Abstract
We present a cost-effective and portable platform based on contact lenses for non-invasively detecting Staphylococcus aureus, which is part of the human ocular microbiome and resides on the cornea and conjunctiva. Using Staphylococcus aureus-specific antibodies and a surface chemistry protocol that is compatible with human tear, contact lenses are designed to specifically capture Staphylococcus aureus. After the bacteria capture on the lens, and right before its imaging, the captured bacteria are tagged with surface-functionalized polystyrene microparticles. These microbeads provide sufficient signal-to-noise ratio for the quantification of the captured bacteria on the contact lens, without any fluorescent labels, by 3D imaging of the curved surface of each lens using only one hologram taken with a lensfree on-chip microscope. After the 3D surface of the contact lens is computationally reconstructed using rotational field transformations and holographic digital focusing, a machine learning algorithm is employed to automatically count the number of beads on the lens surface, revealing the count of the captured bacteria. To demonstrate its proof-of-concept, we created a field-portable and cost-effective holographic microscope, which weighs 77 g, controlled by a laptop. Using daily contact lenses that are spiked with bacteria, we demonstrated that this computational sensing platform provides a detection limit of ~16 bacteria/µL. This contact lens–based wearable sensor can be broadly applicable to detect various bacteria, viruses and analytes in tear using a cost-effective and portable computational imager that might be used even at home by consumers.
Keywords: computational sensing, contact lens-based sensing, holographic sensing, lensfree imaging, mobile sensing, machine learning
Graphical abstract
The human body is the host to several microorganisms, forming the human microbiota.1 These microorganisms live symbiotically in various parts of the human body, including the conjunctiva,2 lungs,3 skin, saliva, gut, and vagina.4 The relationship between the human microbiota and various diseases such as obesity, rheumatoid arthritis, and diabetes has drawn strong attention to the analysis of the human microbiome,5 also motivated by personalized treatments and medicine. Several microorganisms are also found in the ocular microbiota, one of which is Staphylococcus aureus. It is a gram-positive bacterium and is present in different parts of the body, including the conjunctiva, nose, and skin.6–8 It is colonized in approximately 30% of humans.9 It is also a human pathogen, and the carriers of Staphylococcus aureus have a higher risk of infection. It causes a variety of diseases like pneumonia, endocarditis, arthritis, and bacteremia.6
Monitoring of the human microbiome with the current methods is relatively costly and time-consuming. 16S rRNA and whole-genome shotgun metagenomics gene-based sequencing are among the most commonly used techniques for the analysis of microbiome.10,11 16S rRNA sequencing starts with the extraction of the DNA from the isolated sample, and it is followed by polymerase chain reaction (PCR) amplification, cycle sequencing, and database comparison.12 As an additional challenge for monitoring of ocular microbiota, collecting samples from the ocular surface is quite tedious. It involves conjunctival swabs or tear fluid collection8,13 which require laborious sample collection steps as well as relatively expensive and bulky laboratory equipment for the sample analysis.
Here, we present a cost-effective and portable platform that is based on contact lenses for detecting and monitoring Staphylococcus aureus, which is found in the human ocular microbiome. In this study, the detection of Staphylococcus aureus is enabled by a surface functionalized contact lens,14 a lens-free computational imaging setup,15–18 and a machine learning-based algorithm to quantify the amount of bacteria captured on the contact lens. The surface of each contact lens is functionalized by a layer-by-layer (LBL) coating technique19 which is shown to be compatible with the human tear. The LBL coating technique creates biofunctional films on different surface morphologies, and with the appropriate coating material, it enables contact lenses to be worn without any damage to the cornea.14 Seven layers of poly(styrene sulfonate) (PSS) and poly(allylamine hydrochloride) (PAH) coating on a contact lens create a stable structure for binding of the anti-Staphylococcus aureus antibody to the surface of the contact lens. We used antibody attached daily contact lenses for specifically capturing Staphylococcus aureus particles that normally reside on the human cornea and conjunctiva. These surface functionalized contact lenses are expected to be worn for e.g., 12–16 h for continuously capturing Staphylococcus aureus in tear. In this work, however, we spiked daily contact lenses with bacteria and after the capture process, and right before the 3D imaging of the lens surface, we attached 5-µm polystyrene beads to specifically bind to Staphylococcus aureus particles already captured on the lens. Staphylococcus aureus has a diameter of 0.75–1 µm,20 and therefore 5-µm beads make their detection and counting easier using lensfree imaging of the contact lens 3D surface with a single hologram.
