Abstract
Arginine methyltransferases critically regulate cellular homeostasis by modulating the functional outcome of their substrates. The protein arginine methyltransferase 5 (PRMT5) is an enzyme involved in growth and survival pathways promoting tumorigenesis. However, little is known about the biologic function of PRMT5 and its therapeutic potential in multiple myeloma (MM). In the present study, we identified and validated PRMT5 as a new therapeutic target in MM. PRMT5 is overexpressed in patient MM cells and associated with decreased PFS and OS. Either genetic knockdown or pharmacological inhibition of PRMT5 with the inhibitor EPZ015666 significantly inhibited growth of both cell lines and patient MM cells. Furthermore, PRMT5 inhibition abrogated NF-κB signaling. Interestingly, mass spectrometry identified a tripartite motif-containing protein 21 TRIM-21 as a new PRMT5-partner, and we delineated a TRIM21-dependent mechanism of NF-κB inhibition. Importantly, oral administration of EPZ015666 significantly decreased MM growth in a humanized murine model of MM. These data both demonstrate the oncogenic role and prognostic relevance of PRMT5 in MM pathogenesis, and provide the rationale for novel therapies targeting PRMT5 to improve patient outcome.
Introduction
Multiple myeloma (MM) is a lethal malignancy characterized by clonal proliferation of plasma cells in the bone marrow (BM)1. Intrinsic genomic complexity drives a multistep progression from a pre-malignant condition (monoclonal gammopathy of undetermined significance, MGUS) to overt disease2,3. A deeper analyses of the genomic, epigenomic, and proteomic landscape in MM is required to identify novel therapeutic targets3.
An emerging field of investigation is the role of aberrant post-translational modifications (PTMs) in cancer initiation and progression4–7. So far, particular emphasis is on arginine methylation, a PTM catalyzed by protein methyltransferases (PRMTs), which affects key cellular processes via modulation of gene transcription and/or protein function8–9. Among the others, PRMT5 is a major type II PRMT enzyme which catalyzes the symmetric transfer of up to two methyl groups to arginine residues10. Methylation by PRMT5 modulates the biological function of target proteins and is essential to maintain homeostasis in both normal and malignant cells11–13. Interestingly, it is frequently overexpressed in human cancer14–17 and its increased activity has been shown to support cell transformation15,18. To date, several nuclear and cytoplasmic substrates of PRMT5 have been described19: for instance, p53 was recently identified as PRMT5 substrate and PRMT5-mediated arginine methylation in p53 impairs its DNA binding and tumor-suppressor activity20.
In this study, we investigate for the first time the role of PRMT5 in MM pathophysiology. Moreover, we provide the rational framework for its therapeutic targeting by a recently developed selective and orally-bioavailable PRMT5 inhibitor (EPZ015666)21.
Methods
Methods and any associated references are available in the Supplementary methods section.
Results
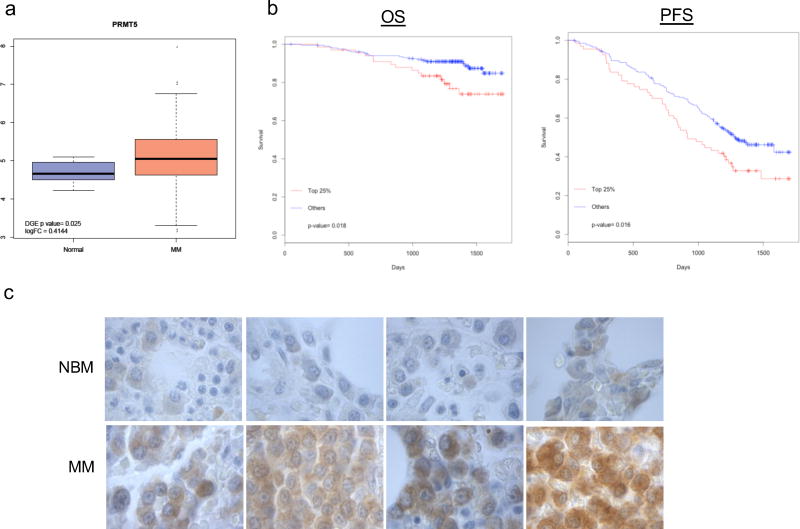
PRMT5 overexpression has clinical impact on MM patient outcome
The first aim of our study was to investigate whether PRMT5 can be a potential novel therapeutic target for MM patients. To this end, CD138+ immunopurified cells from a cohort of newly-diagnosed MM patients (n=320) or healthy donors (n=16) were analyzed by RNA-seq. Interestingly, significant upregulation of PRMT5 was observed in patient MM (pMM) cells (Fig. 1a). Moreover, analysis of PRMT5 expression in transplant eligible MM patients enrolled in the IFM/DFCI 2009 clinical study (NCT01191060)22 showed that higher PRMT5 expression was associated with shorter overall survival (OS, p=0.018) and progression free survival (PFS, p= 0.016) (Fig. 1b). Additional analysis of PRMT5 expression in other independent MM patient datasets (GSE6477, GSE5900 and GSE2658)23–25 showed that upregulation of PRMT5 was associated with disease progression (Supplementary Fig. S1a–b). By qRT-PCR, we found upregulated PRMT5 in MM cell lines as compared to healthy donor plasma cells (Supplementary Fig. S1c). Moreover, both immunohistochemical staining of BM biopsies and western blotting (WB) of PRMT5 indicated its upregulation at protein level in pMM cells (Fig. 1c and Supplementary Fig. S1d). This finding was also confirmed, by WB, in MM cell lines (n=13) as compared to PBMCs (n=3) (Supplementary Fig. S1e). Of note, a parallel increase of cellular symmetric arginine di-methylation (SDMA) substrates was also noted in the same samples (Supplementary Fig. S1e). Overall, these findings provide evidence that PRMT5 is overexpressed in MM patients and portends poor clinical outcome.
