Abstract
Aberrant DNA methylation mediated by deregulation of DNA methyltransferases (DNMT) is a key hallmark of acute myeloid leukemia (AML), yet efforts to target DNMT deregulation for drug development have lagged. We previously demonstrated that upregulation of fatty acid-binding protein 4 (FABP4) promotes AML aggressiveness through enhanced DNMT1-dependent DNA methylation. Here we demonstrate that FABP4 upregulation in AML cells occurs through vascular endothelial growth factor (VEGF) signaling, thus elucidating a crucial FABP4-DNMT1 regulatory feedback loop in AML biology. We show that FABP4 dysfunction by its selective inhibitor BMS309403 leads to downregulation of DNMT1, decrease of global DNA methylation and re-expression of p15INK4B tumor suppressor gene by promoter DNA hypomethylation in vitro, ex vivo and in vivo. Functionally, BMS309403 suppresses cell colony formation, induces cell differentiation, and, importantly, impairs leukemic disease progression in mouse models of leukemia. Our findings highlight AML-promoting properties of the FABP4-DNMT1 vicious loop, and identify an attractive class of therapeutic agents with a high potential for clinical use in AML patients. The results will also assist in establishing the FABP4-DNMT1 loop as a target for therapeutic discovery to enhance the index of current epigenetic therapies.
Introduction
Acute myeloid leukemia (AML) is a highly aggressive hematologic malignancy characterized by the swift uncontrolled growth of immature myeloblasts. It is a lethal disease that lacks effective treatment. Although the precise molecular causes that are responsible for AML development and disease progression are unclear, it seems to result from an interplay of genetic and environmental factors that are largely unidentified. The fatty acid-binding protein (FABP) 4, also known as adipocyte FABP (A-FABP) or aP2, is a master regulator of lipid metabolism and significantly implicated in inflammation and cancers.1, 2 FABP4 is postulated to be predominantly expressed in adipocytes and macrophages and to act as an adipokine.3 FABP4 production can be modulated by environmental factors, including dietary intake,4 which is consistent with the observations that elevation of circulating FABP4 levels is associated with diet-induced obesity.5, 6 FABP4 is also reported to be abundantly expressed in several types of cancer cells.6–9 In agreement with the above expression patterning, FABP4 upregulation in obese host and in leukemia cells enhances AML disease progression in both a cell-autonomous and cell-non-autonomous manner.6 Thus, the specific mechanisms promoting FABP4 expression merit systematic exploration.
Cancer is well known to be a genetic disease. However, mounting evidence suggests that altered epigenetic phenotypes (e.g., aberrant DNA methylation) occur more frequently than genetic mutations, and could serve as an alternative etiological mechanism to a genetic mutation for the identical disease.10 DNA methylation involves the addition of a methyl group to the number 5-carbon of the cytosine pyrimidine ring, which is achieved by a cooperative effect among DNA methyltransferases (DNMTs) 1, 3a and 3b.11, 12 DNMT overexpression is frequent in leukemia cells and aberrant DNA methylation serves as a key hallmark of leukemogenesis.6, 12–14 But efforts to target DNMT deregulation for drug development have lagged. Abnormal DNA methylation is mainly attributed to cell-autonomous signaling, such as microRNAs,11, 14 nucleolin,12 Sp1/NFkB nuclear factor kappa B),15 AML1/ETO,13 ten-eleven translocation (TET) family,16 or cytidine deaminases.17 In support of this idea, our recent discoveries identified cellular FABP4 as an additional cell-autonomous member of the DNA methylation regulator family. More importantly, we demonstrated host/environmental FABP4 as a hitherto unknown cell non-autonomous factor governing the DNA methylation machinery, which acts partially through the IL-6/STAT3 pathway.6 We propose that FABP4 is a novel epigenetic member that can be pharmacologically targeted for AML intervention. Here, using comprehensive molecular, cellular and animal studies, we demonstrate that DNMT1 feeds back to upregulate FABP4 through hyperactive VEGF signaling. We show that BMS309403, a selective FABP4 inhibitor, can induce AML cell differentiation and block AML progression in leukemia-bearing mice. Mechanistic investigations disclose that BMS309403-mediated growth arrest of AML cells takes place through the reduction of DNMT1-dependent DNA methylation. These data suggest that targeting the FABP4-DNMT1 loop is a novel strategy for developing next-generation DNA hypomethylating agents to improve the dismal outcomes of conventional epigenetic compounds.
Materials and Methods
Cell lines, plasmids, reagents and patient samples
The 293T, MV4-11, Kasumi-1, THP-1 and C1498 cell lines were obtained from American Type Culture Collection. The SKNO-1 cell line was kindly provided by Dr. Clara Nervi (University of Rome, Rome, Italy). Cell lines were newly purchased with no further testing for mycoplasma. The primary blasts from AML patients were obtained from the Tumor Tissue/Biospecimen Bank of Mayo Clinic. All patients signed an informed consent document approved by the Mayo Clinic Institutional Review Board before entering the study. Details are in Supplementary Materials and Methods.
Western blotting
The whole cellular lysates were prepared by harvesting the cells in 1 × cell lysis buffer and Western blotting was performed as previously described.6, 11–13, 18 The following antibodies were purchased from Abcam (Cambridge, MA): anti-IL-6 (ab6672, 1:500), and anti-VEGF (ab46154, 1:500). Anti-β-actin (sc-1616, 1:1000) and anti-mouse-DNMT1 (sc-52919, 1:1000) were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-STAT3 (9139, 1:1000) and anti-phospho-STAT3 (Tyr705, 9131, 1:1000) were from Cell Signaling Technology (Danvers, Massachusetts). Anti-human-FABP4 (AF3150, 1:1000) and anti-mouse-FABP4 (AF1443, 1:500) were from R&D systems (Minneapolis, MN) and the anti-human-DNMT1 (M0231L, 1:500) was from New England Biolabs (Ipswich, MA).
DNA Dotblotting
Genomic DNA was purified using DNA Blood/Tissue Kit (QIAGEN, Germantown, MD), denatured and subjected to Dotblotting analysis using a 5mC antibody (39649, 1:2500, Active Motif, Carlsbad, CA) as previously described.6, 11–13, 18 The DNA spotted membrane was stained with 0.02% methylene blue (Sigma) in 0.5 M sodium acetate (pH 5.0) for DNA loading control.