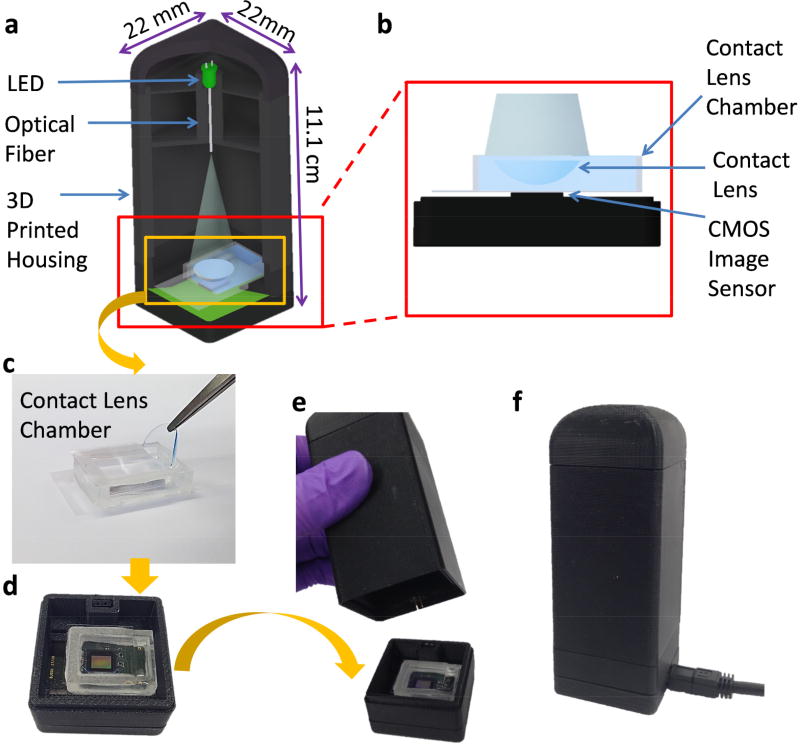
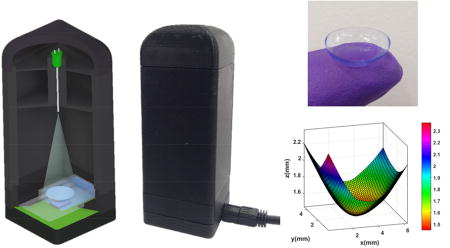
In fact, microscopic imaging of a contact lens is very challenging using traditional microscopy tools because it is large and has a curved 3D structure. Conventional microscopes have a small field of view (FOV) and a limited depth of field, making it extremely difficult and time-consuming to image the 3D surface of a contact lens using traditional imaging approaches. Our computational imaging platform is not only cost-effective and field-portable, but also offers a very large FOV (~30 mm2) and depth of field (> 1cm), which make it ideal for label-free imaging of the 3D surface of a contact lens using a single hologram, without the need for any mechanical scanning or fine alignments (Figure 1). Our lens-free on-chip imaging setup utilizes a multimode-fiber-coupled light emitting diode (LED), which emits partially coherent light at 527 nm. In order to image the contact lenses with their natural curvy shape, they are placed in a custom-designed chamber filled with a phosphate-buffered saline (PBS) solution. An inexpensive complementary metal oxide semiconductor (CMOS) image sensor captures a single hologram of each contact lens, which is then reconstructed to automatically identify the captured micro-beads on the lens surface. This analysis includes a tilt correction algorithm that handles the image distortions caused by the curvature of the contact lenses, a holographic 3D image reconstruction algorithm, as well as a support vector machine (SVM) based learning model that can specifically detect the captured 5-µm beads and digitally separate them from other unwanted particles deposited on the lens surface.
Fig. 1.
(a) 3D schematic illustration and (b) a cross section of the portable lensfree microscope including a contact lens chamber. (c) Contact lens insertion into a PBS filled contact lens chamber. (d) Placement of the chamber on a CMOS image sensor. (e, f) Photographs of the lensfree microscope.
Using daily contact lenses that are surface functionalized and spiked with bacteria, we demonstrated a detection limit of ~16 bacteria/µL with this computational 3D imaging and machine learning approach. The presented wearable sensing framework using surface functionalized contact lenses and computational imaging can be broadly applicable to sense numerous other target bacteria, viruses and analytes in tear samples and might even be suitable for consumer use at home.
