaterials.2017.08.033.
References
- 1.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 2.Mao AS, Shin JW, Mooney DJ. Effects of substrate stiffness and cell-cell contact on mesenchymal stem cell differentiation. Biomaterials. 2016;98:184–191. doi: 10.1016/j.biomaterials.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri O, Koshy ST, Cunha CB, Shin JW, Verbeke CS, Allison KH, Mooney DJ. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Stuart MAC, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WTS. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ. Substrate stress relaxation regulates cell spreading. Nat Commun. 2015;6:6365. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikkhah M, Edalat F, Manoucheri S, Khademhosseini A. Engineering microscale topographies to control the cell-substrate interface. Biomaterials. 2012;33:5230–5246. doi: 10.1016/j.biomaterials.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biggs MJ, Richards RG, Dalby MJ. Nanotopographical modification: a regulator of cellular function through focal adhesions. Nanomedicine. 2010;6:619–633. doi: 10.1016/j.nano.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulangara K, Yang Y, Yang J, Leong KW. Nanotopography as modulator of human mesenchymal stem cell function. Biomaterials. 2012;33:4998–5003. doi: 10.1016/j.biomaterials.2012.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeung T, Georges PG, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 12.Califano JP, Reinhart-King CA. Substrate stiffness and cell area drive cellular traction stresses in single cells and cells in contact. Cell Mol Bioeng. 2010;3:68–75. doi: 10.1007/s12195-010-0102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhart-King CA, Dembo M, Hammer DA. The dynamics and mechanics of endothelial cell spreading. Biophys J. 2005;89:676–689. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubin-Thaler BJ, Hoffman JM, Cai Y, Xenias H, Spielman I, Shneidman AV, David LA, Döbereiner H, Wiggins CH, Sheetz MP. Quantification of cell edge velocities and traction forces reveals distinct motility modules during cell spreading. PLoS One. 2008;3:e3735. doi: 10.1371/journal.pone.0003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isenberg BC, Dimilla PA, Walker M, Kim S, Wong JY. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Bio- phys J. 2009;97:1313–1322. doi: 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadjipanayi E, Mudera V, Brown RA. Close dependence of fibroblast proliferation on collagen scaffold matrix stiffness. J Tissue Eng Regen Med. 2009;3:77–84. doi: 10.1002/term.136. [DOI] [PubMed] [Google Scholar]
- 18.Tilghman RW, Cowan CR, Mih JD, Koryakina Y, Gioeli D, Slack-Davis JK, Blackman BR, Tschumperlin DJ, Parsons JT. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS One. 2010;5:e12905. doi: 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang N, Naruse K, Stamenovic D, Fredberg JJ, Mijailovich SM, Tolić-Nørrelykke IM, Polte T, Mannix R, Ingber DE. Mechanical behavior in living cells consistent with the tensegrity model. Proc Natl Acad Sci U S A. 2001;98:7765–7770. doi: 10.1073/pnas.141199598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo CH, Furukawa K, Suzuki Y, Kasagi N, Ichiki T, Ushida T. A topographically optimized substrate with well-ordered lattice micro-patterns for enhancing the osteogenic differentiation of murine mesenchymal stem cells. Macromol Biosci. 2011;11:938–945. doi: 10.1002/mabi.201000477. [DOI] [PubMed] [Google Scholar]
- 21.Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F, Wang N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker BM, Shah RP, Huang AH, Mauck RL. Dynamic tensile loading improves the functional properties of mesenchymal stem cell-laden nanofiber-based fibrocartilage. Tissue Eng Part A. 2011;17:1445–1455. doi: 10.1089/ten.tea.2010.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi A, Ling QD, Chang Y, Hsu ST, Umezawa A. Physical cues of biomaterials guide stem cell differentiation fate. Chem Rev. 2013;113:3297–3328. doi: 10.1021/cr300426x. [DOI] [PubMed] [Google Scholar]
- 25.Engler AJ, Griffin MA, Sen S, Böonnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissuelike stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice JJ, Martino MM, Laporte LD, Tortelli F, Briquez PS, Hubbell JA. Engineering the regenerative microenvironment with biomaterials. Adv Healthc Mater. 2013;2:57–71. doi: 10.1002/adhm.201200197. [DOI] [PubMed] [Google Scholar]