Bisulfite sequencing
The bisulfite sequencing was performed as previously described.6, 11, 13, 18, 19 Briefly, 2 µg DNA was converted and purified using the EpiTect Bisulfite Kit (59824, QIAQEN). The region (−4 to +247) within the p15INK4B promoter was PCR-amplified from bisulfite-treated DNA. The primers are listed in Supplemental Table. The PCR products were subcloned using the TA Cloning® Kit (450046, Invitrogen) and sequenced by Genewiz (South Plainfield, NJ).
RNA isolation, cDNA preparation and quantitative PCR (qPCR)
RNA was isolated using the miRNeasy Kit (QIAGEN) and reverse transcription for cDNA was performed using the SuperScript® III First-Strand Synthesis System (18080-051, Invitrogen). TaqMan qPCR (4331182, Applied Biosystems) was performed to measure DNMT1 expression, and SYBR-Green qPCR (4309155, Applied Biosystems, Foster City, CA) was used for other genes. The primers are listed in Supplemental Table. The 18S levels were used for normalization and target expression was analyzed using the ΔCT approach.
Cytospin/Wright–Giemsa staining, cell differentiation, wound-healing, and colony-forming assays
All these assays were performed as previously described.6, 11–13, 18, 19 Details are in Supplementary Materials and Methods.
Animal studies
All animal experiments were approved by the Institutional Animal Care and Use Committees of the University of Minnesota and were in accordance with the US National Institutes of Health (NIH) Guide for Care and Use of Laboratory Animals. Details are in Supplementary Materials and Methods.
Statistical analysis
All the graphs were generated using Student’s t-test, but the Kaplan–Meier survival curves were created by log-rank test. Correlation data were acquired using the Pearson’s correlation coefficients. Details are in Supplementary Materials and Methods.
Results
FABP4 and SFAs cooperatively augment leukemia cell colony formation
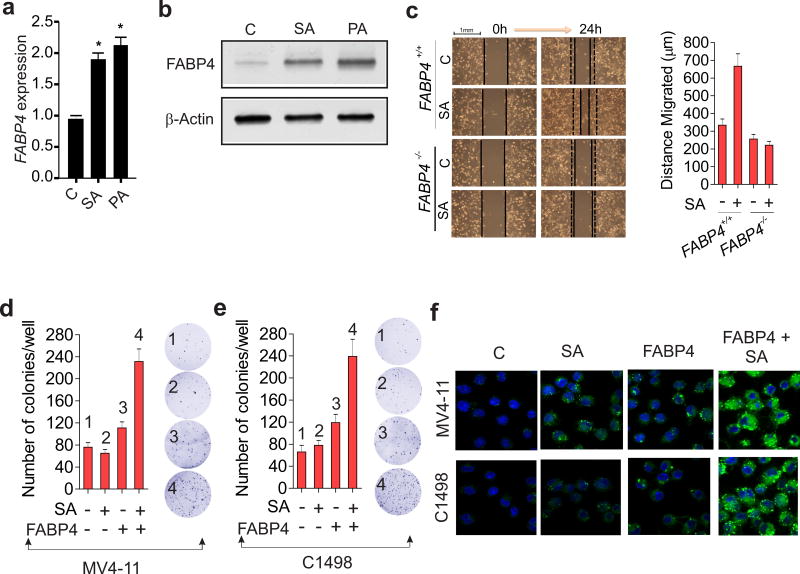
While the serum levels of both saturated fatty acids (SFAs) and FABP4 are high in obesity compared to lean subjects,5, 6 an experimental demonstration of a SFA-FABP4 functional association remains undocumented. To this end, bone-marrow derived cells were exposed to stearic acid (SA) or palmitic acid (PA), which are the common SFAs in high-fat diets. FABP4 expression was significantly increased at both the mRNA (Figure 1a) and protein levels (Figure 1b) in the presence compared to the absence of SA and PA, which support the existence of a causal relationship between SFAs and FABP4. Given the worse prognosis of leukemia in obese people and mice,6 we speculated that the interactions between excess SFAs and higher levels of FABP4 account for a more aggressive form of leukemia. As a proof of concept, we first measured the growth of FABP4 wild-type (FABP4+/+) and knockdown (FABP4−/−) macrophages in the presence or absence of SFAs. We found that FABP4+/+ migrated much faster than FABP4−/− macrophages in the presence of SA, indicating that genetic deficiency of FABP4 curbs the response of the cells to SFA stimulation (Figure 1c). This result suggests that FABP4 is required for SA-augmented cell proliferation, which is aligned with the role of FABP4 as a lipid transporter.1, 2 We then treated MV4-11 or C1498 AML cells with SA alone or with the FABP4 protein, and found that the cells treated with the combination displayed a more robust colony formation than observed with either agent alone (Figure 1 d and e). Confocal microscopy analysis revealed that a combination of SA and FABP4 greatly enhanced lipid accumulation in leukemia cells (Figure 1f). Notably, cellular apoptosis was observed and proliferation was impaired when SA concentrations were above 75 µM (not shown), consistent with previous reports,20, 21 which showed that higher lipid doses could invoke cytotoxicity. Finally, to understand the mechanisms by which SFAs contribute to leukemia growth, we treated MV4-11 cells with suboptimal dose of either stearic acid (SA) alone or plus the FABP4 protein, given that FABP4 is a crucial lipid transporter. We found that, in the presence of the FABP4 protein, SA treatment increased the protein expression and phosphorylation of STAT3, a DNMT1 transactivator, with a concurrent DNMT1 upregulation (Figure S1 a and b). In line with this, the combination of SA with FABP4 protein induced higher levels of DNA methylation compared to single agent groups (Figure S1c), collectively supporting the notion that DNMT1 upregulation by stearic acid could be an important mechanism underlying SFA-promoted leukemia growth. Taken together with the results of our previous studies,6 these data further support a role of FABP4 in AML disease progression.
Figure 1.
FABP4 interacts with SFAs to promote leukemia growth. (a and b) BM cells were isolated from C57BL/6 mice, treated with the indicated fatty acids at 50 µM for 12 hours and subjected to qPCR (a) or Western blotting (b) to measure the changes in FABP4. *P < 0.05. (c) FABP4+/+ or FABP4−/− macrophages were treated with steric acid (50 µM). When the cells grew to confluence, a wound was made and the wound width was measured at 0 or 24 hours. Left: a representative visual analysis of wound-scratch healing assays; right: the migration distance from three biological replicates. (d and e) MV4-11 (d) or C1498 (e) cells were treated with FABP4 or/and stearic acid for 6 hours and subjected to colony-forming assays. Right: a representative visual analysis of colony assays; left: graph indicates the colony number from three biological replicates, mean ± SD. For c–e, data are shown as mean values ± S.D. (f) Lipid droplet formation in MV4-11 and C1498 cells treated with FABP4 or/and stearic acid for 12 hours was analyzed by BODIPY staining (green) and visualized by confocal microscopy. C, control; SA, stearic acid; PA, palmitic acid.