Fig. 1. High levels of PRMT5 in MM patients correlate with poor clinical outcome.
a, Box plot of PRMT5 from RNA-seq analysis data comparing plasma cells from healthy subjects versus newly diagnosed MM patients (P=0.025). b, Kaplan–Meier curves demonstrating association of OS (P= 0.018) and PFS (P= 0.016) with PRMT5 expression, as analyzed by log-rank test in a dataset of 320 newly diagnosed MM patients. c, PRMT5 immunostaining in BM biopsies from healthy individuals (NBM) (n = 4) and MM (n= 4) 250× magnification.
RNAi- or drug-mediated inhibition of PRMT5 decreases MM cell growth and survival in a p53-independent manner
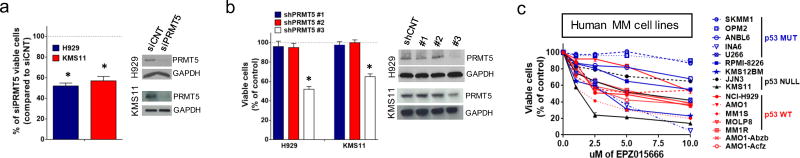
The functional significance of PRMT5 in MM cells was first addressed in RNAi-mediated loss-of-function experiments. Specifically, silencing of PRMT5 by siRNAs decreased proliferation in H929, KMS11, MM1S, U266, RPMI-8226 and AMO1 MM cells (Fig. 2a and Supplementary Fig. S2a), along with induction of apoptotic markers (Supplementary Fig. S2b). Moreover, the anti-proliferative effect by PRMT5 downregulation was further confirmed by lentivirally transduced shRNAs targeting PRMT5 (shPRMT5 #1, #2 and #3) in both H929 (p53wt) and KMS11 (p53null) cells. As shown in Fig. 2b, shPRMT5 #3 impaired proliferation of both MM cell lines, independent of the p53 mutational status. Importantly, decreased proliferation was associated with a decrease of SDMA levels (Supplementary Fig. S2c).
Fig. 2. PRMT5 inhibition impacts MM cell survival.
a, MTT assay of H929 and KMS11 after 72h transfection with siPRMT5 or siCNT. PRMT5 and GAPDH protein levels are shown. b, MTT assay of H929 and KMS11 lentivirally transduced with shPRMT5 (#1, #2 and #3) or shCNT. PRMT5 and GAPDH protein levels are shown. Cell viability was measured on days 6 after selection. c, MTT assay of MM cell lines after EPZ015666 (0 to 10umol/l) treatment. Data are representative of at least 3 independent experiments. * = p<0.05
The effect of PRMT5 inhibition on p53 methylation was next explored in p53 wt MM cells. As expected, PRMT5 silencing by siRNA reduced p53 methylation (Supplementary Fig. S3a), along with increased p53 transcriptional activity (Supplementary Fig. S3b). Despite this effect, either transient or lentiviral p53 silencing did not abrogate the anti-proliferative and pro-apoptotic activity induced by PRMT5 knockdown (Supplementary Fig. S3c–d). These findings indicate a p53-independent anti-MM activity of PRMT5.
EPZ015666 is a specific inhibitor of PRMT521. We next investigated its effect on a large panel of MM cell lines (n=16). Proliferation of MM cell lines treated with increasing concentrations of EPZ015666 was inhibited in a dose-dependent fashion independent of their p53 mutational status (Fig. 2c). Of note, EPZ015666 inhibited proliferation of dexamethasone (MM1R), bortezomib (ABZB), or carfilzomib (ACFZ)-refractory cells and IL-6 dependent INA-6 cells (Fig. 2c). Anti-proliferative activity of EPZ015666 was associated with a parallel decrease of SDMA levels (Supplementary Fig. S2d). Importantly, EPZ015666 treatment significantly inhibited DNA synthesis and clonogenicity and induced apoptosis in H929 and KMS11 cells (Supplementary Fig. S2e–f–g–h). In a parallel analysis, a slight increased proliferation was observed in KMS11 and H929 lentivirally transduced to overexpress PRMT5 (Supplementary Fig. S2i). Moreover, ectopic PRMT5 rescued anti-MM activity of EPZ015666 in KMS11 cells, indicating that drug activity is an on-target effect (Supplementary Fig. S2l). These findings demonstrate a potent anti-proliferative effect of PRMT5 inhibition in MM cells, and provide evidence supporting therapeutic targeting of PRMT5 in MM via the pharmacologic inhibitor EPZ015666.
