Abstract
Ribonuclase P (RNase P) is an essential metallo-endonuclease that catalyzes 5′ precursor-tRNA (ptRNA) processing and exists as an RNA-based enzyme in bacteria, archaea, and eukaryotes. In bacteria, a large catalytic RNA and a small protein component assemble to recognize and accurately cleave ptRNA and tRNA-like molecular scaffolds. Substrate recognition of ptRNA by bacterial RNase P requires RNA-RNA shape complementarity, intermolecular base pairing, and a dynamic protein-ptRNA binding interface. To gain insight into the binding specificity and dynamics of the bacterial protein-ptRNA interface, we report the backbone and side chain 1H, 13C, and 15N resonance assignments of the hyperthermophilic Thermatoga maritima RNase P protein in solution at 318 K. Our data confirm the formation of a stable RNA recognition motif (RRM) with intrinsic heterogeneity at both the N- and C-terminus of the protein, consistent with available structural information. Comprehensive resonance assignments of the bacterial RNase P protein serve as an important first step in understanding how coupled RNA binding and protein-RNA conformational changes give rise to ribonucleoprotein function.
Keywords: NMR resonance assignment, Thermatoga maritima, RNase P, protein-tRNA binding, ribonucleoprotein, TALOS-N prediction
Biological Context
Ribonucleoprotein complexes (RNPs) assemble into defined tertiary architectures and undergo dynamic interactions to catalyze and mediate fundamental cellular reactions. Ribonuclease P (RNase P) is an essential ribonucleoprotein complex that catalyzes the conversion of precursor tRNA (ptRNA) into functional tRNA, facilitating the removal of the 5′ leader RNA sequence (Guerrier-Takada et al. 1983). The RNA-based form of Ribonuclease P is a multi-turnover ribozyme found in almost all organisms yet relies on one or more protein subunits for increased catalytic efficiency and enzymatic function in vivo. Structural and biochemical studies of the RNase P have proven to be fundamental to our understanding of catalytic RNA structure and function, as well as RNA-based gene regulatory processes (see reviews: (Kazantsev and Pace 2006; Hartmann et al. 2009; Liu and Altman 2010; Mondragon 2013; Klemm et al. 2016)). However, the mechanism of action for this essential ribozyme and the multifunction roles of RNase P protein components, in bacteria and higher organisms, remain unresolved and incomplete (Klemm et al. 2016; Lemieux et al. 2016; Gopalan et al. 2017; Martin and Reiter 2017). Specifically, elucidating the conformational transitions that give rise to the formation of the RNA-based active site and defining the molecular details of how the RNase P protein components orient diverse RNA substrates will be required to better understand how dynamic RNP assemblies function as enzymes. The RNA-based form of RNase P serves as an ideal system to gain insight into the origins of RNA catalysis and to better understand the evolution of RNP complexes.
In bacteria, the RNase P holoenzyme is composed of a large RNA (~400 nucleotides), a small protein subunit (~120 amino acids), and at least two catalytically important metal ions. Crystallographic studies of the bacterial RNase P RNA alone and as a holoenzyme-product complex show how the active site is globally organized and how shape-dependent recognition occurs for the ptRNA substrate (Krasilnikov et al. 2003; Krasilnikov et al. 2004; Kazantsev et al. 2005; Torres-Larios et al. 2005; Reiter et al. 2010). In addition, biochemical studies show that the protein component can increase the enzyme’s functionality and alter substrate recognition properties to facilitate RNase P activation (Crary et al. 1998; Kurz et al. 1998; Niranjanakumari et al. 1998; Buck et al. 2005a; Buck et al. 2005b; Lin et al. 2016; Niland et al. 2016; Liu et al. 2017; Niland et al. 2017). At the molecular level, the P protein associates with three universally conserved regions of the catalytic RNA via basic amino acids and a bacterially conserved electropositive-rich α-helix (Stams et al. 1998; Buck et al. 2005a; Reiter et al. 2010). Along with this key RNA-protein interaction, the formation of the RNP complex enables protein-mediated conformational transitions that help to optimally position, align, and discriminate RNA substrates adjacent to the enzyme active site (Crary et al. 1998; Buck et al. 2005a; Buck et al. 2005b; Christian et al. 2006; Sun et al. 2006; Hsieh and Fierke 2009; Koutmou et al. 2010; Sun et al. 2010; Reiter et al. 2012; Guenther et al. 2013). Thus, although RNase P function likely varies from different organisms, protein stabilization of the highly conserved P RNA sub-domain structure and protein recognition of the 5′ ptRNA leader serve as key general properties of the RNase P holoenzyme.