DNMT1 induces ectopic expression of FABP4 in leukemia cells
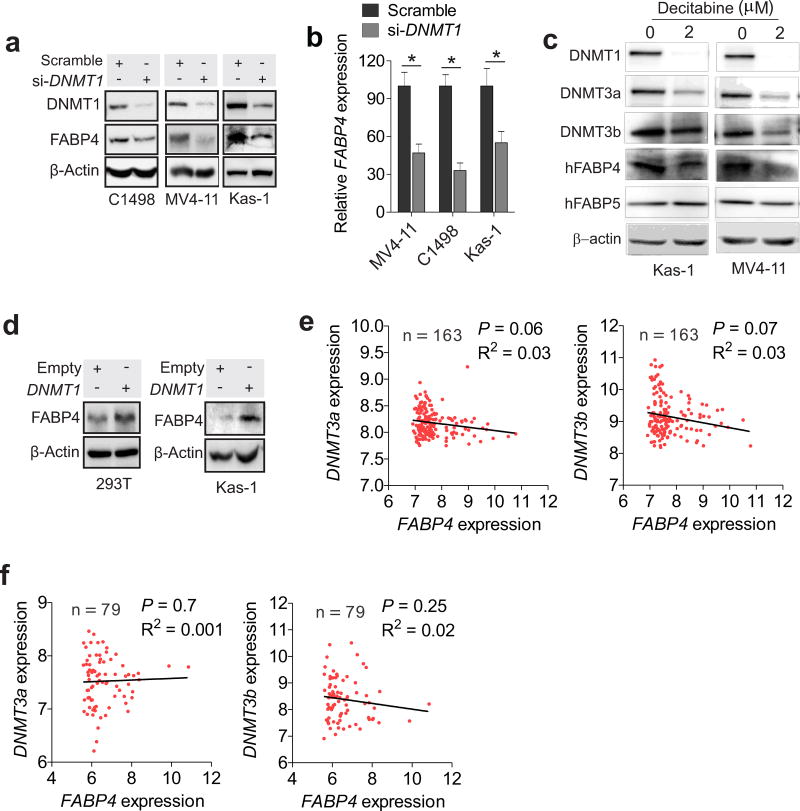
To understand the molecular mechanisms underlying FABP4 upregulation, we focused on DNMT1, because FABP4 expression parallels DNMT1 expression in leukemia cells and in obesity.6 In accordance, shRNA-triggered DNMT1 knockdown in C1498, MV4-11 and Kasumi-1 cells resulted in an apparent decrease in FABP4 expression (Figure 2 a and b). Consistently, exposure of leukemia cells to decitabine, a classical DNMT1 inhibitor,22 led to substantial downregulation of FABP4, but not FABP5 (Figure 2c). To further validate DNMT1 in FABP4 regulation, we transfected 293T or Kasumi-1 cells with DNMT1 expression vectors, and initially verified the promoting properties of DNMT1 overexpression (Figure S2a) in DNA methylation (Figure S2b) and cell migration (Figure S2 c and d), supporting our previous studies showing that DNMT1 knockdown impairs clonogenic potential of AML cells.6 As expected, enforced DNMT1 expression led to a concurrent increase in FABP4 levels (Figure 2d; Figure S2e). Because a cooperation among DNMT1, 3a and 3b determines DNA methylation status,11, 12 we further analyzed GEO datasets containing 242 AML patients,23 but did not find a parallel association between FABP4 with DNMT3a or DNMT3b (Figure 2 e and f), providing further support of DNMT1 in governing FABP4 gene expression. Considering a positive role of FABP4 in regulating the DNMT1 gene,6 these results suggest that DNMT1 and FABP4 are engaged in a reciprocal regulatory loop in AML cells.
Figure 2.
DNMT1 positively regulates VEGF and FABP4 expression. (a and b) C1498, MV4-11 or Kasumi-1 cells were transfected with DNMT1 siRNA and subjected to Western blotting (a) or qPCR (b). (c) Western blotting in MV4-11 or Kasumi-1 cells treated with 2 µM decitabine for 72 hours. (d) 293T or Kasumi-1 cells were transfected with DNMT1 expression vectors and subjected to Western blotting. (e and f) Spearman correlation analysis for the mRNA expression of FABP4 and DNMT3a or DNMT3b in AML patients (GSE12417; GPL570, e; GPL96, f). The Y or X-axis represents the fold change in maximum expression. For Western blotting and qPCR, the experiments were performed 3 times independently; Data are shown as mean values ± SD, *P < 0.05, **P < 0.01.
VEGF mediates DNMT1-upregulated FABP4 expression
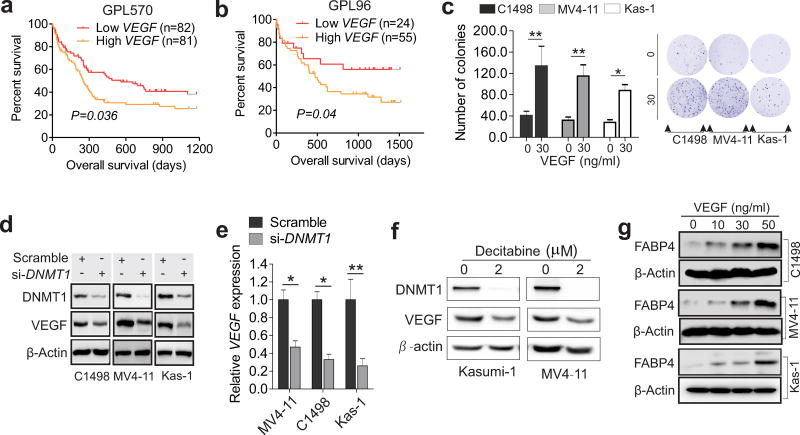
Because limited numbers of CpG within 3kb up- or down-stream of the transcription start site of human or mouse FABP4 promoter were identified (not shown), we reasoned that DNMT1 might not directly regulate FABP4 by promoter methylation. We targeted vascular endothelial growth factor (VEGF), because a) VEGF is an important obesity-leukemia associated molecule;24–28 b) compared to lean subjects, VEGF is expressed at a higher level in serum (Figure S3a) and BM (Figure S3b) from leukemia-bearing obese mice, in line with more aggressive leukemia growth in obese patients and mice;6 c) the analysis of GSE1241729 showed that AML patients with higher VEGF levels displayed shorter survival times compared with those with lower VEGF (Figure 3 a and b). These in vivo observations were experimentally verified by showing that treatment with VEGF recombinant protein largely increased colony number of C1498, MV4-11 and Kasumi-1 cells (Figure 3c). To this end, we manipulated DMNT1 expression in AML or 293T cells, and observed that VEGF levels were decreased by DNMT1 knockdown (Figure 3 d and e), but increased by DNMT1 overexpression (Figure S3 c and d). Consistently, exposure of MV4-11 and Kasumi-1 cells to decitabine inhibited VEGF expression (Figure 3f). Collectively, these data support a positive regulatory role of DNMT1 in VEGF expression.