EPZ015666 inhibits growth of CD138+ patient MM cells and overcomes the protective effect of the huBMM
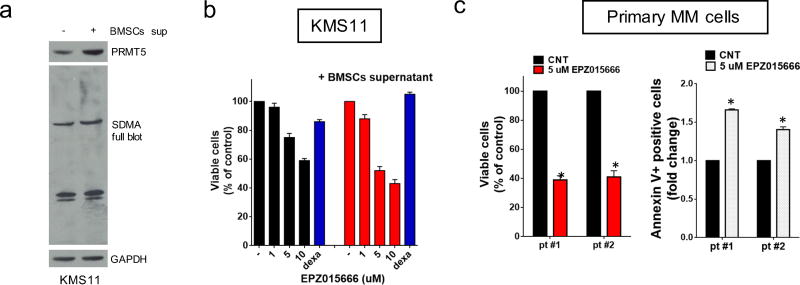
The human bone marrow microenvironment (huBMM) exerts a protective role on MM cells26. Here we show by WB analysis that expression of both PRMT5 and SDMA in H929 and KMS11 is increased after culture with BM stromal cells-conditioned medium (BMSCs-cm) (Fig. 3a and Supplementary Fig. S4a), implicating PRMT5 in huBMM-mediated pro-MM survival signaling. Importantly, EPZ015666 exerted its anti-proliferative effect in H929 and KMS11 even in the presence of BMSCs-cm (Fig. 3b and Supplementary Fig. S4b).
Fig. 3. EPZ015666 inhibits patients MM cell growth and overcomes protective effects of the huBMM.
a, WB of PRMT5, SDMA, and GAPDH in KMS11 cultured alone or in the presence of BMSCs-supernatant. b, MTT assay of KMS11 treated with EPZ015666, alone or in the presence of BMSCs supernatant. Dexamethasone was used as a positive control. c. Left panel: Cell growth analysis of CD138+ pMM cells (n=2) cultured on a BMSCs monolayer for 6 days, with or without 5 umol/l of EPZ015666. Right panel: pMM cells (n=2) were cultured in the presence of BMMCs. Apoptosis was assessed by Annexin-V staining. Data are representative of at least 3 independent experiments.* = p<0.05
Pro-survival signaling and oncogene-dependency could differ in CD138+ pMM cells compared to MM cell lines. We show that EPZ015666 also inhibited growth in pMM cells (n=2) cultured on a BMSCs monolayer, and also induced apoptosis of pMM cells (n=2) within bone marrow mononuclear cell cultures (Fig. 3c). Importantly, EPZ015666 did not affect the viability of health PBMCs, even though SDMA expression was decreased (Supplementary Fig. S4c–d).
EPZ015666 inhibits NF-κB signaling by triggering an IKKbeta (IKKβ) inhibitor-like gene signature
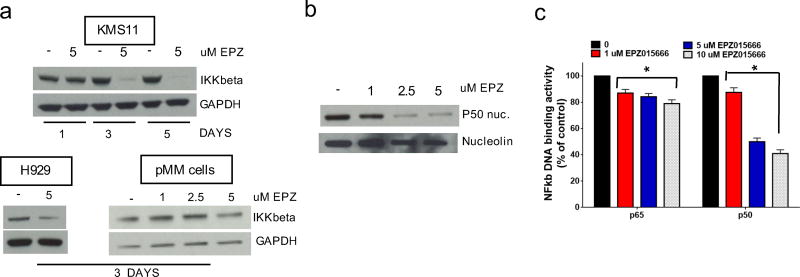
Molecular perturbations induced by EPZ015666 were next investigated at the transcriptome level. Unsupervised hierarchical clustering segregated samples based on treatment identified a coherent transcriptome modulation induced by EPZ015666 (data not shown). Gene set enrichment analysis (GSEA) was performed to identify biological pathways affected by EPZ015666 in KMS11 MM cells. Importantly, an open-ended enrichment analysis including the Hallmark gene sets collection available from Molecular Signature Database (MSigBD)27 revealed that the transcriptional signature of NF-κb-dependent genes (Hallmark_TNFA_Signaling_via_NF-kB) strongly correlated with genes downregulated by EPZ015666 [NES= −3.15, FDR = <0.0001] (Supplementary Figure S5a and Supplementary table S1). Genes which were differentially-expressed after EPZ015666 treatment were analyzed by Ingenuity pathway Analysis (IPA). In particular, Upstream Regulator Analysis identified NF-κB1 as a transcriptional regulator of the observed gene expression changes, and predicted its inactivation after EPZ015666 treatment (data not shown).
