Abstract
Staphylococcus aureus is a widespread and persistent pathogen of humans and livestock. The bacterium expresses a wide variety of virulence proteins, many of which serve to disrupt the host’s innate immune system from recognizing and clearing bacteria with optimal efficiency. The extracellular adherence protein (Eap) is a multidomain protein that participates in various protein-protein interactions that inhibit the innate immune response, including both the complement system (Woehl et al. 2014) and Neutrophil Serine Proteases (NSPs) (Stapels et al. 2014).
The third domain of Eap, Eap3, is an ~11 kDa protein that was recently shown to bind complement component C4b (Woehl et al. 2017) and therefore play an essential role in inhibiting the classical and lectin pathways of complement (Woehl et al. 2014). Since structural characterization of Eap3 is still incomplete, we acquired a series of 2D and 3D NMR spectra of Eap3 in solution. Here we report the backbone and side-chain 1H, 15N, and 13C resonance assignments of Eap3 and its predicted secondary structure via the TALOS-N server. The assignment data have been deposited in the BMRB data bank under accession number 27087.
Keywords: NMR assignment, Staphylococcus aureus, virulence protein, extracellular adherence protein (Eap)
Biological context
In the earliest phase of bacterial infection, the human body relies on the rapid recognition and removal of pathogens by the innate immune system. The innate immune system is comprised of an arsenal of protective barriers (e.g., skin), but also blood-borne proteins (e.g., the complement system) and phagocytic cells that get activated by the presence of non-self biomaterial. Activation of the complement system is essential for initiation of the inflammatory response and recruitment of immune cells to the site of infection (Ricklin et al. 2010). There are three pathways of complement activation and they depend on binding of pattern recognition proteins to pathogen-specific surface structures. Once triggered, all these pathways (classical, lectin, or alternative) lead to assembly of C3 convertase enzymes that can cleave complement component 3 (C3) into its bioactive proteolytic fragments C3a, an anaphylatoxin, and C3b, which opsonizes cellular surfaces. In the presence of increasing levels of surface-bound C3b, C3 convertases undergo a switch in their specificity to allow for cleavage of complement component C5. This process drives generation of C5a, which triggers inflammation and recruitment of phagocytic cells, and C5b, which is responsible for formation of the terminal complement complex.
It has become increasingly clear that excessive or unregulated complement activation can lead to damage of healthy host cells and tissues, and therefore development of various diseases (Ricklin et al. 2010; Ricklin and Lambris 2013). Furthermore, recruitment of phagocytes such as neutrophils to the site of inflammation can be highly damaging to otherwise healthy tissues since they contain subcellular granules replete with chymotrypsin-like Neutrophil Serine Proteases (NSPs) and Myeloperoxidase (MPO) (Amulic et al. 2012; Nauseef 2007; Nauseef 2014), which generate reactive oxidants (e.g. HOCl). Characterization of novel classes of inhibitors that target key proteins within this innate anti-bacterial repertoire may therefore open new directions for development of anti-inflammatory therapies.
Staphylococcus aureus is a widespread and persistent pathogen of humans and livestock. The bacterium causes numerous infections of varying severity, including skin abscesses, endocarditis, and bacteremia (Archer 1998; Lowy 1998). Numerous studies have shown S. aureus secretes an array of virulence proteins whose activities block the central events required for bacterial opsonization by complement components and subsequent phagocytosis by neutrophils (Garcia et al. 2016; Kim et al. 2012; Lambris et al. 2008; Spaan et al. 2013). Furthermore, novel classes of S. aureus secreted proteins, such as the Extracellular Adherence Protein (Eap) family (Stapels et al. 2014) and Staphylococcal Peroxidase Inhibitor (SPIN) proteins (de Jong et al. 2017) have recently been identified as nanomolar-affinity inhibitors of NSPs and MPO, respectively.
Eap is a secreted multi-domain protein with a mass of 50–70 kDa depending on the number ~100 residue repeating domains found in its various isoforms (Geisbrecht et al. 2005). Eap contributes to the overall virulence of S. aureus by blocking both the classical and lectin complement pathways (Woehl et al. 2014) and Neutrophil Serine Proteases (NSPs) (Stapels et al. 2014). Crystal structures of the second domain of Eap (Eap2), along with two homologs EapH1 and EapH2, revealed that the individual domains are characterized by a beta-grasp type fold (Geisbrecht et al. 2005). Within the context of the multi-domain protein, these individual repeats are connected to the adjacent domains through a short (~10 residue) linker.