Results and Discussion
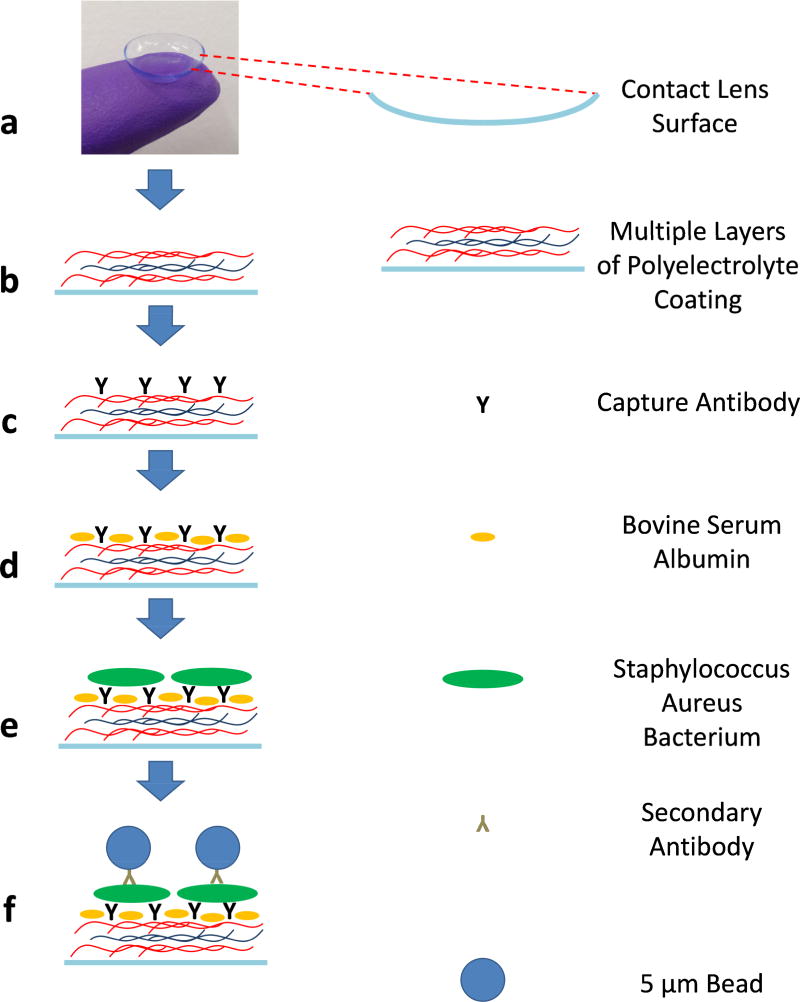
Acuvue Moist 1-day contact lenses have an inherently ionic surface structure. Etafilcon A of these contact lenses is negatively charged21 at the physiological pH so that the innermost layer of the positively charged polyelectrolyte, PAH, can bind to the contact lens surface electrostatically. In a similar fashion, the chemical functionalization of the contact lens surface was performed with an alternative LBL deposition of the positively charged polyelectrolyte, PAH, and the negatively charged polyelectrolyte, PSS, by using the electrostatic interaction between them. The outermost layer was coated with the positively charged PAH to enable the electrostatic interaction between the functionalized contact lens surface and the anti-Staphylococcus aureus antibody, which is negatively charged at the physiological pH. These seven alternating layers of coating on the contact lens surface created a stable interface for the successive antibody binding and bead-based immunoassay formation (Figure 2). Importantly, previous work showed that this polyelectrolyte coating did not exhibit cell toxicity.14
Fig. 2.
Schematic illustration of the contact lens surface functionalization and bead-based immunoassay steps.
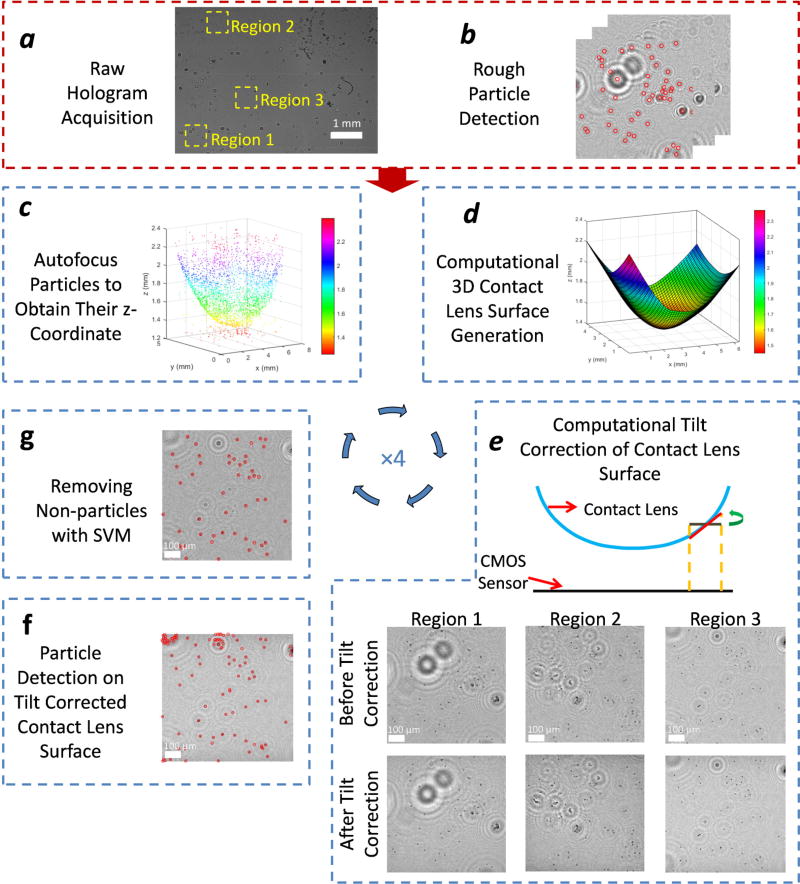
As detailed in the Methods section, the digital 3D surface mapping of each contact lens under test provides the rotation angle for each sub-region on the lens surface that we need for field rotations. Figure 3-d shows the digitally reconstructed 3D surface of a contact lens. The color map indicates the distance of each point on the contact lens surface from the CMOS image sensor plane. In our experiments, we observed a maximum tilt of approximately 20° at the corners of the contact lens with respect to the CMOS sensor. After the tilt correction of each sub-region of interest, we create a set of images, each of which is tangential to the contact lens surface. By digitally stitching all these tilt-corrected small tiles, we obtained an in-focus image of the contact lens surface, covering approximately 31 mm2 on the lens surface, which is larger than the active area of the CMOS imager, as expected. In Figure 3-e, the reconstructions of some of these sub-regions are shown without any tilt correction and after tilt correction. We can clearly observe that the 5-µm beads are in focus, as desired, over the entire FOV after the tilt correction, whereas in the regular reconstructions, they are out of focus at the edges of each region of interest. This phenomenon is more obvious if one looks at the corners of the contact lens; for example, in regions 1 and 2, the impact of tilt corrections is more apparent compared to region 3 (Figure 3).