The NF-κB pathway plays a central role in the pathophysiology of MM28. Previous observations identified a NF-κB gene signature by integrating genes co-expressed in MM cell lines with those modulated after IKKβ inhibition29. Interestingly, a significant downregulation of the depicted NF-κB signature was found in EPZ015666-treated cells (Supplementary Fig. S5b). Downregulation was further confirmed by RT-PCR in 9 of 11 genes in the signature (NF-κBIA, RELB, TNFAIP3, NF-κBIE, MALT1, BIRC3, CD74, ILRG2 and NF-κB2) in KMS11 treated with EPZ015666 (Supplementary Fig. S5c). Moreover, consistent with a IKKβ inhibitor-like activity, IKKβ protein expression significantly decreased in KMS11, H929 and pMM cells exposed to EPZ015666 (Fig. 4a). Accordingly, WB revealed a concentration-dependent decrease of NF-κB p50 subunit within the nuclear fraction in KMS11 MM cells (Fig. 4b). Indeed, measurement of NF-κB DNA-binding activity in nuclear extracts from treated cells showed a decrease of DNA binding by complexes containing p65 and p50 subunits (Fig. 4c). Of note, inhibition of NF-κB by EPZ015666 was observed only in MM cells, and was not detected in healthy PBMCs (Supplementary Fig. S5d). These findings, including in silico inference and orthogonal validation, identify and confirm that inhibition of NF-κB pathway is a molecular consequence of PRMT5 inhibition.
Fig. 4. Inhibition of NF-κB-dependent transcription in MM cell lines by EPZ015666.
a, WB of IKKβ and GAPDH in KMS11,H929 and pMM cells treated with EPZ015666. b, WB of p50 subunit and Nucleolin in nuclear extracts of EPZ01566-treated KMS11. c, Nuclear extracts were assayed by ELISA for induction of p65 and p50 DNA binding after EPZ015666 treatment. * = p<0.05
Functional interaction of PRMT5 with the E3 ubiquitin-ligase TRIM21
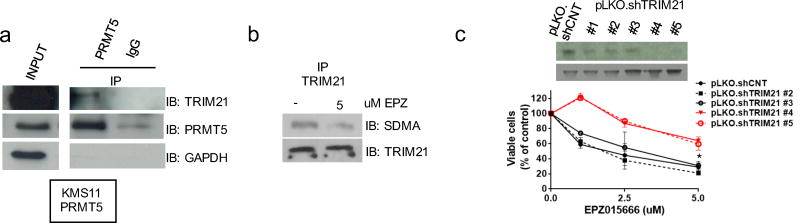
Based on above findings, our next aim was to delineate the functional mechanism of PRMT5 inhibition on NF-κB signaling. Indeed, this functional interaction likely accounts, at least in part, for the anti-MM activity of EPZ015666. Thus, we next identified PRMT5-interacting proteins in MM cells. Specifically, we analyzed proteins in immunocomplexes which co-precipitated with PRMT5 in PRMT5-overexpressing (KMS11-PRMT5) cells. PRMT5-associated proteins were submitted to mass spectrometry (MS). As expected, a large number of novel or established partners were identified. Among these, the E3 ubiquitin ligase TRIM21 (also known as Ro52) was further studied, since it was a high affinity PRMT5-binding partner present only in PRMT5-associated complexes. The PRMT5-TRIM21 interaction was further confirmed by WB analysis of PRMT5-immunoprecipitated proteins (Fig. 5a). Moreover, arginine methylation of TRIM21 in lysates from KMS11 cells was decreased after treatment with EPZ015666 (Fig.5b), indicating the functional significance of PRMT5/TRIM21 interaction. Of note, stable silencing of TRIM21 in MM cell lines significantly abrogated the anti-proliferative effect of EPZ015666 (Fig. 5c). Overall, these findings show that TRIM21 function is regulated by PRMT5-induced arginine methylation, and provide the rationale to explore the role of TRIM21 in mediating the anti-proliferative activity of PRMT5 inhibition.
Fig. 5. PRMT5 methylates TRIM21.
a, KMS11 were stably transfected to overexpress PRMT5. Cell lysates were immunoprecipitated with control-IgG or PRMT5, and then immunoblotted with TRIM21, PRMT5, and GAPDH antibodies. Input levels are shown. b, KMS11 were treated with or without 5 umol/l of EPZ015666 for 48h. Cell lysates were then immunoprecipitated with TRIM21 antibody and immunoblotted for TRIM21 and SDMA. c, MTT assay of KMS11 lentivirally transduced with pLKO.shTRIM21 (#1, #2, #3, #4 and #5) or p.LKO.shCNT and treated with EPZ015666. TRIM21 protein levels are shown.