Recent work on Eap domains 3 and 4 reported their interaction with C4b and their ability to inhibit classical and lecitin pathways (Woehl et al. 2014; Woehl et al. 2017). While the individual domains bound C4b with KD ~40 µM, the construct containing both domains, Eap34, bound C4b with KD = 525 nM. The binding affinity is even higher for Eap, KD = 185 nM. However, the structural features responsible for these differences in binding affinity are not completely characterized. Moreover, while the secondary structure for Eap4 has been reported by NMR (Woehl et al. 2016), Eap3 and Eap4 share only 58.9% similarity as reported in Geisbrecht et al. (2005).
To further enhance our current structure/function understanding of Eap interaction with C4b and other targets, we investigated herein the secondary structural features of Eap3 in the free form in solution. After assigning the backbone and side-chain 1H, 15N, and 13C resonances of Eap3, we predicted the secondary structure using TALOS-N server along with the observed chemical shifts.
Methods and Experiments
Protein expression and purification
Eap3 was overexpressed using the method described by Geisbrecht et al. (2006), following subcloning of a DNA fragment encoding its sequence into the Sal1 and Not1 sites of the prokaryotic expression vector pT7HMT. This vector encodes an N-terminal affinity His-tag that is used for Ni-affinity chromatography purification of the protein, but which can be removed by digestion with Tobacco Etch Virus (TEV) protease. After cleavage, the recombinant form of Eap3 contained an additional “Gly-Ser-Thr” sequence at the N-terminus. The plasmid containing this DNA fragment was further verified by sequencing and transformed into Escherichia coli BL21(DE3) cells.
Both uniformly 15N and 13C/15N double-labeled Eap3 proteins were overexpressed in minimal medium (M9) enriched with 15NH4Cl and 13C-glucose as described in the protocol by Woehl et al. (2016). The purified protein yield from 1 L of E. coli culture was in the range of 5–10 mg for both 15N and 13C/15N double-labeled Eap3. The samples for NMR experiments contained 0. 5 – 1.0 mM uniformly 15N or 13C/15N double-labeled Eap3 protein in 50 mM sodium phosphate buffer (pH 6.5) containing 5 % (v/v) D2O (used as a lock solvent). The purity and mass of the labelled protein was verified using mass spectrometry (Ultra Flex III TOF, Bruker Daltonics) prior to NMR data acquisition.
NMR spectroscopy
Nuclear magnetic spectroscopy spectra were acquired at 25°C on a Varian 500VNMR System (Agilent Technologies) equipped with a 5 mm triple-resonance inverse detection pulse gradient cold probe operating at 499.84 MHz for 1H frequency, and on a Bruker 800 MHz Avance III spectrometer equipped with a TCI cryo-probe. Backbone resonance assignments were achieved following standard procedure (Whitehead et al. 1997) using 2D 1H-15N HSQC and 3D HNCO, HN(CA)CO, HN(CO)CA, HNCA, CBCA(CO)NH and HNCACB spectra. The following NMR spectra were collected for side chain assignments: 2D 13C HSQC and 3D TOCSY-HSQC, NOESY-HSQC, and 15N-edited NOESY (mixing time 100 ms). All NMR spectra were processed using NMRPipe (Delagio et al. 1995), and analyzed with CARA (http://www.nmr.ch) (Keller 2004). The 1H chemical shift assignments were referenced by using 2,2-dimethy-2-silapentane-5-sulphonic acid (DSS) at 25°C as a standard. The 13C and 15N chemical shift were referenced indirectly to DSS, using the absolute frequency ratios.