Fig. 3.
Schematic diagram of the image reconstruction and processing steps.
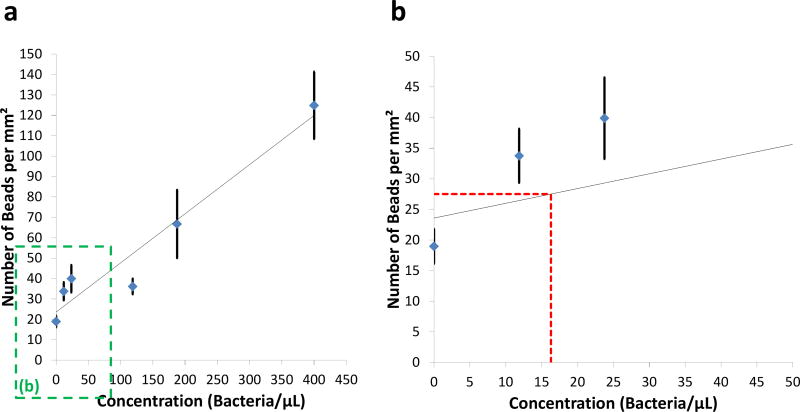
To explore the detection limit of our platform, five solutions of Staphylococcus aureus at different concentrations were applied after the surfaces of the contact lenses were functionalized as described in the Methods section. At each concentration of bacteria, we used three individual contact lenses to test the repeatability of our approach. After the incubation of the streptavidin-coated antibody conjugated 5-µm beads, the contact lenses were imaged using our lens-free on-chip microscope, and the 5-µm beads were automatically counted using an SVM-based algorithm described in the Methods section. Figure 4 shows the number of the detected beads on contact lens surfaces for different concentrations of Staphylococcus aureus. A detection limit of 16.3 bacteria/µL was achieved using our platform, based on µ+3σ of our control samples, where µ refers to the mean and σ is the standard deviation. Previous results reported that the human eye can contain increased concentrations of bacteria (e.g., >10 cfu/µL) even for asymptomatic individuals.8 Considering the fact that under a bacterial infection the concentration of bacteria in tear is expected to be significantly higher than this baseline, we believe that our sensing limit is relevant for the detection of such infections. A further improvement in our sensing performance can potentially be achieved by increasing our training data size and the number of the spatial features used in the SVM-based particle detection algorithm, which can help us reduce the rate of false positives. Convolutional neural network based deep learning approaches could also be utilized to further advance our results, which is left as future work.
Fig. 4.
(a) The number of the detected beads on the contact lens surface per mm2 as a function of the bacteria concentration. (b) Zoomed in version of (a), red dashed line refers to µ+3σ of the control samples.
The presented work makes use of contact lenses as the basis of a computational sensing platform. Although not demonstrated in this work since we did not experiment with human samples, we believe that this wearable method is more convenient for patients when compared to e.g., conjunctival swabs and tear collection. We also believe that this method should increase the efficiency of sample collection because the contact lens will be worn throughout the day for e.g., 12–16 h and it will be continuously in contact with the human tear.
Conclusions
We demonstrated a proof-of-concept platform that can potentially monitor and non-invasively analyze the human ocular microbiome in a cost-effective manner. This platform can automatically detect and enumerate Staphylococcus aureus particles that are captured on contact lenses. It includes a field-portable lens-free microscope, a custom-made contact lens chamber, surface functionalized contact lenses, and an automated holographic image reconstruction and processing algorithm that also utilizes machine learning. We have achieved a detection limit of 16.3 CFU/µL. We believe that this platform might serve as a promising tool for the analysis and monitoring of the human ocular microbiome and can be broadly applicable to other target bacteria, viruses and analytes that can be sensed using wearable and flexible substrates, including but not limited to contact lenses.