EPZ015666 triggers TRIM21-mediated autophagic proteolysis of IKKβ
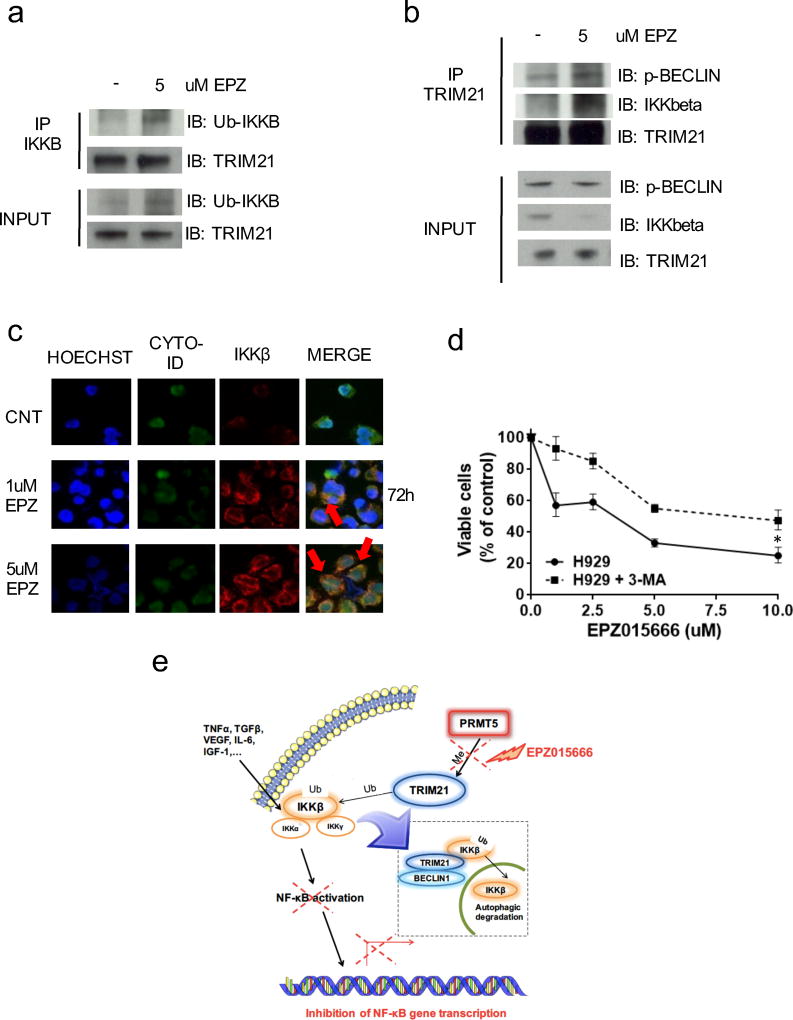
Previous findings showed that TRIM21 monoubiquitinates IKKβ30, and that this modification triggers its degradation via selective autophagy31,32. Specifically, proteins in the TRIM family may selectively induce degradation of targeted proteins due to sequence-specific recognition and formation of an autophagic complex, which shuttles the cargo protein into the autophagosome33. Here we hypothesized that the interaction of TRIM21 with PRMT5 blocks TRIM21-mediated degradation of IKKβ, allowing activation of NF-κB signaling. Indeed, consistent with our hypothesis, increased IKKβ monoubiquitination (Fig. 6a) and formation of IKKβ-TRIM21-pBECLIN1 autophagic complexes (Fig. 6b) was detected after treatment of MM cells with EPZ015666. Moreover, fluorescence microscopy analysis confirmed co-localization of IKKβ in the autophagosome after treatment with EPZ015666 (Fig. 6c and Supplementary Fig. S6a). Conversely, exposure of MM cells to 3-methyladenine (3-MA), an inhibitor of autophagosome formation, rescued EPZ015666-induced growth inhibition in KMS11 and H929 (Fig.6d and Supplementary Fig. S6b). At protein level, 3-MA prevented the degradation of endogenous IKKβ (Supplementary Fig. S6c). Taken together, these data indicate that PRMT5-mediated TRIM21 methylation has a key regulatory function of NF-κB signaling in MM cells mediated via inhibition of autophagic proteolysis of IKKβ (Fig. 6e). Indeed, direct inhibition of the PRMT5/TRIM21 axis mediates anti-MM activity of EPZ015666.
Fig. 6. PRMT5 inhibition by EPZ015666 induces autophagic proteolysis of IKKβ.
a–b, H929 were treated with EPZ015666 for 72h in the presence of 0.5 mmol/l of 3-MA. Cell lysates were immunoprecipitated with: (a) IKKβ antibody, and then immunoblotted for Ubiquitin and TRIM21 or (b) TRIM21 antibody, and then immunoblotted for phospho-Beclin1, IKKβ, and TRIM21. c, H929 were treated with EPZ015666. After treatment, samples dual stained with Cyto-ID Green dye and IKKβ antibody were analyzed by fluorescence microscopy. Arrows indicate colocalization of IKKβ in the autophagosome. d, MTT assay of H929 treated with EPZ015666 in presence or absence of 0.5 mmol/l of 3-MA. e, Proposed model of PRMT5/TRIM21/IKKB axis in MM. Data are representative of at least two independent experiments. * = p<0.05
Oral EPZ015666 treatment inhibits tumor growth of MM xenografts in NOD SCID mice
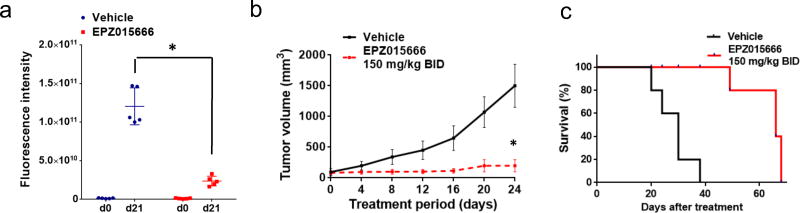
In vivo anti-MM activity of EPZ015666 was next investigated in NOD SCID mice bearing subcutaneous GFP+-KMS11 xenografts. Mice received twice-daily oral treatment with 150 mg/kg EPZ015666 for 21 days. Importantly, this treatment resulted in a significant inhibition of tumor growth (Fig. 7a–b and Supplementary Fig. S7a). Kaplan-Meier curves showed a significantly prolonged survival in the EPZ015666-treated group (log-rank analysis p=0.0001), with a median survival of 30 days in control mice versus 66 days in the EPZ015666 treated cohort (Fig. 7c). WB analysis of tumors retrieved from EPZ015666-treated mice confirmed in vivo target inhibition by EPZ015666, assessed by decreased SDMA substrates (Supplementary Fig S7b). These data demonstrate that PRMT5 inhibition by EPZ015666 inhibits in vivo growth of MM xenografts and prolongs survival without adverse toxicity profile, providing the preclinical rationale for its clinical evaluation in MM.