Figure 3.
VEGF mediates DNMT1-associated FABP4 overexpression. (a and b) Kaplan-Meier estimate for overall survival in the whole population of AML patients (GSE12417), in which high VEGF was compared to low VEGF (log-rank test). (c) Colony-forming assays for C1498, MV4-11 and Kasumi-1 cells treated with the VEGF protein. Graph shows the colony number from 3 independent experiments. (d and e) Western blotting (d) or qPCR (e) of C1498, MV4-11 and Kasumi-1 cells transfected with DNMT1 siRNA. (f) Western blotting of Kasumi-1 and MV4-11 cells treated with 2 µM decitabine for 72 hours. (g) Western blotting of C1498, MV4-11 and Kasumi-1 cells treated with VEGF recombinant protein (30 ng/ml). For Western blotting and qPCR analyses, the experiments were performed 3 times independently; Data are shown as mean values ± SD, *P < 0.05, **P < 0.01; All treatments are 48 hours unless otherwise indicated; si, siRNA; Kas-1, Kasumi-1.
To validate the role of VEGF in FABP4 regulation, C1498, MV4-11 and Kasumi-1 AML cells were incubated with the VEGF protein. We found that FABP4 expression was increased in a dose-dependent manner (Figure 3g, Figure S4a), consistent with a previous report showing that FABP4 is induced by VEGFA in endothelial cells.30 Further, treatment with the VEGF protein rescued FABP4 downregulation driven by DNMT1 depletion (Figure S4b), but attenuation of both DNMT1 and VEGF induced a more pronounced suppression of FABP4 production (Figure S4c). Finally, by analysis of GSE12417, we identified a significant positive correlation between VEGF and FABP4 expression in AML patients (n = 242) (Figure S5). Thus, our data suggest that VEGF mediates, at least partially, the DNMT1-FABP4 feedback loop in AML cells.
Pharmacological inactivation of FABP4 reduces DNMT1-dependent DNA methylation and restores the epigenetically-silenced p15INK4B expression
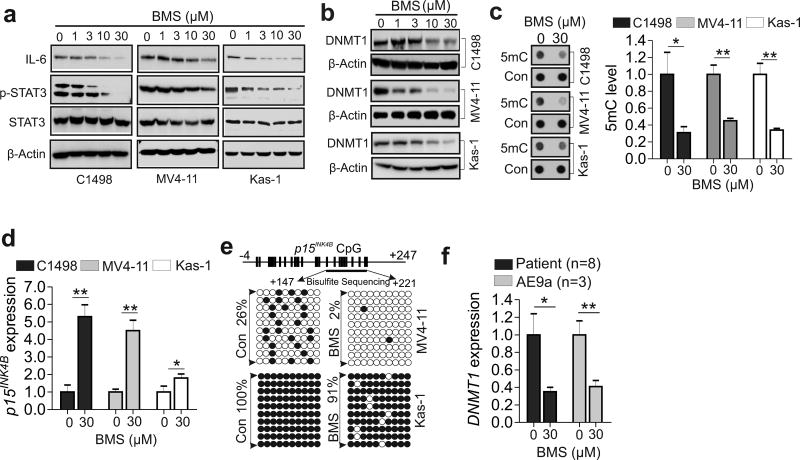
Considering our current and previous findings6 demonstrating the existence of an FABP4-DNMT1 loop in AML disease progression, pharmacological targeting FABP4 might be a valid approach to treat AML. BMS309403 has been reported to be a biphenyl azole inhibitor that specifically binds and inactivates FABP4.31 Consistently, BMS309403 treatment specifically suppressed FABP4+/+ macrophage migration while leaving the FABP4−/− macrophages largely unaffected (Figure S6 a and b), indicating that BMS309403 inhibitory effects were FABP4-dependent. Further, in support of the role of FABP4 in controlling the IL-6-STAT3-DNMT1 cascade,6 exposure of AML cell lines to BMS309403 resulted in IL-6 reduction, STAT3 dephosphorylation and DNMT1 downregulation (Figure 4 a and b), followed by global DNA demethylation (Figure 4c). Notably, BMS309403-induced DNMT1 downregulation (Figure S7 a and b) and DNA demethylation (Figure S7 c and d) were rescued by the addition of the FABP4 protein into the culture medium, substantiating the idea that BMS309403-induced DNA hypomethylation is not off-target, but specific to FABP4 function.
Figure 4.
Pharmacological inhibition of FABP4 induces DNA hypomethylation and restores the epigenetically silenced p15INK4N. (a and b) Western blotting of C1498, MV4-11 or Kasumi-1 cells exposed to BMS. (c) C1498, MV4-11 or Kasumi-1 cells were treated with BMS and the genomic DNA was subjected to Dotblotting. Left, representative images of dots; right, graph shows quantification of dot intensity. (d) qPCR of C1498, MV4-11 or Kasumi-1 cells treated with BMS. (e) Bisulfite sequencing of the p15INK4B promoter in MV4-11 or Kasumi-1 cells treated with BMS. Results of 10 clones are presented. Vertical bars indicate CpG locations, arrows indicate the bisulfite sequencing region, open circles show unmethylated CpG, and solid circles show methylated CpGs. (f) qPCR of human patient (n = 8) or mouse (n = 3) AML primary cells treated with BMS. The experiments were performed three times independently and data are shown as mean values ± SD, *P < 0.05, **P < 0.01. All treatments are 48 hours unless otherwise indicated; Con, control; BMS, BMS309403; Kas-1, Kasumi-1.