Materials and Methods
The procedure for the automated detection and quantification of Staphylococcus aureus starts with the functionalization of the surface of a contact lens, and it is followed by a bead-based immunoassay on the lens surface to achieve specificity and sensitivity. After the capture of the target bacteria, each contact lens under test is placed in a custom-designed sample holder, and its lens-free hologram is taken using our portable on-chip microscope. Each hologram is then rapidly reconstructed to reveal a microscopic image of the 3D surface of the contact lens, which is then analyzed using an automated machine-learning algorithm to estimate the count of Staphylococcus aureus captured on the lens surface.
Materials
Poly(sodium 4-styrenesulfonate) (PSS; Mw ~200000 g mol−1) (product no. 561967), bovine serum albumin (BSA) (product no. B4287), phosphate buffered saline (PBS pH 7.4) (product no. P3813), and TweenR20 (product no. P9416) were purchased from Sigma–Aldrich. Poly(allylamine hydrochloride) (PAH; Mw ~120000–200000 g mol−1) (product no. 43092) was purchased from Alfa Aesar. Anti-Staphylococcus aureus antibody (product no. ab73962) and Anti-Staphylococcus aureus antibody biotin (product no. ab35192) were purchased from Abcam. Contact lenses (Acuvue Moist 1-day) were purchased from Contact Lenses Canada. Staphylococcus aureus bacteria (product no. 27660) were purchased from American Type Culture Collection (ATCC). Sodium chloride (NaCl) (product no. SX0420) was purchased from Millipore Sigma. Streptavidin coated 5-µm polystyrene beads (product no. SVP-50-5) were purchased from Spherotech Inc. Acrylic glass (PMMA) (product no. 8560K354) was purchased from McMaster-Carr. The LED (product no. C503B-GAN-CB0F0791-ND) was purchased from Digikey and the multimode fiber (product no. FG105LCA) was purchased from Thorlabs. The coverslips were purchased from Fisher Scientific. Reagent grade water was used throughout the experiments.
Design of the portable lens-free on-chip microscope
Our lens-free microscope consists of an LED that emits green light with a peak wavelength of 527 nm, a multimode fiber (105 µm core diameter), a CMOS image sensor with a pixel size of 1.67 µm (MT9J003STM/STC, ON Semiconductor), a custom-made contact lens holder, and a 3D printed housing that holds all the components (Figure 1). The design of the housing was done using Autodesk Inventor Professional, and it was printed using a 3D printer and acrylonitrile butadiene styrene (ABS) material (Dimensions Elite, Stratasys).
The top part of the contact lens chamber is a 22×22 mm No.1 coverslip (thickness: 120 µm), and the bottom part is formed by a 24×35 mm No.0 coverslip (thickness of 70 µm). The rectangular sidewalls of the chamber were prepared using a laser cutting device with PMMA (thickness of 6 mm). The coverslips and PMMA were glued together using an epoxy (Figure 1-c). After the bead-based immunoassay, each contact lens was placed inside the PBS-filled contact lens chamber for holographic imaging. This sample holder provides an environment for the contact lenses to retain their natural 3D structure and curvature while enabling us to image them by preventing surface fluctuations that may occur at the liquid–air interface. The holder was then placed directly on the top of the CMOS image sensor. The fiber-coupled LED provided sufficient spatial coherence to capture a single hologram that contains all the microparticles captured over the 3D surface of the contact lens. The exposure time of each hologram was ~50 ms, which is short enough to avoid any problems related to a potential shift in the contact lens position within the sample holder. For illumination, a hole was drilled on the LED package, and a multi-mode fiber was inserted into that hole and fixed with an optical glue.22 This fiber-coupled LED was then placed approximately 5 cm away from the CMOS sensor plane. The LED was powered by the CMOS sensor board.
Contact lens surface functionalization
After unpacking the daily contact lenses, we washed them by dipping each one of the lenses into a PBS solution (10 mM, pH 7.4) to equilibrate the polymer surface of the contact lenses. The surface was then functionalized using the LBL deposition of polyelectrolytes, PAH, and PSS. First, the contact lenses were dipped into cationic polyelectrolyte, PAH (5 mg mL−1 in 0.5 M NaCl) for 15 min. Then, they were washed with 0.5 M NaCl solution three times. Next, the contact lenses were dipped into anionic polyelectrolyte, PSS (5 mg mL−1, in 0.5 M NaCl), for 15 min. Then, they were washed again with 0.5 M NaCl solution three times. The same PAH and PSS deposition steps were repeated until seven layers of polyelectrolyte coating (i.e., PAH-PSS-PAH-PSS-PAH-PSS-PAH) were created on the surface of each contact lens (Figure 2-b).