The hyperthermophilic Thermatoga maritima RNase P undergoes reversible folding and has optimal activity at 50 °C (Paul et al. 2001; Buck et al. 2005a; Buck et al. 2005b). The T. maritima RNase P holoenzyme serves as excellent system to probe the dynamics of protein-ptRNA binding. Both the RNA and protein components of T. maritima RNase P are highly structured and both represent ideal biomolecules to examine how large RNA-protein complexes are assembled and become functional enzymes. Here, we report the 1H, 13C, and 15N resonance assignments of the T. maritima RNase P protein. This study will help to illuminate the conformational transitions of a bacterial RNase P protein in the reaction mechanism, from substrate recognition to facilitating the stabilization of the active site. Complete resonance assignments of the backbone and side chain residues will allow us to probe the dynamics of protein-ptRNA binding and define the motional properties associated with bacterial RNase P holoenzyme activation.
Methods and experiments
Sample preparation
The protein sample was prepared using the BL21 Gold Escherichia coli strain and derived from a pGEX4Ta vector that contained the rnpA gene from T. maritima and an N-terminal glutathione S-transferase (GST) fusion protein. The optimized expression protocol utilized M9 minimal media containing 15NH4Cl and 13C6-D-glucose (Cambridge Isotope Laboratories). A 3 mL starter cell culture was grown at 310 K (225 rpm) with 150ug/mL ampicillin for 20 hours and was used to inoculate 50 mL M9 media (200 ug/mL ampicillin). This 50 mL culture was grown at 310 K (225 rpm) for 12–14 hours and subsequently transferred into 2 L M9 media for 8–10 hours. When OD600 reached 1.0–1.2, temperature was reduced to 303 K and 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce expression 10–14 hours. Cells were subsequently harvested by centrifugation at 5000 g for 15 min, flash cooled in liquid nitrogen, and stored at −80°C until use.
15N,13C-labeled RNase P protein purification followed the previous protocol (Krivenko et al. 2002) with the following modifications. Pellets were thawed on ice and fully resuspended in an appropriate volume of lysis buffer (50 mM Tris-HCl pH 7.5, 4 mM EDTA, 5% (v/v) glycerol, 0.1% (v/v) NP-40 and cOmplete™ protease inhibitor cocktail (Roche)). The cell suspension was lysed on ice by sonication for 13 cycles of 60 second bursts followed by 1 min pauses to prevent overheating. The lysate was separated by centrifugation at 28,000 g and 4 °C for 30 min. and the filtered lysate was treated with 800 NIH units of thrombin per 40 mL for overnight at room temperature. In this step, the GST was cleaved from the RNase P protein, resulting in a Gly-Ser sequence at the N-terminus.
The digested 15N, 13C-labeled sample was purified following a protein denaturation - renaturation strategy that was previously developed (Paul et al. 2001; Buck et al. 2005a; Buck et al. 2005b). To denature the protein, lysate dilution buffer (50 mM Tris-HCl pH7.5, 4 mM EDTA, 8 M urea) was added into the sample to make a final concentration to 5 M urea. Sample was loaded on to a pre-equilibrated 15S cation exchange column (50 mM Tris-HCl pH 7.5, 0.2 mM EDTA, 5 M urea) and subsequently eluted with a linear gradient (50 mM Tris-HCl pH 7.5, 0.2 mM EDTA, 5 M urea, 2 M NaCl). Appropriate fractions containing the denatured 15N, 13C-labeled RNase P protein were dialyzed against 4 L buffer of 50 mM Tris-HCl pH 7.5, 0.2 mM EDTA, 1 M NaCl for 1–2 days. After the protein was renatured, the sample solution was diluted to approximately 200mM NaCl and a second 15S cation exchange chromatography step was applied in the absence of urea, with a pre-equilibration buffer (50mM Tris-HCl pH7.5, 0.2mM EDTA) and an elution gradient containing 50mM Tris-HCl pH7.5, 0.2mM EDTA, and 3M NaCl. Highly pure 15N, 13C-labeled RNase P protein fractions were pooled, concentrated, and extensively dialyzed against 2 L buffer (50 mM Tris-HCl pH 7.5, 0.2 mM EDTA). A final dialysis against 4 liters of 20 mM sodium phosphate pH 6.0, 80 mM NaCl, 8 mM sodium sulfate solution was performed prior to NMR sample preparation. Protein purity was verified by SDS-PAGE.