Fig. 7. EPZ015666 decreased MM tumor growth in vivo.
Anti-tumor activity induced by oral administration of EPZ015666 or vehicle against GFP+-KMS11 cell xenografts: a, quantification of fluorescence intensity in the ROI of each tumor of mice treated with EPZ015666 or control, and representation of the mean intensity for each group. b, Average tumor volume of each group measured by electronic caliper ± s.d. is shown. * = p<0.05. c, Right panel: Kaplan–Meier curves of EPZ015666-treated mice as compared to controls (log-rank test, P<0.05). Survival was evaluated from the first day of treatment until death or sacrifice. * = p<0.05
Discussion
In the present study, we identified PRMT5 as a prognostic marker and promising therapeutic target in MM. Our data suggest that overexpression of PRMT5 plays a significant role of in MM pathogenesis, since it progressively increases during MM progression, as well as correlates with clinical outcome. We show MM dependence on PRMT5, since genetic knockdown via RNAi-approaches inhibits growth and induces apoptosis of both MM cell lines and patient MM cells. These effects are observed in both p53wt and p53null MM cell lines, suggesting that the oncogenic role of PRMT5 does not rely on p53 status in MM.
To explore the potential clinical translation of PRMT5 inhibition, we examined the anti-MM activity of a recently described PRMT5 inhibitor EPZ015666. Importantly, proliferation of both p53wt and p53null MM cells was inhibited by this agent. Previous reports have shown that p53 is a direct target of PRMT5, which alters its target gene specificity by arginine methylation20. Consistent with these findings, we show that p53 transcriptional activity was enhanced in H929 p53wt cells by either si-PRMT5 or EPZ015666 treatment. However, our data show that activation of p53 transcriptional activity is not essential for growth inhibition triggered by downregulation of PRMT5, since genetic knockdown of p53 by shRNAs in AMO1 (p53wt) did not decrease sensitivity to EPZ015666. Among p53 mutated cells, only SKMM1 and OPM-2 MM cells are insensitive to EPZ015666; these cell lines have a p53 mutation on codon 175, which represents a hotspot gain-of-function mutation possibly involved in resistance to EPZ015666. This mutation is rarely observed in MM patients34,35, and therefore has not been further studied here. Altogether, we found that EPZ015666 treatment decreased DNA synthesis, clonogenicity and induced apoptosis in MM cells, without impacting normal donor PBMCs, suggesting a favorable therapeutic index.
The interplay between MM cells and their huBMM drives disease progression through promotion of pro-survival and drug resistance pathways26. Notably, we here observed an increase of PRMT5 expression after culture of MM cells in BMSC-conditioned media. This data suggests involvement of PRMT5 in huBMM-mediated MM cell protective signaling, and a potential direct effect of huBMM stimuli on PRMT5 transcription, which is currently the object of ongoing studies. Importantly, EPZ015666 retains its anti-MM activity against either MM cell lines or patient MM cells cultured with BMSCs-conditioned media or with adhesion to BMSCs, indicating that EPZ015666 can overcome drug resistance due to the interplay of tumor cells with their huBMM26.