Bead-based immunoassay and related experimental procedures
200 µL of 10 µg/mL captured antibody anti-Staphylococcus aureus solution in 10 mM PBS (pH 7.4) was incubated on the contact lenses, which were previously coated with polyelectrolytes as detailed earlier (Figure 2-c). After this incubation for 16 h, they were washed with 10 mM PBS at pH 7.4 to remove the excess antibody from the contact lenses. Then, the lens surface was blocked by a buffer (1% BSA in 10 mM PBS), incubated for 2 h to reduce the non-specific binding of the antibody attached polystyrene beads (Figure 2-d). The contact lenses were then washed once with 10 mM PBS at pH 7.4.
To mimic the daily use of the contact lenses, 200 µL solution of 4% (v/v) formaldehyde fixed Staphylococcus aureus (at different concentrations) in 10 mM PBS (pH 7.4) was incubated on the contact lenses for 16 h (Figure 2-e). During this incubation period, Staphylococcus aureus would adhere to the antibody coated contact lens surface. After this incubation, the contact lenses were washed once with 10 mM PBS (pH 7.4). Then, at the testing phase (i.e., after each contact lens captured the Staphylococcus aureus particles on its surface) the 200-µL bead and antibody mixture was incubated on the contact lenses for 2 h (Figure 2-f). To prepare these streptavidin-coated 5-µm beads, they were mixed with 27 µg/mL biotin-anti-Staphylococcus aureus in a separate tube in 10 mM PBS (pH 7.4) for 1 h. The strong interaction between streptavidin and biotin enables antibody-conjugated microbeads to specifically attach to Staphylococcus aureus. This approach provided specificity, helping us correlate the number of bacteria with the number of microbeads. Finally, the contact lenses were washed thrice with a washing buffer (10 mM PBS, 0.1% BSA, and 0.05% Tween R 20 at pH 7.4) to remove the excess beads before their imaging using the holographic on-chip microscope.
Automated analysis of the holograms of contact lenses
We prepared an image processing algorithm (Figure 3) for the automated detection and counting of Staphylococcus aureus captured on contact lenses. This algorithm starts with the holographic reconstruction of the 3D contact lens surface and uses an SVM-based learning algorithm for label-free classification of the captured particles on the lens surface. First, a rough estimate of all the particles on a contact lens was made by reconstructing its lens-free hologram at all the possible object planes with a vertical spacing of 5 µm as shown in Figure 3-b. The possible 5-µm bead candidates were then detected from each reconstructed amplitude image by a simple threshold. In order to have a better accuracy in the axial position estimation of each particle, an autofocusing algorithm using the Tamura coefficient was employed on these potential particle candidates.23–27 The resulting x-y-z positions of these beads were then selected as the sampled points on the contact lens 3D surface (Figure 3-c). A physical constraint based on this initially estimated 3D shape of the contact lens was also applied to remove possible detection artifacts and false positives due to unbound floating particles in the sample holder. A locally-weighted linear regression was then performed on these points to digitally generate a 3D map of each contact lens surface that is under test, as shown in Figure 3-d. This step is important to properly image and count the 5-µm beads captured on the surface of the contact lenses, and that is why we first reconstructed the 3D contact lens surface digitally, and then employed a tilt correction algorithm28,29 to smaller regions of interest on the contact lens surface to digitally bring each region of interest in focus. There are different approaches to image tilted objects such as extended-focus imaging (EFI)30,31 and rotational field transformations.28,29 In this work, we used the latter approach, which is based on the angular spectrum method. This tilt correction algorithm involves two Fast Fourier Transform (FFT) operations and one interpolation operation. In essence, with the knowledge of the complex optical field at one of the planes, we can obtain the complex field at another rotated plane by using this tilt correction algorithm. In order to perform this operation, each lens-free hologram of a contact lens was digitally divided into 192 equal tiles (i.e., smaller regions of interest) and each tile was processed separately. These tiles were reconstructed at their corresponding heights, where their centers intersected with the computationally generated 3D contact lens surface. The natural curvature of the contact lenses prevents each tile to be completely in-focus, making the edges of the tiles out of focus. To mitigate this, the reconstructed tiles were rotated using the tilt correction algorithm so that they became tangent to the computationally generated contact lens 3D surface (Figure 3-e). For each tile, we calculated the rotation angle based on the reconstructed 3D contact lens surface.
To further eliminate false positives, we employed an SVM based learning algorithm26 to distinguish 5-µm beads from other non-specifically bound particles on the lens surface that appear in the reconstructed and tilt-corrected holographic images. Each object’s size, intensity and ratio of the Tamura coefficient at the focus plane with respect to four other planes were fed into an SVM algorithm forming a set of six features. The SVM was then trained on approximately 3,000 particles, where 1,200 of them were 5-µm beads, all of which were manually labeled, forming our training data. Figure 3-g shows the blind detection of the target beads captured on the contact lens surface after the SVM was successfully trained. The steps shown in Figure 3-c to Figure 3-g were repeated three more times for refining the detection results.