NMR spectroscopy
All NMR experiments were conducted using 400 μM 15N, 13C-RNase P protein in 20 mM sodium phosphate pH 6.0, 80 mM NaCl, 50 μM 4, 4-dimethyl-4-silapentane-sulfonate (DSS) at 318 K with 5 % (v/v) D2O in a 3 mm Norell® select series NMR tube unless noted. NMR spectra were acquired on Bruker Avance-III 600MHz, 800MHz and 900 MHz spectrometers, equipped with cryogenic probes, as well as Varian VNMRS 600 and 800 MHz spectrometer equipped with a cryogenic probe. For backbone assignments, (1H, 15N) heteronuclear single quantum coherence (HSQC) and a set of traditional triple-resonance experiments were performed, including HNCO, HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HN(CA)CO. Side-chain assignments were carried out using three-dimensional (H)C(CCO)NH, H(CCCO)NH, (H)CCH-TOCSY, H(C)CH-TOCSY, 15N-NOESY-HSQC, 13C-NOESY-HSQC and (1H, 13C) HSQC for aliphatic and aromatic groups, respectively. The 13C-NOESY-HSQC and aliphatic and aromatic optimized 2D 13C HSQC experiments were obtained in 95% (v/v) D2O. 1H chemical shifts were referenced with respect to internal DSS, and 13C and 15N chemical shifts were referenced indirectly using nuclei-specific gyromagnetic ratios (Wishart et al. 1995). Spectra were processed with Topspin 3.5.7 (Bruker Inc.) and NMRPipe (Delaglio et al. 1995), and analyzed by using CARA (Keller 2004) and Sparky (Goddard and Kneller).
Assignments and data deposition
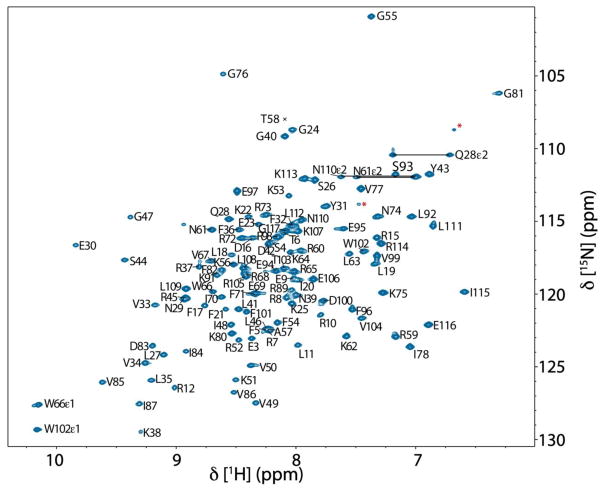
Excluding the N-terminal non-native glycine and serine, 110 of 114 non-proline residues were assigned in the (1H, 15N) HSQC spectrum, which corresponds to 96.5% of the backbone amides (Figure 1). The amide groups of T2, L13, R14 and K90 remain unassigned due to peak overlap, solvent exchange, or intermediate exchange broadening on the millisecond timescale. 99.1% of both Cα and Cβ resonances were assigned, while backbone C′ were 97.4% assigned. In addition, the side-chain assignments of Cγ, Cδ and Cε resonances, respectively, are 78.5%, 77.8% and 57.1% complete. Regarding the completeness of protons, 99.2% of Hα, 99.0% of Hβ, along with 91.8%, 84.8% and 50.6% of Hγ, Hδ and Hε resonances, respectively, were unambiguously assigned.
Fig. 1.
900 MHz (1H, 15N) HSQC spectrum of 400 μM T. maritima RNase P protein in 20 mM sodium phosphate (pH 6.0), 80 mM NaCl, 50 μM DSS, 8 mM sodium sulfate, and 5 % D2O at 318 K. Assigned residues are indicated using single letter codes. Side-chain NH2 resonances of asparagine and glutamine are connected by horizontal lines. Signals labeled by red asterisks likely derive from arginine εNH resonances while those non-labeled are likely from minor population of the structural conformations in the sample. The assigned resonances with only a black cross indicate the position of the residue below the spectrum’s intensity levels. Numbering of T. maritima RNase P protein corresponds to the gene sequence RNPA_THEMA (UniProtKB - Q9X1H4).