To date, multiple oncogenic pathways have been described which are directly affected by PRMT519. By transcriptome analysis, we here identified a new interplay between PRMT5 with the NF-κB survival pathway which could account, at least part, for MM dependence on PRMT5. Specifically, we found downregulated NF-κB transcriptional signatures in MM cells exposed to EPZ015666. Additional validation of this outcome was carried out by qRT-PCR, analysis of NF-κB DNA binding activity, and immunoblotting. Of note, our findings overlapped with those obtained by Annunziata et al. after MM cell exposure to an IKKβ inhibitor29, and our protein analysis showed markedly decreased IKKβ expression in MM cell lines and pMM cells after treatment with EPZ015666. Hence, we hypothesized a mechanism involving IKKβ degradation. To address this point, we searched by MS for direct interactors linking PRMT5 to IKKβ; and importantly, this analysis identified the E3 ubiquitin (Ub)-ligase TRIM21. This interaction was further validated by detection of TRIM21 protein in PRMT5 immunocomplexes. TRIM family proteins are known to regulate different cellular processes, and their altered activity can sustain carcinogenesis36. Specifically, a large body of evidence has characterized their role in selective autophagy: by acting as autophagy receptors, they allow the formation of the autophagy platform or “TRIMosome”, promoting the selective degradation of tagged-cargo proteins32,33. As for other TRIM proteins, TRIM21 acts as autophagy receptor37. Importantly, it inhibits NF-κB signaling by driving direct IKKβ monoubiquitination and its subsequent degradation through an autophagy-mediated mechanism30,31. In our studies, EPZ015666 decreased TRIM21 SDMA and increased monoubiquitination of IKKβ, consistent with PRMT5-mediated regulation of TRIM21 functions. Indeed, arginine methylation, along with other PTMs, has been shown to modulate autophagy receptor activity38,39. Importantly, we here show that EPZ015666 promotes formation of IKKβ-TRIM21-pBECLIN1 autophagic complexes, as assessed by fluorescence microscopy and WB. Of note, both the RNAi-mediated silencing of TRIM21 and the inhibition of autophagosome formation by 3-methyladenine abrogates the anti-MM activity of EPZ015666. Together, these findings indicate that the anti-MM activity of EPZ015666 is due to inhibition of canonical NF-κB signaling. Mechanistically, they indicate that PRMT5/NF-κB interplay takes place via TRIM21, a newly-identified interactor of PRMT5 which governs selective autophagy-mediated turn-over of IKKβ.
Finally, we demonstrated a potent in vivo growth-inhibitory activity of EPZ015666 against human MM xenografts in NOD SCID mice, without attendant toxicities to normal tissues. Indeed, oral administration of EPZ015666 resulted in a complete inhibition of tumor-growth, associated with target inhibition and its sequelae in tumors harvested from treated mice. Further studies using in vivo models recapitulating the huBMM40,41 will better define the effects of PRMT5 inhibition in the BM milieu: indeed, our prior studies have shown that NF-κB regulates adhesion molecule expression on MM cells and BM cells, as well as modulates transcription of MM growth and survival factors such as interleukin-642.
Altogether, our findings define the role of PRMT5 in MM pathogenesis, and provide the preclinical rationale for clinical trials targeting PRMT5 with EPZ015666 to improve patient outcome in MM.
Supplementary Material
Acknowledgments
This work has been supported by a grant from NIH P50-100707 (KCA,NCM), RO-1 CA050947 (KCA), and RO-1 CA 178264 (TH) and VA merit grant (I01BX001584) (NCM) and partially by the Italian Association for Cancer Research (AIRC) with “Special Program for Molecular Clinical Oncology–5 per mille”, 2010/15 and its Extension Program” No. 9980, 2016/18 (PI: PT); and also by “Innovative Immunotherapeutic Treatments of Human Cancer” Multi Unit Regional No. 16695 (cofinanced by AIRC and the CARICAL foundation), 2015/18 (PI: PT). KCA is an American Cancer Society Clinical Research Professor.
Footnotes
Authorship Contributions
A.G., T.H. and K.C.A. designed research and wrote the manuscript; A.G., G.B., M.F., T.H., E.M. and N.A. performed the in vitro experiments and analyzed the data; MKS analyzed gene expression and RNA-seq data; R.C. performed IHC staining; J.Q. provided reagents and analytic tools; Y.-T.T. and K.C.A. provided MM patients samples; A.G. and E.M. designed, performed and analyzed in vivo experiments; N.C.M., P.T. and P.T. provided critical evaluation of experimental data and edited the manuscript.
Conflicts of Interest:
N.C.M. serves on advisory boards to Millennium, Celgene, and Novartis. K.C.A. serves on advisory boards Celgene, Millennium and Gilead Sciences and is a Scientific founder of OncoPep and C4 Therapeutics. All other authors declare no competing financial interests.
Supplementary information is available at Leukemia’s website.