Results
Extent of Assignments and Data Deposition
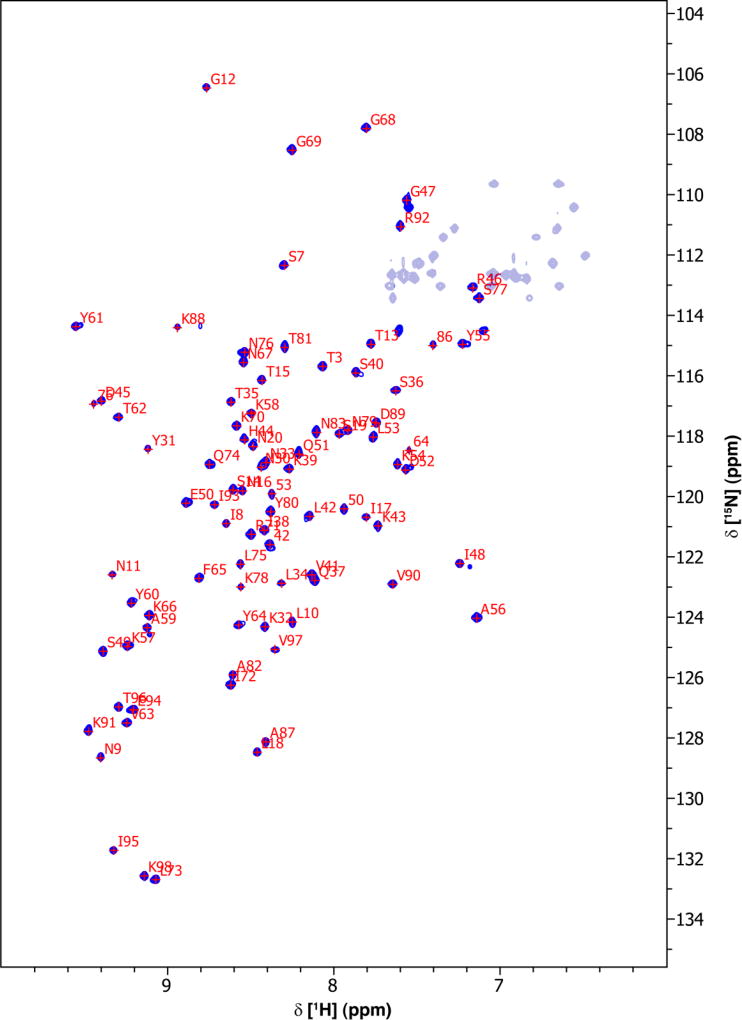
2D 1H-15N HSQC measurement of Eap3 resulted in a well-dispersed spectrum (Fig. 1). Amino acid numbering is based upon the Eap3 sequence, with an extra “Gly-Ser-Thr” sequence at the N-terminus. A total of 93% of backbone 1H and 15N resonances of 96 non-proline residues, and 71% of the expected 13Cβ resonances have been unambiguously assigned based on a standard set of triple resonance spectra described above. Some of the backbone amide residues that could not be assigned include G1 and S2. G1 and S2 lie in a loop region of the artifactual N-terminus as a result of the subcloning procedure.
Fig. 1.
2D 1H-15N HSQC spectrum of 0.7 mM 13C/15N-labeled Eap3 recorded at 298 K, pH 6.5 on a Bruker 800 MHz Avance III spectrometer equipped with a TCI cryoprobe. Sequence specific assignments of backbone amide groups are indicated by single letter residue name and sequence number.
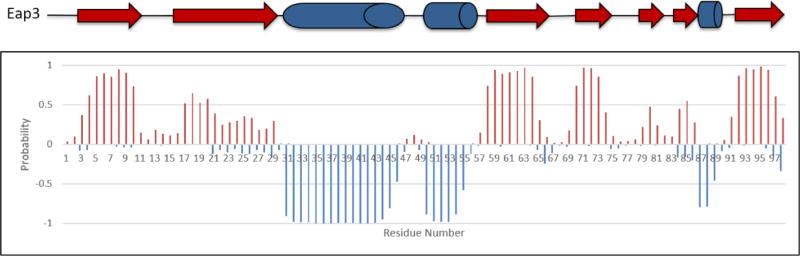
The secondary structure elements of Eap3 were predicted by the TALOS-N program (Shen et al. 2013) using the resonance assignments of 13Cα, 13Cβ, and 13C’ (Fig. 2). This prediction is in agreement with the secondary structures observed in EapH1, EapH2, and Eap2, as determined by X-ray crystallography (Geisbrecht et al. 2005). The chemical shift assignments have been deposited in BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 27087.
Fig. 2.
Secondary structure prediction for the Eap3 domain based on the TALOS-N program using obtained chemical shift values. β-strand probabilities are given by positive values, α-helices are given by negative values, and loop regions are given by values approximately from −0.3 to 0.3. The TALOS-N prediction of Eap3 with α-helices shown as cylinders and β-sheets shown as arrows is presented at the top.
Acknowledgments
This research was funded by awards from the National Institutes of Health to B.V.G. (GM121511 and AI111203). We are thankful to Dr. Justin Douglas of University of Kansas for helping with NMR spectroscopy. We also thank Center of Biomedical Research Excellence in Protein Structure and Function (COBRE-PSF) at The University of Kansas (NIH grant P30 GM110761) for support in NMR studies.