- 27.Hinderer S, Layland SL, Schenke-Layland K. ECM and ECM-like materials — biomaterials for applications in regenerative medicine and cancer therapy. Adv Drug Deliv Rev. 2016;97:260–269. doi: 10.1016/j.addr.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotech. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 29.Geckil H, Xu F, Zhang X, Moon SJ, Demirci U. Engineering hydrogels as extracellular matrix mimics. Nanomedicine. 2010;5:469–484. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, Mick R, Janmey PA, Furth EE, Wells RG. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1147–G1154. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 31.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Moore SW, Keller RE, Koehl MA. The dorsal involuting marginal zone stiffens anisotropically during its convergent extension in the gastrula of Xenopus laevis. Development. 1995;121:3131–3140. doi: 10.1242/dev.121.10.3131. [DOI] [PubMed] [Google Scholar]
- 34.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease, Cold Spring Harb. Perspect Biol. 2011;3:005058. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and longterm cellular responses to dynamic mechanics. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 37.Caliari SR, Perepelyuk M, Cosgrove BD, Tsai SJ, Lee GY, Mauck RL, Wells RG, Burdick JA. Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci Rep. 2016;6:21387. doi: 10.1038/srep21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stowers RS, Allen SC, Suggs LJ. Dynamic phototuning of 3D hydrogel stiffness. Proc Natl Acad Sci U S A. 2015;112:1953–1958. doi: 10.1073/pnas.1421897112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kloxin AM, Tibbitt MW, Anseth KS. Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nat Protoc. 2010;5:1867–1887. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caliari SR, Perepelyuk M, Soulas E, Lee GY, Wells RG, Burdick JA. Gradually softening hydrogels for modeling hepatic stellate cell behavior during fibrosis regression. Integr Biol. 2016;8:720–728. doi: 10.1039/c6ib00027d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Previtera ML, Trout KL, Verma D, Chippada U, Schloss RS, Langrana NA. Fibroblast morphology on dynamic softening of hydrogels. Ann Biomed Eng. 2012;40:1061–1072. doi: 10.1007/s10439-011-0483-2. [DOI] [PubMed] [Google Scholar]
- 43.Cameron AR, Frith JE, Cooper-White JJ. The influence of substrate creep on mesenchymal stem cell behaviour and phenotype. Biomaterials. 2011;32:5979–5993. doi: 10.1016/j.biomaterials.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Cameron AR, Frith JE, Gomez GA, Yap AS, Cooper-White JJ. The effect of time-dependent deformation of viscoelastic hydrogels on myogenic induction and Rac1 activity in mesenchymal stem cells. Biomaterials. 2014;35:1857–1868. doi: 10.1016/j.biomaterials.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 45.Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H, Lippens E, Duda GN, Mooney DJ. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webber RE, Shull KR. Strain dependence of the viscoelastic properties of alginate hydrogels. Macromolecules. 2004;37:6153–6160. [Google Scholar]
- 47.Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ. Substrate stress relaxation regulates cell spreading. Nat Commun. 2015;6:6364. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutierrez E, Tkachenko E, Besser A, Sundd P, Ley K, Danuser G, Ginsberg MH, Groisman A. High refractive index silicone gels for simultaneous total internal reflection fluorescence and tractiosn force microscopy of adherent cells. PLoS One. 2011;6:e23807. doi: 10.1371/journal.pone.0023807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boudou T, Legant WR, Mu A, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, Chen CS. A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng Part A. 2012;18:910–919. doi: 10.1089/ten.tea.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huh D, Hamilton GA, Ingber DE. From three-dimensional cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakar MS, Neal D, Boudou T, Borochin MA, Li Y, Weiss R, Kamm RD, Chen CS, Asada HH. Formation and optogenetic control of engineered 3D skeletal muscle bioactuators. Lab Chip. 2012;12:4976–4985. doi: 10.1039/c2lc40338b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, Jiang LT, Okada R, Fu J. UV-Modulated substrate rigidity for multiscale study of mechanoresponsive cellular behaviors. Langmuir. 2012;28:10789–10796. doi: 10.1021/la300978x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie J, Zhang Q, Zhu T, Zhang Y, Liu B, Xu J, Zhao H. Substrate stiffness-regulated matrix metalloproteinase output in myocardial cells and cardiac fibroblasts: implications for myocardial fibrosis. Acta Biomater. 