TSGs (e.g., p15INK4B) are frequently silenced epigenetically, and TSG-silencing predicts worse survival in leukemia.6, 13 We used p15INK4B as readout to determine whether BMS309403 changes promoter DNA methylation, because a literature review suggests that hypermethylation of the p15INK4B gene is very common (> 50%), and is independently correlated with poor prognosis in patients with AML.32–35 Our results indicated that p15INK4B expression was upregulated in Kasumi-1, MV4-11 and C1498 cells in the presence compared to the absence of BMS309403 (Figure 4d). The bisulfite sequencing11–13 disclosed that p15INK4B promoter methylation was decreased by BMS309403 in MV4-11 (26% versus 2%) and Kasumi-1 (100% versus 91%) cells (Figure 4e), in agreement with the effects of siRNA-mediated FABP4 downregulation.6 To determine the therapeutic relevance of BMS309403 in human AML, we treated human patient (n = 8) or mouse (n = 3) primary cells with BMS309403 for 48 hours. Consistent with its ability to strongly disrupt the FABP4-DNMT1 loop in cell lines, BMS309403 markedly suppressed DNMT1 expression (Figure 4f), lowered global DNA methylation (Figure S8a) and restored p15INK4B expression (Figure S8b) in these primary cells. Bisulfite sequencing revealed DNA hypomethylation of the p15INK4 promoter (20% versus 14%) in BMS309403-treated patient cells (Figure S8c). These data support the idea that BMS309403 possesses DNA hypomethylating activity.
Treatment with BMS309403 alters the behavior of leukemia cells in vitro and ex vivo
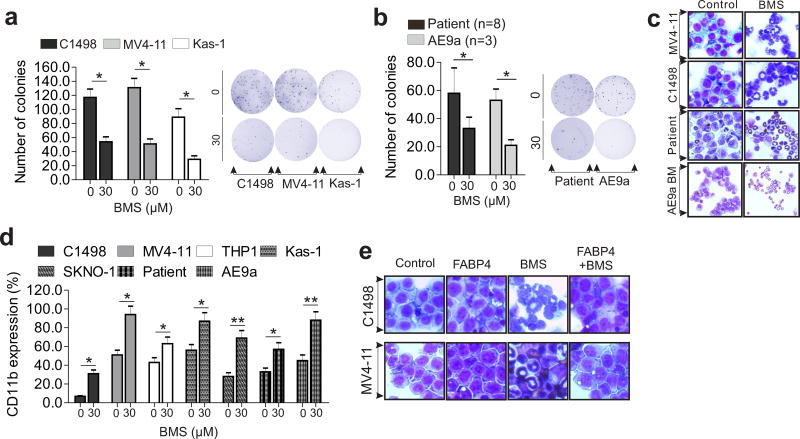
Given the contribution of FABP4 deregulation to leukemia growth, we sought to examine the response of leukemia cells to BMS309403 treatment. Exposure of AML cell lines and primary cells to BMS309403 did not significantly affect apoptosis as assessed by Annexin V-FITC staining (not shown), but greatly attenuated the clonogenic potential compared to control cells (C1498, 59% inhibition; MV4-11, 51% inhibition; Kasumi-1, 67% inhibition; patient, 44% inhibition; AE9a, 61% inhibition; Figure 5 a and b). Further, BMS309403-treated leukemia cells underwent granulocytic differentiation (Figure 5c). Because CD11b serves as a cell differentiation biomarker,14 we assessed CD11b levels by FACS in the above BMS309403-treated cells. Data indicated that CD11b expression was significantly upregulated by BMS309403 (C1498, 6.74% versus 31.0%; THP1, 43% versus 63%; Kasumi-1, 56% versus 87%; SKNO-1, 28% versus 69%; patient, 33% versus 57%; AE9a, 45.3% versus 87.7%; Figure 5d). These results suggest that BMS309403 impairs the proliferative rate of leukemia cells by inducing cell differentiation, but not by promoting apoptotic cell death. Finally, BMS309403-induced cell differentiation could be disrupted by adding the FABP4 protein into culture medium (Figure 5e), supporting that the added FABP4 might bind and sequentially reduce the biological effects of BMS309403, a further confirmation of FABP4 specificity.
Figure 5.
FABP4 blockade by its inhibitor impairs leukemia growth in vitro and ex vivo. (a and b) Colony-forming assays of C1498, MV4-11 and Kasumi-1 (a) or human patient (n = 8) and mouse (n = 3) AML primary cells (b) treated with BMS. Right, graphs show quantification of colony number; left, representative images of colonies. (c) Visual analysis of representative images of Wright-Giemsa-stained cytospins of MV4-11, C1498 or human patient and mouse AML primary cells treated with BMS (30 µM) for 96 hours (original magnification ×400). (d) Graphs show quantification of CD11b+ expression in AML cells treated with BMS for 96 hours. (e) Representative images of Wright-Giemsa-stained cytospins of C1498 or MV4-11 cells treated with the mouse/human FABP4 protein (200 ng/ml) only, BMS (30 µM) only or the FABP4 protein plus BMS for 96 hours (magnification × 400). The experiments were performed three times independently and data are shown as mean values ± SD, *P < 0.05, **P < 0.01; Kas-1, Kasumi-1.