Footnotes
The authors declare no conflicts of interest.
References
- Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- Archer GL. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- de Jong NWM, Ramyar KX, Guerra FE, Nijland R, Fevre C, Voyich JM, McCarthy AJ, Garcia BL, van Kessel KPM, van Strijp JAG, Geisbrecht BV, Haas PJA. Immune evasion by a staphylococcal inhibitor of myeloperoxidase. Proc Natl Acad Sci. 2017;35:9439–9444. doi: 10.1073/pnas.1707032114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Garcia BL, Zwarthoff SA, Rooijakkers SHM, Geisbrecht BV. Novel Evasion Mechanisms of the Classical Complement Pathway. J. Immunol. 2016;197:2051–2060. doi: 10.4049/jimmunol.1600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht BV, Hamaoka BY, Perman B, Zemla A, Leahy DJ. The crystal structures of EAP domains from Staphylococcus aureus reveal an unexpected homology to bacterial superantigens. J. Biol. Chem. 2005;280:17243–17250. doi: 10.1074/jbc.M412311200. [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Bouyan S, Pop M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expr. Purif. 2006;46:23–32. doi: 10.1016/j.pep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Keller R. The Computer Aided Resonance Assignment Tutorial. 2004. [Google Scholar]
- Kim HK, Thammavongsa V, Schneewind O, Missiakas D. Recurrent Infections and Immune Evasion Strategies of Staphylococcus aureus. Curr. Opin. Microbiol. 2012;15:92–99. doi: 10.1016/j.mib.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambris JD, Ricklin D, Geisbrecht BV. Complement Evasion by Human Pathogens. Nat. Rev. Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy FD. Staphylococcus aureus Infections. N. Engl. J. Med. 1998;339:550–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Nauseef WM. How Human Neutrophils Kills and Degrade Microbes: an Integrated View. Immunol. Rev. 2007;219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- Nauseef WM. Myeloperoxidase in Human Neutrophil Host Defense. Cell Microbiol. 2014;16:1146–1155. doi: 10.1111/cmi.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD. Compement in Immune and Inflammatory Disorders: Pathophysiological Mechanisms. J. Immunol. 2013;190:3831–3838. doi: 10.4049/jimmunol.1203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a Key System for Immune Surveillance and Homeostasis. Nat. Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan AN, Surewaard BGJ, Nijland R, van Strijp JAG. Neutrophils versus Staphylococcus aureus: a Biological Tug of War. Annu. Rev. Microbiol. 2013;67:629–650. doi: 10.1146/annurev-micro-092412-155746. [DOI] [PubMed] [Google Scholar]
- Stapels DA, Ramyar KX, Bischoff M, von Köckritz-Blickwede M, Milder FJ, Ruyken M, Eisenbeis J, McWhorter WJ, Herrmann M, van Kessel KP, Geisbrecht BV, Rooijakkers SH. Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc Natl Acad Sci. 2014;111:13187–192. doi: 10.1073/pnas.1407616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead B, Craven CJ, Waltho JP. Double and Triple Resonance NMR Methods for Protein Assignment. In: Reid DG, editor. Protein NMR Techniques. Methods in Molecular Biology. Vol. 60. Humana Press; 1997. [DOI] [PubMed] [Google Scholar]
- Woehl JL, Stapels DAC, Garcia BL, Ramyar KX, Keightley A, Ruyken M, Syriga M, Sfyroera G, Weber AB, Zolkiewski M, Ricklin D, Lambris JD, Rooijakkers SHM, Geisbrecht BV. The Extracellular Adherence Protein from Staphylococcus aureus Inhibits the Classical and Lectin Pathways of Complement by Blocking Formation of the C3 Pro-Convertase. J. Immunol. 2014;193:6161–6171. doi: 10.4049/jimmunol.1401600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehl JL, Takahashi D, Herrera AI, Geisbrecht BV, Prakash O. 1H, 15N, and 13C resonance assignments of Staphylococcus aureus extracellular adherence protein domain 4. Biomol NMR Assign. 2016;10:301–305. doi: 10.1007/s12104-016-9688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehl JL, Ramyar KX, Katz BB, Walker JK, Geisbrecht BV. The Structural Basis for Inhibition of the Classical and Lectin Complement Pathways by S. aureus Extracellular Adherence Protein. Protein Sci. 2017;26:1595–1608. doi: 10.1002/pro.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]