2014;10:2463–2472. doi: 10.1016/j.actbio.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 55.Goffin JM, Pittet P, Csucs G, Lussi JW, Meister JJ, Hinz B. Focal adhesion size controls tension-dependent recruitment of α-smooth muscle actin to stress fibers. J Cell Biol. 2006;172:259–268. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen KDQ, Megone WV, Konga D, Gautrot JE. Ultrafast diffusioncontrolled thiol-ene based crosslinking of silicone elastomers with tailored mechanical properties for biomedical applications. Polym Chem. 2016;7:5281–5293. [Google Scholar]
- 57.Berg OVD, Nguyen LTT, Teixeira RFA, Goethals F, Özdilek C, Berghmans S, Prez FED. Low modulus dry silicone-gel materials by photoinduced thiol-ene chemistry. Macromolecules. 2014;47:1292–1300. [Google Scholar]
- 58.Cole MA, Bowman CN. Synthesis and characterization of thiol-ene functionalized siloxanes and evaluation of their crosslinked network properties. J Polym Sci Part A Polym Chem. 2013;51:1749–1757. doi: 10.1002/pola.26245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi KM, Rogers JA. A photocurable poly(dimethylsiloxane) chemistry designed for soft lithographic molding and printing in the nanometer regime. J Am Chem Soc. 2003;125:4060–4061. doi: 10.1021/ja029973k. [DOI] [PubMed] [Google Scholar]
- 60.Xue L, Zhang Y, Zuo Y, Diao S, Zhang J, Feng S. Preparation and characterization of novel UV-curing silicone rubber via thiol-ene reaction. Mater Lett. 2013;106:425–427. [Google Scholar]
- 61.Hoyle CE, Lowe AB, Bowman CN. Thiol-click chemistry: a multifaceted toolbox for small molecule and polymer synthesis. Chem Soc Rev. 2010;39:1355–1387. doi: 10.1039/b901979k. [DOI] [PubMed] [Google Scholar]
- 62.Lowe AB. Thiol-ene “click” reactions and recent applications in polymer and materials synthesis: a first update. Polym Chem. 2014;5:4820–4870. [Google Scholar]
- 63.Buxboim A, Rajagopal K, Brown AE, Discher DE. How deeply cells feel: methods for thin gels. J Phys Condens Matter. 2010;22:190301–195108. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Q, Qu F, Han B, Wang C, Li H, Mauck RL, Han L. Micromechanical anisotropy and heterogeneity of the meniscus extracellular matrix. Acta Biomater. 2017;54:356–366. doi: 10.1016/j.actbio.2017.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin DC, Shreiber DI, Dimitriadis EK, Horkay F. Spherical indentation of soft matter beyond the Hertzian regime: numerical and experimental validation of hyperelastic models. Biomech Model Mechanobiol. 2009;8:345–358. doi: 10.1007/s10237-008-0139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dimitriadis EK, Horkay F, Maresca J, Kachar B, Chadwick RS. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys J. 2002;82:2798–2810. doi: 10.1016/S0006-3495(02)75620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kubo H, Jaleel N, Kumarapeli A, Berretta RM, Bratinov G, Shan X, Wang H, Houser SR, Margulies KB. Increased cardiac myocyte progenitors in failing human hearts. Circulation. 2008;118:649–657. doi: 10.1161/CIRCULATIONAHA.107.761031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bowers SLK, Baudino TA. Analyzing cell-cell Interactions in 3-dimensional adhesion assays. Methods Mol Biol. 2013;1066:29–43. doi: 10.1007/978-1-62703-604-7_3. [DOI] [PubMed] [Google Scholar]
- 69.Han B, Chery DR, Yin J, Lu XL, Lee D, Han L. Nanomechanics of layer-by-layer polyelectrolyte complexes: a manifestation of ionic cross-links and fixed charges. Soft Matter. 2016;12:1158–1169. doi: 10.1039/c5sm01430a. [DOI] [PubMed] [Google Scholar]
- 70.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast Adaptation and Stiffness Matching To soft elastic substrates. Biophys J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lajiness JD, Conway SJ. The dynamic role of cardiac fibroblasts in development and disease. J Cardiovasc Trans Res. 2012;5:739–748. doi: 10.1007/s12265-012-9394-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lajiness JD, Conway SJ. Origin, development, and differentiation of cardiac fibroblasts. J Mol Cell Cardiol. 2014;70:2–8. doi: 10.1016/j.yjmcc.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac fibrosis. Circ Res. 2016;118:1021–1040. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan D, Takawale A, Lee J, Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease, Fibrogenes. Tissue Repair. 2012;5:15. doi: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