Treatment with BMS309403 markedly suppresses in vivo leukemia growth
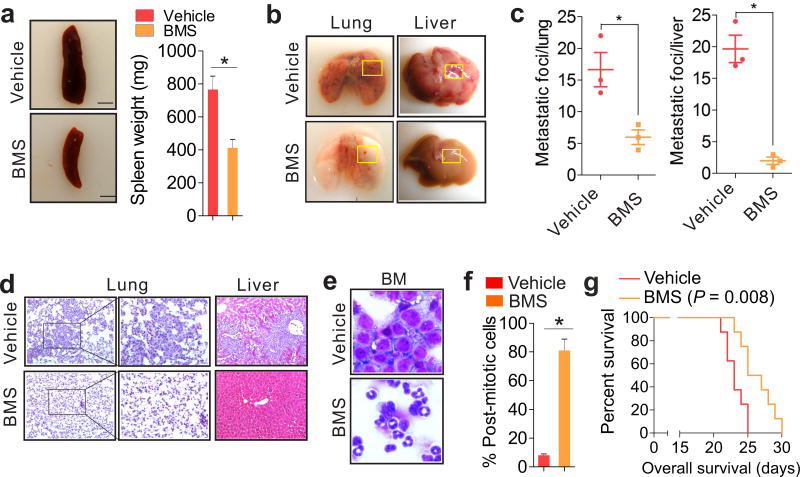
To explore the emerging roles of FABPs in leukemia therapy, we initially analyzed publicly accessible raw database GSE12417.29 No significant association of FABP1, FABP2, FABP3, FABP5 and FABP7 upregulation with poor patient survival was observed (Figure S9). Although FABP6 levels displayed positive association with patient outcomes, the role of FABP6 in cancer is not known. Given the cell autonomous and non-autonomous roles of FABP4 in leukemia progression,6 we mainly tested the therapeutic potential of FABP4 in vivo. Because C57BL/6 mice have baseline FABP4 (~20 ng/ml) in circulation and leukemia cells were affected even at 10 ng/ml of FABP4 protein,6 these mice were given one dose of BMS309403 (5 mg/kg) intraperitoneally 12 hours prior to cell injection so that the baseline activity of FABP4 could be inhibited. The second injection was given 3 days after the intravenous injection of C1498 (0.5 × 106) cells. After administration of 5 mg/kg three times over 3-days, an additional 10 mg/kg was given twice over 3-days. The leukemia-bearing mice injected with vehicle only were used as controls. Note that the selection of BMS309403 dosages and treatment strategy was based on evidence from previous studies.36–38 Consistent with its inhibitory effects on AML cells in vitro and ex vivo, BMS309403 administration markedly lowered the number of WBCs (Figure S10a) and reversed splenomegaly (766 ± 82 versus 412 ± 52 mg; Figure 6a). The quantification of metastatic foci revealed that the mice treated with BMS309403 exhibited fewer nodules and smaller lesion areas in lungs (16.7 ± 4.7 versus 6.1 ± 2.2) and livers (19.7 ± 3.8 versus 2.0 ± 1.0) (Figure 6 b and c). In addition, H&E stained sections of lungs and livers displayed less infiltration of blasts into these organs (Figure 6d) compared with vehicle control, indicating less aggressive leukemia growth in lungs and livers. The reduction of Ki-67 positive cells argued for anti-proliferative effects of BMS309403 on leukemia cells (Figure S10b). Moreover, BMS309403 therapy efficiently impaired BM clonogenic potential (Figure S10c) and rescued granulocytic differentiation of myeloid cells (Figure 6e). In fact, the majority of BM cells from BMS309403-treated mice were post-mitotic cells containing metamyelocytes, bands and segmented neutrophils (73% increase; Figure 6f). BMS309403-induced cell differentiation in vivo was further demonstrated by the higher levels of CD11b expression in BM from BMS309403-treated than those from vehicle-treated leukemia-bearing mice (27.3% versus 3.72%; Figure S10d). Accordingly, the survival of BMS309403-treated leukemia-bearing mice was longer than that of the control group (Figure 6g).
Figure 6.
FABP4 inactivation in vivo achieves leukemia remission. C1498 cells (0.5 × 106) were intravenously injected into 4–6 weeks old C57BL/6 mice. Twelve hours prior to cell injection, a BMS (5 mg/kg) was initiated by a one-time dose. After an additional 3 doses of BMS were administered, another 10 mg/kg was given twice. (a) Photos show representative visual analysis and the graph indicates the calculated weight of spleens (scale bars, 5.0 mm). (b) External views of lungs and livers. (c) Graphs show quantifications of tumor nodules growing on lungs or livers. (d) H&E staining of lung or liver sections (original magnification ×200, ×400) from boxed areas in b. (e) Wright-Giemsa-stained BM (original magnification ×400). (f) Graph shows quantification of post-mitotic cells from (e). (g) Effects of BMS on survival of leukemia-bearing mice (n = 6) were determined by the Kaplan-Meier estimate (log-rank test). Note, the survival time is from the start of leukemia cell injection. In a–f, n = 3; BMS, BMS309403. Data are shown as mean values ± SD, *P < 0.05.
To mimic human AML therapy, we first engrafted C57BL/6 mice (n = 8) with C1498 cells, and began BMS309403 treatment when the WBC accounts were significantly higher in leukemia cell-engrafted mice than in normal mice. Consistently, compared to the vehicle group, BMS309403 treatment greatly suppressed the development of leukemic disease, as indicated by a significant decrease of WBCs (Figure S11a) and spleen weight (655 ± 76 versus 243 ± 35 mg; Figure S11b). Giemsa–stained BM and H&E-stained sections of spleen, lung and liver from BMS309403-treated mice showed no significant infiltration of leukemic blasts, which were readily detectable in BM and organs of vehicle-treated subjects (Figure S11c). In agreement with the pathological phenotype, BMS309403-treated mice had a significantly longer survival time (Figure S11d). We did not observe obvious alterations in mouse body weight, food intake or mobility (not shown), suggesting that the side effects of BMS309403 are likely to be minimal in the current settings. Mechanistically, the antileukemic effects of BMS309403 in vivo were mediated by its DNA hypomethylating activities, because BM from BMS309403-treated mice had decreased DNMT1 expression (Figure S12a), reduced global DNA methylation (Figure S12b) and upregulated methylation-silenced p15INK4B (Figure S12c). Thus, lowering or inhibiting the circulating FABP4 might represent a promising approach in AML management.
Discussion
In this study, we demonstrate that DNMT1 deregulation accounts for FABP4 abnormality in AML cells, and this process appears to be mediated by VEGF signaling. We show that FABP4 is a novel druggable candidate for leukemia intervention. We present evidence showing that BMS309403, a selective FABP4 inhibitor, blocks leukemia growth in vitro, ex vivo and in vivo mechanistically by reversing aberrant DNA methylation and re-activating epigenetically-silenced p15INK4B. Together with our recent report6 showing that FABP4 augments AML disease progression through an epigenetic abnormality in AML cells, these findings further underscore the role of FABP4 in leukemia progression and epigenetic abnormality, elucidate a crucial and vicious FABP4-DNMT1 loop in leukemia biology (Figure S13), highlight the promise and merit of targeting the FABP4-DNMT1 loop to treat leukemia, and discover a novel type of DNA hypomethylating agent that mechanistically differs from the classical DNA methylation inhibitors.
Several recent studies have highlighted the importance of FABP4 acting as a master gene in controlling lipid metabolism and cancer cell proliferation.6–9 Although our findings suggest a causal relationship between SFAs and FABP4 production, the molecular basis of FABP4 deregulation remains elusive, particularly in the context of leukemia cells. The simultaneous upregulation of both DNMT1 and FABP4 in obesity and leukemia cells raises the possibility that DNMT1 could reciprocally modulate the FABP4 gene. In fact, forced DNMT1 expression increased, whereas DNMT1 inactivation reduced, FABP4 production. Mechanistically, DNMT1 regulates FABP4 in a methylation-independent manner, because fewer CpGs were identified at the FABP4 promoter. Actually, our data suggest that DNMT1 modulates FABP4 partially through VEGF signaling, a key pathogenic factor in obesity and leukemia,6, 24–28 because ectopic DNMT1 expression enhanced, whereas DNMT1 depletion inhibited, VEGF expression. Treatment with the VEGF protein significantly upregulated FABP4 and rescued the repressive effects of DNMT1 attenuation on FABP4 expression. Given the significant contribution of FABP4 to DNMT1 regulation,6 we conclude that FABP4 and DNMT1 shape a positive regulatory circuit in leukemia cells, in which FABP4 regulates DNMT1 expression through the IL-6/STAT3 axis, whereas DNMT1 controls FABP4 by VEGF signaling (Figure S13).