Acknowledgments
The authors acknowledge the National Institutes of Health (NIH, R21EB023115). The Ozcan Research Group at UCLA acknowledges the support of NSF Engineering Research Center (ERC, PATHS-UP), the Army Research Office (ARO; W911NF-13-1-0419 and W911NF-13-1-0197), the ARO Life Sciences Division, the National Science Foundation (NSF) CBET Division Biophotonics Program, the NSF Emerging Frontiers in Research and Innovation (EFRI) Award, the NSF EAGER Award, NSF INSPIRE Award, NSF Partnerships for Innovation: Building Innovation Capacity (PFI:BIC) Program, Office of Naval Research (ONR), the Howard Hughes Medical Institute (HHMI), Vodafone Americas Foundation, the Mary Kay Foundation, Steven & Alexandra Cohen Foundation, and KAUST. This work is based upon research performed in a laboratory renovated by the National Science Foundation under Grant No. 0963183, which is an award funded under the American Recovery and Reinvestment Act of 2009 (ARRA). The authors also acknowledge Drs. Hatice Ceylan Koydemir and Hyou-Arm Joung of UCLA for helpful discussions regarding the bead-based immunoassay, Yichen Wu of UCLA for help in the assembly of the light source and data processing.
References
- 1.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI. The Human Microbiome Project: Exploring the Microbial Part of Ourselves in a Changing World. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong Q, Brulc JM, Iovieno A, Bates B, Garoutte A, Miller D, Revanna KV, Gao X, Antonopoulos DA, Slepak VZ, Shestopalov VI. Diversity of Bacteria at Healthy Human Conjunctiva. Invest. Ophthalmol. Visual Sci. 2011;52:5408–5413. doi: 10.1167/iovs.10-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck JM, Young VB, Huffnagle GB. The Microbiome of the Lung. Transl. Res. 2012;160:258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium THMP. Structure, Function and Diversity of the Healthy Human Microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biteen JS, Blainey PC, Cardon ZG, Chun M, Church GM, Dorrestein PC, Fraser SE, Gilbert JA, Jansson JK, Knight R, Miller JF, Ozcan A, Prather KA, Quake SR, Ruby EG, Silver PA, Taha S, Engh G, Weiss PS, Wong GCL, et al. Tools for the Microbiome: Nano and Beyond. ACS Nano. 2016;10:6–37. doi: 10.1021/acsnano.5b07826. [DOI] [PubMed] [Google Scholar]
- 6.Dayan GH, Mohamed N, Scully IL, Cooper D, Begier E, Eiden J, Jansen KU, Gurtman A, Anderson AS. Staphylococcus Aureus: The Current State of Disease, Pathophysiology and Strategies for Prevention. Expert Rev. Vaccines. 2016;15:1373–1392. doi: 10.1080/14760584.2016.1179583. [DOI] [PubMed] [Google Scholar]
- 7.Miller D, Iovieno A. The Role of Microbial Flora on the Ocular Surface. Curr. Opin. Allergy Clin. Immunol. 2009;9:466–470. doi: 10.1097/ACI.0b013e3283303e1b. [DOI] [PubMed] [Google Scholar]
- 8.Larkin DFP, Leeming JP. Quantitative Alterations of the Commensal Eye Bacteria in Contact Lens Wear. Eye. 1991;5:70–74. doi: 10.1038/eye.1991.14. [DOI] [PubMed] [Google Scholar]
- 9.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus Aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clooney AG, Fouhy F, Sleator RD, Driscoll AO, Stanton C, Cotter PD, Claesson MJ. Comparing Apples and Oranges?: Next Generation Sequencing and Its Impact on Microbiome Analysis. PLoS One. 2016;11:e0148028. doi: 10.1371/journal.pone.0148028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014;12:87, 1–12. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel JB. 16S RRNA Gene Sequencing for Bacterial Pathogen Identification in the Clinical Laboratory. Mol. Diagn. 2001;6:313–321. doi: 10.1054/modi.2001.29158. [DOI] [PubMed] [Google Scholar]
- 13.Willcox MDP. Characterization of the Normal Microbiota of the Ocular Surface. Exp. Eye Res. 2013;117:99–105. doi: 10.1016/j.exer.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Mak WC, Cheung KY, Orban J, Lee C-J, Turner APF, Griffith M. Surface-Engineered Contact Lens as an Advanced Theranostic Platform for Modulation and Detection of Viral Infection. ACS Appl. Mater. Interfaces. 2015;7:25487–25494. doi: 10.1021/acsami.5b08644. [DOI] [PubMed] [Google Scholar]