Among the side-chain residues, L13, R44, R89 and K51 contain three or more unassigned side-chain aliphatic resonances that were not observable or were potentially in the overlapped areas in spectra. In total, 741 1H, 547 13C and 115 15N chemical shifts are assigned for the 14.2 kDa protein. The 13C and 15N chemical shifts were internally referenced using 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), according to the literature (Markley et al. 1998). The chemical shift data has been deposited at the Biological Magnetic Resonance Data Bank (BMRB) with the accession number 27307.
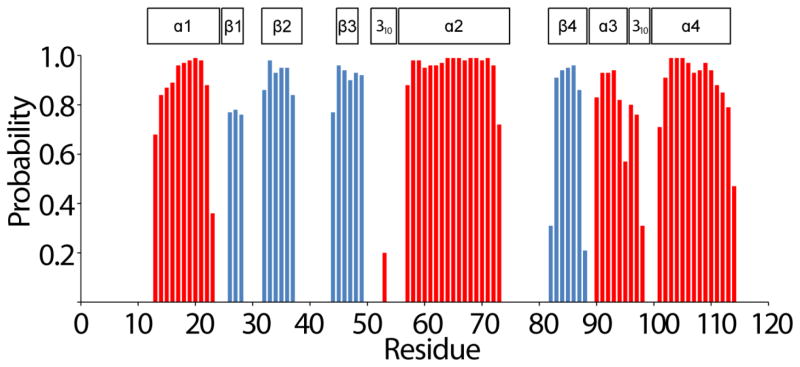
The secondary structure of T. maritima RNase P protein in solution was predicted by uploading the HN, N, Cα, Hα, Cβ, C′, H chemical shifts to the TALOS-N webserver (Shen and Bax 2013). The comparison between the secondary structure from the prediction and the secondary structure of the crystal structure (PDB: 1NZ0) is illustrated in Figure 2. These data show a high consistency, which indicate that the solution conformation of the protein at ≥ 0.4 mM is similar to the 1NZ0 structure. This result provides strong confidence in the resonance assignments of the T. maritima RNase P protein and will enable a comprehensive analysis of the motional properties associated with protein-ptRNA binding interface.
Fig. 2.
TALOS-N prediction of secondary structure elements of T. maritima RNase P protein derived from HN, N, Hα, Cα, Cβ and C′ chemical shifts. The secondary structure probability is reflected by the height of the bars (blue β-strand, red α-helices). For comparison, the positions of secondary structure elements according to the crystal structure (PDB entry 1ZN0) are indicated above the amino acid residues.
Acknowledgments
This work was supported in part by grants for NMR instrumentation from the NSF (0922862), NIH (S10 RR025677) and Vanderbilt University. BPB acknowledges support from the NIGMS (T32GM007347). NJR acknowledges support from Vanderbilt University and Marquette University, as well as funding from the American Heart Association (14GRNT20380334) and the NIH (GM120572). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. a Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNase P RNA by protein. Nat Struct Mol Biol. 2005b doi: 10.1038/nsmb1004. [DOI] [PubMed] [Google Scholar]
- Christian EL, Smith KM, Perera N, Harris ME. The P4 metal binding site in RNase P RNA affects active site metal affinity through substrate positioning. RNA. 2006;12:1463–1467. doi: 10.1261/rna.158606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crary SM, Niranjanakumari S, Fierke CA. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry. 1998;37:9409–9416. doi: 10.1021/bi980613c. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: Available from: https://http://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]
- Gopalan V, Jarrous N, Krasilnikov AS. Chance and necessity in the evolution of RNase P. RNA. 2017 Sep 29; doi: 10.1261/rna.063107.117. pii: rna.063107.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther UP, Yandek LE, Niland CN, Campbell FE, Anderson D, Anderson VE, Harris ME, Jankowsky E. Hidden specificity in an apparently nonspecific RNA-binding protein. Nature. 2013;502:385–388. doi: 10.1038/nature12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A. The making of tRNAs and more - RNase P and tRNase Z. Progress in molecular biology and translational science. 2009;85:319–368. doi: 10.1016/S0079-6603(08)00808-8. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Fierke CA. Conformational change in the Bacillus subtilis RNase P holoenzyme--pre-tRNA complex enhances substrate affinity and limits cleavage rate. RNA. 2009;15:1565–1577. doi: 10.1261/rna.1639409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci U S A. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Pace NR. Bacterial RNase P: a new view of an ancient enzyme. Nat Rev Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- Keller RLJ. The Computer Aided Resonance Assignment Tutorial. Cantina verlag: The Swiss Federal Institute of Technology Zürich, Switzerland; 2004. [Google Scholar]