While pharmaceutical approaches to target FABP4 for improved metabolic health are under investigation, no study has been initiated to exploit FABP4 as a therapeutic target in leukemia. Although many leukemia patients (≤ 20%) achieve remission with initial therapies, including decitabine and azacitidine, most patients, particularly elderly adults, are not cured. Accumulating evidence indicates that DNMT1 gene abundance is essential to leukemia progression, but few agents target the key regulators of the DNMT1 gene, particularly from a metabolic standpoint. The isocitrate dehydrogenase (IDH) is a metabolic enzyme that is mutated in multiple cancers.39 IDH mutations alter genomic DNA methylation, thereby serving as potential drug targets. In fact, an inhibitor for IDH mutation has been developed and shown anticancer activity against glioma and leukemia,40, 41 but it acts independently of DNA methylation.40 Because FABP4 is a molecular linker of irregular lipid metabolism and aggressive leukemia by virtue of altered DNA methylation, pharmacological targeting of the FABP4-DNMT1 loop represents a viable strategy to accomplish DNA hypomethylation. Indeed, treatment with BMS309403 suppressed DNMT1 expression, reduced global DNA methylation and reactivated p15INK4B in AML cell lines and primary cells, thus leading to disruption of cellular clonogenic frequencies and induction of cell differentiation. Importantly, BMS309403 administration in leukemia-bearing mice decreased leukemic burden and prolonged survival duration, which mechanistically took place through the BMS309403-restored DNA hypomethylation content in AML cells. Notably, this is a proof of principle study to further support how the FABP4-DNMT1 loop is important to leukemia, and to address whether pharmacological intervention of a lipid-relevant molecule could restore the perturbed epigenetic landscapes that are the hallmarks of cancers. The dosages and schedule used for BMS309403 administration require further optimization before clinical application. Overall, our studies bridge a knowledge gap concerning the causes of AML progression, reveal the pivotal importance of the FABP4-DNMT1 regulatory loop in AML pathogenesis and establish FABP4 inactivation as distinct therapeutic approach to treat leukemia and potentially, other types of cancer, mechanistically by reversing epigenetic deregulations.
Supplementary Material
Acknowledgments
This work was supported partially by The Hormel Foundation, National Cancer Institute (Bethesda, MD) grants R01CA149623, R01CA177679, R01CA180986, R21CA155915, R03CA186176, and the Predolin Foundation. We thank Dr. Dong-Er Zhang for providing AE9a model, Dr. Jill Suttles for providing FABP4-deficient mice and Dr. Clara Nervi for providing the SKNO-1 cell line.
Footnotes
Conflict of interest: The authors state no conflict of interest
Author contributions
S.J.L. and B.L. designed research; F.Y., N.S., J.X.P., N.Z., and Y.W.Z. performed research; A.A. and M.R.L. provided the leukemia patient samples; S.J.L., B.L., M.R.L., F.Y., A.M. B., and A.A. analyzed data and wrote the paper; S.J.L. conceived ideas and oversaw the entire research project.
References
- 1.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nature reviews Drug discovery. 2008 Jun;7(6):489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annual review of nutrition. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 3.Furuhashi M, Saitoh S, Shimamoto K, Miura T. Fatty Acid-Binding Protein 4 (FABP4): Pathophysiological Insights and Potent Clinical Biomarker of Metabolic and Cardiovascular Diseases. Clinical Medicine Insights Cardiology. 2014;8(Suppl 3):23–33. doi: 10.4137/CMC.S17067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Rao E, Zeng J, Hao J, Sun Y, Liu S, et al. Adipose Fatty Acid Binding Protein Promotes Saturated Fatty Acid-Induced Macrophage Cell Death through Enhancing Ceramide Production. J Immunol. 2017 Jan 15;198(2):798–807. doi: 10.4049/jimmunol.1601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terra X, Quintero Y, Auguet T, Porras JA, Hernandez M, Sabench F, et al. FABP 4 is associated with inflammatory markers and metabolic syndrome in morbidly obese women. European journal of endocrinology / European Federation of Endocrine Societies. 2011 Apr;164(4):539–547. doi: 10.1530/EJE-10-1195. [DOI] [PubMed] [Google Scholar]
- 6.Yan F, Shen N, Pang JX, Zhang YW, Rao EY, Bode AM, et al. Fatty acid-binding protein FABP4 mechanistically links obesity with aggressive AML by enhancing aberrant DNA methylation in AML cells. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2016 Dec 02; doi: 10.1038/leu.2016.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature medicine. 2011;17(11):1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancke K, Grubeck D, Hauser N, Kreienberg R, Weiss JM. Adipocyte fatty acid-binding protein as a novel prognostic factor in obese breast cancer patients. Breast cancer research and treatment. 2010 Jan;119(2):367–367. doi: 10.1007/s10549-009-0577-9. [DOI] [PubMed] [Google Scholar]
- 9.Lee D, Wada K, Taniguchi Y, Al-Shareef H, Masuda T, Usami Y, et al. Expression of fatty acid binding protein 4 is involved in the cell growth of oral squamous cell carcinoma. Oncology reports. 2014 Mar;31(3):1116–1120. doi: 10.3892/or.2014.2975. [DOI] [PubMed] [Google Scholar]
- 10.Hitchins MP. Constitutional epimutation as a mechanism for cancer causality and heritability? Nature reviews Cancer. 2015 Oct;15(10):625–634. doi: 10.1038/nrc4001. [DOI] [PubMed] [Google Scholar]
- 11.Yan F, Shen N, Pang J, Xie D, Deng B, Molina JR, et al. Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell death & disease. 2014;5:e1413. doi: 10.1038/cddis.2014.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen N, Yan F, Pang J, Wu LC, Al-Kali A, Litzow MR, et al. A nucleolin-DNMT1 regulatory axis in acute myeloid leukemogenesis. Oncotarget. 2014 Jul 30;5(14):5494–5509. doi: 10.18632/oncotarget.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao XN, Yan F, Lin J, Gao L, Lu XL, Wei SC, et al. AML1/ETO cooperates with HIF1alpha to promote leukemogenesis through DNMT3a transactivation. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2015 Mar 2; doi: 10.1038/leu.2015.56. [DOI] [PubMed] [Google Scholar]