- 15.Greenbaum A, Luo W, Su T-W, Göröcs Z, Xue L, Isikman SO, Coskun AF, Mudanyali O, Ozcan A. Imaging without Lenses: Achievements and Remaining Challenges of Wide-Field on-Chip Microscopy. Nat. Methods. 2012;9:889–895. doi: 10.1038/nmeth.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tseng D, Mudanyali O, Oztoprak C, Isikman SO, Sencan I, Yaglidere O, Ozcan A. Lensfree Microscopy on a Cellphone. Lab Chip. 2010;10:1787–1792. doi: 10.1039/c003477k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozcan A, McLeod E. Lensless Imaging and Sensing. Annu. Rev. Biomed. Eng. 2016;18:77–102. doi: 10.1146/annurev-bioeng-092515-010849. [DOI] [PubMed] [Google Scholar]
- 18.Göröcs Z, Ozcan A. On-Chip Biomedical Imaging. IEEE Reviews in Biomedical Engineering. 2013;6:29–46. doi: 10.1109/RBME.2012.2215847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decher G. Fuzzy Nanoassemblies: Toward Layered Polymeric Multicomposites. Science. 1997;277:1232–1237. [Google Scholar]
- 20.Hugo WB. An Introduction to Microbiology: Pharmaceutical Monographs. Butterworth-Heinemann; 2014. [Google Scholar]
- 21.Santos L, Rodrigues D, Lira M, Oliveira MECDR, Oliveira R, Vilar EY-P, Azeredo J. The Influence of Surface Treatment on Hydrophobicity, Protein Adsorption and Microbial Colonisation of Silicone Hydrogel Contact Lenses. Contact Lens & Anterior Eye. 2007;30:183–188. doi: 10.1016/j.clae.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Feizi A, Zhang Y, Greenbaum A, Guziak A, Luong M, Lok Chan RY, Berg B, Ozkan H, Luo W, Wu M, Wu Y, Ozcan A. Rapid, Portable and Cost-Effective Yeast Cell Viability and Concentration Analysis Using Lensfree on-Chip Microscopy and Machine Learning. Lab Chip. 2016;16:4350–4358. doi: 10.1039/c6lc00976j. [DOI] [PubMed] [Google Scholar]
- 23.Memmolo P, Paturzo M, Javidi B, Netti PA, Ferraro P. Refocusing Criterion via Sparsity Measurements in Digital Holography. Opt. Lett. 2014;39:4719–4722. doi: 10.1364/OL.39.004719. [DOI] [PubMed] [Google Scholar]
- 24.Memmolo P, Miccio L, Paturzo M, Caprio GD, Coppola G, Netti PA, Ferraro P. Recent Advances in Holographic 3D Particle Tracking. Adv. Opt. Photon. 2015;7:713–755. [Google Scholar]
- 25.Merola F, Miccio L, Memmolo P, Paturzo M, Grilli S, Ferraro P. Simultaneous Optical Manipulation, 3-D Tracking, and Imaging of Micro-Objects by Digital Holography in Microfluidics. IEEE Photonics J. 2012;4:451–454. [Google Scholar]
- 26.Wu Y-C, Shiledar A, Li Y-C, Wong J, Feng S, Chen X, Chen C, Jin K, Janamian S, Yang Z, Ballard ZS, Göröcs Z, Feizi A, Ozcan A. Air Quality Monitoring Using Mobile Microscopy and Machine Learning. Light: Sci. Appl. 2017;6:e17046. doi: 10.1038/lsa.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Wang H, Wu Y, Tamamitsu M, Ozcan A. Edge Sparsity Criterion for Robust Holographic Autofocusing. Opt. Lett. 2017;42:3824–3827. doi: 10.1364/OL.42.003824. [DOI] [PubMed] [Google Scholar]
- 28.Matsushima K. Formulation of the Rotational Transformation of Wave Fields and Their Application to Digital Holography. Appl. Opt. 2008;47:D110–D116. doi: 10.1364/ao.47.00d110. [DOI] [PubMed] [Google Scholar]
- 29.Greenbaum A, Zhang Y, Feizi A, Chung P-L, Luo W, Kandukuri SR, Ozcan A. Wide-Field Computational Imaging of Pathology Slides Using Lens-Free on-Chip Microscopy. Sci. Transl. Med. 2014;6:267ra175–267ra175. doi: 10.1126/scitranslmed.3009850. [DOI] [PubMed] [Google Scholar]
- 30.Ferraro P, Paturzo M, Memmolo P, Finizio A. Controlling Depth of Focus in 3D Image Reconstructions by Flexible and Adaptive Deformation of Digital Holograms. Opt. Lett. 2009;34:2787–2789. doi: 10.1364/OL.34.002787. [DOI] [PubMed] [Google Scholar]
- 31.Paturzo M, Ferraro P. Creating an Extended Focus Image of a Tilted Object in Fourier Digital Holography. Opt. Express. 2009;17:20546–20552. doi: 10.1364/OE.17.020546. [DOI] [PubMed] [Google Scholar]