- Klemm BP, Wu N, Chen Y, Liu X, Kaitany KJ, Howard MJ, Fierke CA. The Diversity of Ribonuclease P: Protein and RNA Catalysts with Analogous Biological Functions. Biomolecules. 2016:6. doi: 10.3390/biom6020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, Zahler NH, Kurz JC, Campbell FE, Harris ME, Fierke CA. Protein-precursor tRNA contact leads to sequence-specific recognition of 5′ leaders by bacterial ribonuclease P. Journal of Molecular Biology. 2010;396:195–208. doi: 10.1016/j.jmb.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Xiao Y, Pan T, Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, Mondragón A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- Krivenko AA, Kazantsev AV, Adamidi C, Harrington DJ, Pace NR. Expression, purification, crystallization and preliminary diffraction analysis of RNase P protein from Thermotoga maritima. Acta crystallographica Section D, Biological crystallography. 2002;58:1234–1236. doi: 10.1107/s0907444902007965. [DOI] [PubMed] [Google Scholar]
- Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- Lemieux B, Laterreur N, Perederina A, Noel JF, Dubois ML, Krasilnikov AS, Wellinger RJ. Active Yeast Telomerase Shares Subunits with Ribonucleoproteins RNase P and RNase MRP. Cell. 2016;165:1171–1181. doi: 10.1016/j.cell.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HC, Zhao J, Niland CN, Tran B, Jankowsky E, Harris ME. Analysis of the RNA Binding Specificity Landscape of C5 Protein Reveals Structure and Sequence Preferences that Direct RNase P Specificity. Cell Chemical Biology. 2016;23:1271–1281. doi: 10.1016/j.chembiol.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Altman S. Ribonuclease P. Springer Science+Business Media, LLC; New York: 2010. [Google Scholar]
- Liu X, Chen Y, Fierke CA. Inner-sphere Coordination of Divalent Metal Ion with Nucleobase in Catalytic RNA. Journal of the American Chemical Society. 2017 Nov 8; doi: 10.1021/jacs.7b08755. [DOI] [PMC free article] [PubMed]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. Journal of Biomolecular NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- Martin WJ, Reiter NJ. Structural Roles of Noncoding RNAs in the Heart of Enzymatic Complexes. Biochemistry. 2017;56:3–13. doi: 10.1021/acs.biochem.6b01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon A. Structural studies of RNase P. Annual Review of Biophysics. 2013;42:537–557. doi: 10.1146/annurev-biophys-083012-130406. [DOI] [PubMed] [Google Scholar]
- Niland CN, Anderson DR, Jankowsky E, Harris ME. The contribution of the C5 protein subunit of Escherichia coli ribonuclease P to specificity for precursor tRNA is modulated by proximal 5′ leader sequences. RNA. 2017;23:1502–1511. doi: 10.1261/rna.056408.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niland CN, Zhao J, Lin HC, Anderson DR, Jankowsky E, Harris ME. Determination of the Specificity Landscape for Ribonuclease P Processing of Precursor tRNA 5′ Leader Sequences. ACS Chemical Biology. 2016;11:2285–2292. doi: 10.1021/acschembio.6b00275. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc Natl Acad Sci USA. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Lazarev D, Altman S. Characterization of RNase P from Thermotoga maritima. Nucleic Acids Research. 2001;29:880–885. doi: 10.1093/nar/29.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman AK, Mondragon A. The bacterial ribonuclease P holoenzyme requires specific, conserved residues for efficient catalysis and substrate positioning. Nucleic Acids Research. 2012 doi: 10.1093/nar/gks744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. Journal of Biomolecular NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stams T, Niranjanakumari S, Fierke CA, Christianson DW. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science. 1998;280:752–755. doi: 10.1126/science.280.5364.752. [DOI] [PubMed] [Google Scholar]
- Sun L, Campbell FE, Yandek LE, Harris ME. Binding of C5 Protein to P RNA Enhances the Rate Constant for Catalysis for P RNA Processing of Pre-tRNAs Lacking a Consensus G(+1)/C(+72) Pair. Journal of Molecular Biology. 2010;395:1019–1037. doi: 10.1016/j.jmb.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Campbell FE, Zahler NH, Harris ME. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. The EMBO journal. 2006;25:3998–4007. doi: 10.1038/sj.emboj.7601290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. Journal of Biomolecular NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]