- 14.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009 Jun 18;113(25):6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Liu Z, Xie Z, Pang J, Yu J, Lehmann E, et al. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood. 2008 Feb 15;111(4):2364–2373. doi: 10.1182/blood-2007-08-110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013 Oct 24;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation? Genes & development. 2010 Oct 01;24(19):2107–2114. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen N, Yan F, Pang J, Zhao N, Gangat N, Wu LC, et al. Inactivation of receptor tyrosine kinases reverts aberrant DNA methylation in acute myeloid leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017 Jul 18; doi: 10.1158/1078-0432.CCR-17-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan F, Shen N, Pang J, Molina JR, Yang P, Liu S. The DNA Methyltransferase DNMT1 and Tyrosine-Protein Kinase KIT Cooperatively Promote Resistance to 5-Aza-2'-deoxycytidine (Decitabine) and Midostaurin (PKC412) in Lung Cancer Cells. The Journal of biological chemistry. 2015 Jul 24;290(30):18480–18494. doi: 10.1074/jbc.M114.633693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung KC, Park CH, Hwang YH, Rhee HS, Lee JH, Kim HK, et al. Fatty acids, inhibitors for the DNA binding of c-Myc/Max dimer, suppress proliferation and induce apoptosis of differentiated HL-60 human leukemia cell. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2006 Jan;20(1):122–127. doi: 10.1038/sj.leu.2404022. [DOI] [PubMed] [Google Scholar]
- 21.Malhi H, Bronk SF, Werneburg NW, Gores GJ. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. The Journal of biological chemistry. 2006 Apr 28;281(17):12093–12101. doi: 10.1074/jbc.M510660200. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Liu S, Xie Z, Blum W, Perrotti D, Paschka P, et al. Characterization of in vitro and in vivo hypomethylating effects of decitabine in acute myeloid leukemia by a rapid, specific and sensitive LC-MS/MS method. Nucleic acids research. 2007;35(5):e31. doi: 10.1093/nar/gkl1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008 Nov 15;112(10):4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casalou C, Fragoso R, Nunes JF, Dias S. VEGF/PLGF induces leukemia cell migration via P38/ERK1/2 kinase pathway, resulting in Rho GTPases activation and caveolae formation. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2007 Jul;21(7):1590–1594. doi: 10.1038/sj.leu.2404668. [DOI] [PubMed] [Google Scholar]
- 25.Frankel AE, Gill PS. VEGF and myeloid leukemias. Leukemia research. 2004 Jul;28(7):675–677. doi: 10.1016/j.leukres.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Dias S, Hattori K, Zhu Z, Heissig B, Choy M, Lane W, et al. Autocrine stimulation of VEGFR-2 activates human leukemic cell growth and migration. The Journal of clinical investigation. 2000 Aug;106(4):511–521. doi: 10.1172/JCI8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loebig M, Klement J, Schmoller A, Betz S, Heuck N, Schweiger U, et al. Evidence for a relationship between VEGF and BMI independent of insulin sensitivity by glucose clamp procedure in a homogenous group healthy young men. PloS one. 2010;5(9):e12610. doi: 10.1371/journal.pone.0012610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Ambrosi J, Catalan V, Rodriguez A, Ramirez B, Silva C, Gil MJ, et al. Involvement of serum vascular endothelial growth factor family members in the development of obesity in mice and humans. The Journal of nutritional biochemistry. 2010 Aug;21(8):774–780. doi: 10.1016/j.jnutbio.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nature medicine. 2010 Aug;16(8):903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harjes U, Bridges E, McIntyre A, Fielding BA, Harris AL. Fatty acid-binding protein 4, a point of convergence for angiogenic and metabolic signaling pathways in endothelial cells. The Journal of biological chemistry. 2014 Aug 15;289(33):23168–23176. doi: 10.1074/jbc.M114.576512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sulsky R, Magnin DR, Huang Y, Simpkins L, Taunk P, Patel M, et al. Potent and selective biphenyl azole inhibitors of adipocyte fatty acid binding protein (aFABP) Bioorganic & medicinal chemistry letters. 2007 Jun 15;17(12):3511–3515. doi: 10.1016/j.bmcl.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 32.Shimamoto T, Ohyashiki JH, Ohyashiki K. Methylation of p15(INK4b) and E-cadherin genes is independently correlated with poor prognosis in acute myeloid leukemia. Leukemia research. 2005 Jun;29(6):653–659. doi: 10.1016/j.leukres.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Matsuno N, Hoshino K, Nanri T, Kawakita T, Mitsuya H, Asou N. Transcriptional repression of the p15 gene predicts the clinical outcome of acute myeloblastic leukemia with intermediate and adverse cytogenetics. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2004 Jun;18(6):1146–1148. doi: 10.1038/sj.leu.2403362. [DOI] [PubMed] [Google Scholar]
- 34.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Methylation of p15INK4B is common, is associated with deletion of genes on chromosome arm 7q and predicts a poor prognosis in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2003 Sep;17(9):1813–1819. doi: 10.1038/sj.leu.2403054. [DOI] [PubMed] [Google Scholar]
- 35.Deneberg S, Grovdal M, Karimi M, Jansson M, Nahi H, Corbacioglu A, et al. Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010 May;24(5):932–941. doi: 10.1038/leu.2010.41. [DOI] [PubMed] [Google Scholar]
- 36.Lee MY, Li H, Xiao Y, Zhou Z, Xu A, Vanhoutte PM. Chronic administration of BMS309403 improves endothelial function in apolipoprotein E-deficient mice and in cultured human endothelial cells. British journal of pharmacology. 2011 Apr;162(7):1564–1576. doi: 10.1111/j.1476-5381.2010.01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan H, Cheng CC, Kowalski TJ, Pang L, Shan L, Chuang CC, et al. Small-molecule inhibitors of FABP4/5 ameliorate dyslipidemia but not insulin resistance in mice with diet-induced obesity. Journal of lipid research. 2011 Apr;52(4):646–656. doi: 10.1194/jlr.M012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neviani P, Santhanam R, Trotta R, Notari M, Blaser BW, Liu S, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer cell. 2005 Nov;8(5):355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 39.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010 Dec 14;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013 May 3;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013 May 3;340(6132):622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.