References
- 1.Anderson KC. Progress and Paradigms in Multiple Myeloma. Clin Cancer Res. 2016;22(22):5419–27. doi: 10.1158/1078-0432.CCR-16-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi G, Richardson PG, Anderson KC. Promising therapies in multiple myeloma. Blood. 2015;126(3):300–10. doi: 10.1182/blood-2015-03-575365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohguchi H, Hideshima T, Bhasin MK, Gorgun GT, Santo L, Cea M, et al. The KDM3A-KLF2-IRF4 axis maintains myeloma cell survival. Nat Commun. 2016;7:10258. doi: 10.1038/ncomms10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amodio N, Stamato MA, Gulla AM, Morelli E, Romeo E, Raimondi L, et al. Therapeutic Targeting of miR-29b/HDAC4 Epigenetic Loop in Multiple Myeloma. Mol Cancer Ther. 2016;15(6):1364–75. doi: 10.1158/1535-7163.MCT-15-0985. [DOI] [PubMed] [Google Scholar]
- 6.Hideshima T, Qi J, Paranal RM, Tang W, Greenberg E, West N, et al. Discovery of selective small-molecule HDAC6 inhibitor for overcoming proteasome inhibitor resistance in multiple myeloma. Proc Natl Acad Sci U S A. 2016;113(46):13162–7. doi: 10.1073/pnas.1608067113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amodio N, D'Aquila P, Passarino G, Tassone P, Bellizzi D. Epigenetic modifications in multiple myeloma: recent advances on the role of DNA and histone methylation. Expert Opin Ther Targets. 2017;21(1):91–101. doi: 10.1080/14728222.2016.1266339. [DOI] [PubMed] [Google Scholar]
- 8.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9(10):1208–15. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 9.Copeland RA. Molecular pathways: protein methyltransferases in cancer. Clin Cancer Res. 2013;19(23):6344–50. doi: 10.1158/1078-0432.CCR-13-0223. [DOI] [PubMed] [Google Scholar]
- 10.Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72(11):2041–59. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351(6278):1214–8. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13(1):37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 14.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26(15):3558–69. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Chitnis N, Nakagawa H, Kita Y, Natsugoe S, Yang Y, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5(3):288–303. doi: 10.1158/2159-8290.CD-14-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu D, Gur M, Zhou Z, Gamper A, Hung MC, Fujita N, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun. 2015;6:8419. doi: 10.1038/ncomms9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B, Dong S, Zhu R, Hu C, Hou J, Li Y, et al. Targeting protein arginine methyltransferase 5 inhibits colorectal cancer growth by decreasing arginine methylation of eIF4E and FGFR3. Oncotarget. 2015;6(26):22799–811. doi: 10.18632/oncotarget.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alinari L, Mahasenan KV, Yan F, Karkhanis V, Chung JH, Smith EM, et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood. 2015;125(16):2530–43. doi: 10.1182/blood-2014-12-619783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci. 2011;36(12):633–41. doi: 10.1016/j.tibs.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10(12):1431–9. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 21.Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11(6):432–7. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 22.Attal M, Lauwers-Cances V, Hulin C, Leleu X, Caillot D, Escoffre M, et al. Lenalidomide, Bortezomib, and Dexamethasone with Transplantation for Myeloma. N Engl J Med. 2017;7(4):e554. doi: 10.1056/NEJMoa1611750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chng WJ1, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67(7):2982–9. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 24.Zhan F1, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109(4):1692–700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanamura I, Huang Y, Zhan F, Barlogie B, Shaughnessy J. Prognostic value of cyclin D2 mRNA expression in newly diagnosed multiple myeloma treated with high-dose chemotherapy and tandem autologous stem cell transplantations. Leukemia. 2006;20(7):1288–90. doi: 10.1038/sj.leu.2404253. [DOI] [PubMed] [Google Scholar]
- 26.Kawano Y, Roccaro AM, Azzi J, Ghobrial IM. Multiple Myeloma and the immune microenvironment. Curr Cancer Drug Targets. 2017 doi: 10.2174/1568009617666170214102301. [DOI] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T, et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277(19):16639–47. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 29.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12(2):115–30. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada K, Niida M, Tanaka M, Kamitani T. Ro52-mediated monoubiquitination of IKK{beta} down-regulates NF-{kappa}B signalling. J Biochem. 2009;146(6):821–32. doi: 10.1093/jb/mvp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niida M, Tanaka M, Kamitani T. Downregulation of active IKK beta by Ro52-mediated autophagy. Mol Immunol. 2010;47(14):2378–87. doi: 10.1016/j.molimm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12(9):836–41. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 33.Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014;30(4):394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chng WJ, Price-Troska T, Gonzalez-Paz N, Van Wier S, Jacobus S, Blood E, et al. Clinical significance of TP53 mutation in myeloma. Leukemia. 2007;21(3):582–4. doi: 10.1038/sj.leu.2404524. [DOI] [PubMed] [Google Scholar]
- 35.Lode L, Eveillard M, Trichet V, Soussi T, Wuilleme S, Richebourg S, et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica. 2010;95(11):1973–6. doi: 10.3324/haematol.2010.023697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatakeyama S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem Sci. 2017 doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015;210(6):973–89. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Yang P, Tian E, Zhang H. Arginine methylation modulates autophagic degradation of PGL granules in C. elegans. Mol Cell. 2013;52(3):421–33. doi: 10.1016/j.molcel.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16(6):495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 40.Calimeri T, Battista E, Conforti F, Neri P, Di Martino MT, Rossi M, et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia. 2011;25(4):707–11. doi: 10.1038/leu.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tassone P, Neri P, Carrasco DR, Burger R, Goldmacher VS, Fram R, et al. A clinically relevant SCID-hu in vivo model of human multiple myeloma. Blood. 2005;106(2):713–6. doi: 10.1182/blood-2005-01-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hideshima T, Chauhan D, Kiziltepe T, Ikeda H, Okawa Y, Podar K, et al. Biologic sequelae of I{kappa}B kinase (IKK) inhibition in multiple myeloma: therapeutic implications. Blood. 2009;113(21):5228. doi: 10.1182/blood-2008-06-161505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.